Abstract
Free full text

Sharing and community curation of mass spectrometry data with GNPS
Abstract
The potential of the diverse chemistries present in natural products (NP) for biotechnology and medicine remains untapped because NP databases are not searchable with raw data and the NP community has no way to share data other than in published papers. Although mass spectrometry techniques are well-suited to high-throughput characterization of natural products, there is a pressing need for an infrastructure to enable sharing and curation of data. We present Global Natural Products Social molecular networking (GNPS, http://gnps.ucsd.edu), an open-access knowledge base for community wide organization and sharing of raw, processed or identified tandem mass (MS/MS) spectrometry data. In GNPS crowdsourced curation of freely available community-wide reference MS libraries will underpin improved annotations. Data-driven social-networking should facilitate identification of spectra and foster collaborations. We also introduce the concept of ‘living data’ through continuous reanalysis of deposited data.
Introduction
Natural products (NPs) from marine and terrestrial environments, including their inhabiting microorganisms, plants, animals, and humans, are routinely analyzed using mass spectrometry. However a single mass spectrometry experiment can collect thousands of MS/MS spectra in minutes1 and individual projects can acquire millions of spectra. These datasets are too large for manual analysis. Further, comprehensive software and proper computational infrastructure are not readily available and only low-throughput sharing of either raw or annotated spectra is feasible, even among members of the same lab. The potentially useful information in MS/MS datasets can thus remain buried in papers, laboratory notebooks, and private databases, hindering retrieval, mining, and sharing of data and knowledge. Although there are several NP databases — Dictionary of Natural Products2, AntiBase3 and MarinLit4 — that assist in dereplication (identification of known compounds), these resources are not freely available and do not process mass spectrometry data. Conversely, mass spectrometry databases including Massbank5, Metlin6, mzCloud7, and ReSpect8 host MS/MS spectra but limit data analyses to several individual spectra or a few LC-MS files. While Metlin and mzCloud provide a spectrum search function, unfortunately, their libraries are not freely available.
Global genomics and proteomics research has been facilitated by the development of integral resources such as the National Center for Biotechnology Information (NCBI) and UniProt KnowledgeBase (UniProtKB), which provide robust platforms for data sharing and knowledge dissemination9,10. Recognizing the need for an analogous community platform to effectively share and analyze natural products MS data, we present the Global Natural Products Social Molecular Networking (GNPS, available at gnps.ucsd.edu). GNPS is a data-driven platform for the storage, analysis, and knowledge dissemination of MS/MS spectra that enables community sharing of raw spectra, continuous annotation of deposited data, and collaborative curation of reference spectra (referred to as spectral libraries) and experimental data (organized as datasets).
GNPS provides the ability to analyze a dataset and to compare it to all publically available data. By building on the computational infrastructure of the University of California San Diego (UCSD) Center for Computational Mass Spectrometry (CCMS), GNPS provides public dataset deposition/retrieval through the Mass Spectrometry Interactive Virtual Environment (MassIVE) data repository. The GNPS analysis infrastructure further enables online dereplication6,11–13, automated molecular networking analysis14–21, and crowdsourced MS/MS spectrum curation. Each dataset added to the GNPS repository is automatically reanalyzed in the next monthly cycle of continuous identification (see Living Data by Continuous Analysis below). Each of these tens of millions of spectra in GNPS datasets is matched to reference spectral libraries to annotate molecules and to discover putative analogs (Fig. 1a). From January 2014 to November 2015, GNPS has grown to serve 9,267 users from 100 countries (Fig. 1b), with 42,486 analysis sessions that have processed more than 93 million spectra as molecular networks from a quarter million LC-MS runs. Searches against a combined catalog of over 221,000 MS/MS reference library spectra from 18,163 compounds (Supplementary Table 1) are possible, and GNPS has matched almost one hundred million MS/MS spectra in all public and private search jobs using an estimated 84,000 compute hours.
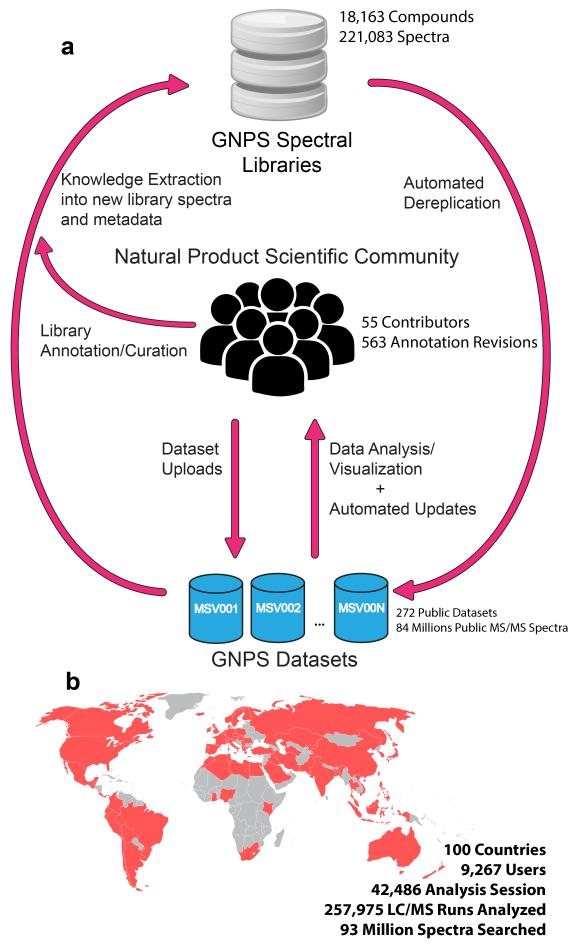
(a) Representation of interactions between the natural product community, GNPS spectral libraries, and GNPS datasets. At present 221,083 MS/MS spectra from 18,163 unique compounds are used for the search in the GNPS. These include both 3rd party libraries such as MassBank, ReSpect, and NIST, as well as spectral libraries created for GNPS (GNPS-Collections) and spectra from the natural product community (GNPS-Community). GNPS spectral libraries grow through user contributions of new identifications of MS/MS spectra. To date, 55 community members have contributed 8,853 MS/MS spectra from 5,568 unique compounds (30.5% of the unique compounds available). In addition, on-going curation efforts have already yielded 563 annotation updates for library spectra. The utility of these libraries is to dereplicate compounds (recognition previously characterized and studied known compounds), in both public and private data. This dereplication process is performed on all public datasets and results are automatically reported, thus enabling users to query all datasets/organisms/conditions. Automatic reanalysis of all public data creates a virtuous cycle in which contributions to libraries can be matched to all public data. Combined with molecular networking (Fig. 3), this automatic reanalysis empowers community members to identify analogs that can then be added to GNPS spectral libraries. (b) The GNPS platform has grown to serve a global user base of 9,200+ users from 100 countries.
GNPS Spectral Libraries
GNPS spectral libraries enable dereplication, variable dereplication (approximate matches to spectra of related molecules), and identification of spectra in molecular networks. GNPS has collected available MS/MS spectral libraries relevant to NPs (which also include other metabolites and molecules), including MassBank5, ReSpect8 and NIST22 (Table 1, Fig. 2a, and Supplementary Table 1). Altogether, these third party libraries total 212,230 MS/MS spectra representing 12,694 unique compounds (Fig. 2b). While this combined collection of reference spectra, provides a starting point for dereplication, only 1.01% of all spectra public GNPS datasets has been matched to this collection, indicating insufficient chemical space coverage. Although the NP community is working to populate this “missing” chemical space, there is no way to report discoveries of chemistries in an easily verifiable and reusable format.
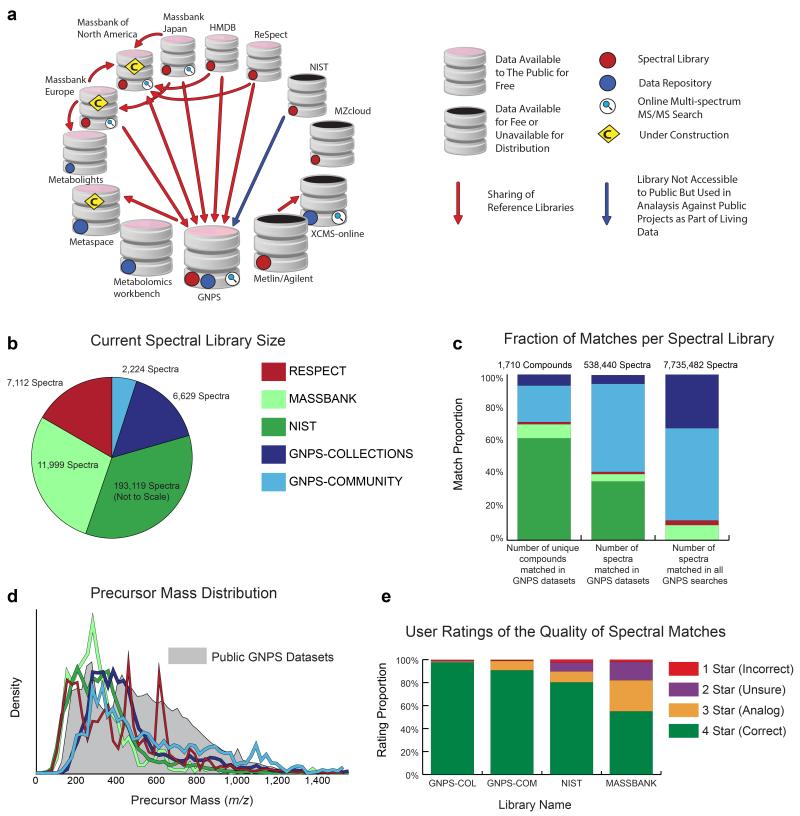
(a) The computational resources of the metabolomics and natural products community fall into two main categories: i) Reference collections (red dots) of MS/MS spectral libraries and ii) Data Repositories (blue dots)designed to publicly share raw mass spectrometry data associated with research projects. Reference collection resources are contributors and aggregators of reference MS/MS spectra, some of which also include data analysis tools, e.g. online multi-spectrum MS/MS search (magnifying glass icon). Several resources have aggregated MS/MS spectra from various reference collections so that the analysis tools at a respective resource can leverage more of the community efforts to annotate data (red and blue arrows). GNPS has imported all freely available reference collections (>221,000 MS/MS spectra) and makes them available for online analyses. GNPS and several other resources provide both reference MS/MS spectra and data in an open and free manner to the public (pink caps). (b) Comparison of spectral library sizes of available libraries (MassBank, ReSpect, and NIST) and GNPS libraries; GNPS-Collections includes newly acquired spectra from synthetic or purified compounds and GNPS-Community includes all community-contributed spectra. (c) Searching all public GNPS datasets revealed that Massbank/ReSpect/NIST libraries matched to 1,217 unique compounds, with GNPS libraries increasing unique compound matches by 41% (corresponding to 29% of total unique matches) with an accompanying 4% increase in spectral library size. Overall, GNPS libraries increase the total number of spectra matched in public datasets by 144% (59% of total public MS/MS matches) and spectra matches across all GNPS public and private data by 767% (88% of all MS/MS matches). (d) The distribution of precursor masses in all GNPS public datasets is shown in gray and compared to the precursor mass distributions of Massbank, ReSpect, NIST, and GNPS libraries. Though GNPS libraries have a combined size that is smaller than MassBank/ReSpect/NIST, GNPS libraries have a higher proportion of molecules in the higher m/z range and therefore complement the proportionately lower precursor mass molecules in other libraries. (e) The quality of spectrum matches obtained by searching against the available spectral libraries is assessed by user ratings (1 to 4 stars see Supplementary Table 6) of continuous identification results. User ratings of 2.5+ stars for 98%+ of GNPS library matches compares favorably with the 90% mark for NIST matches, whose high marks demonstrate how important these 3rd party libraries still are to the GNPS platform. We note that the lower mark for NIST matches does not suggest lower quality spectra. It is more likely explained by its higher emphasis on lower precursor mass molecules with spectra that have fewer peaks and are generally harder to match.
Table 1
Metabolomics and Natural Products MS/MS Computational Resources Overview
| Summary | Data repository* | Reference collections^ | Open online data analysis& | Reference | |
|---|---|---|---|---|---|
| GNPS | Natural products and metabolomics crowdsourced analysis platform with public reference libraries, public data repository and living data | Yes, with automated reanalysis, minimal required metadata (220 w/MS2, 274 total) | Yes, open access, crowdsourced curation | Can search any number of files, analog searches and molecular networking (G,J,E,NA,R,H,N) | |
| Reference Collections | |||||
| MassBank Japan | The first public large scale database for metabolomics reference spectra. | Yes, open access | Can search up to one file at a time (J) | 5 | |
| MassBank Europe | European counterpart of massbank japan. This public reference spectral library is under construction to include draft structures. | Yes, open access | Can search up to one file at a time (J,E) | ||
| MassBank North America | North American public spectral library warehouse and distribution database. | Yes, open access | Can search up to one file at a time (G,J,NA,R,H) | ||
| ReSpect | Public reference library for plant metabolites. | Yes, open access | Can search single spectrum (R) | 8 | |
| HMDB | Public reference library for human metabolites. | Yes, open access | Can search single spectrum (H) | 55 | |
| XCMS-online/Metlin | Reference library for metabolomics. Can be searched but the library is commercial and not available for public redistribution. | Yes, no reanalysis (10 w/MS2, 23 total) | Yes, not freely available | Can search any number of files up to 25Gb (Mt) | 6 |
| NIST/EPA/NIH | Reference libraries for metabolomics. Accessible through purchase but not available for redistribution. | Yes, not freely available | |||
| mzCloud | A metabolomics search engine and reference library. The library is not available to the scientific community. | Yes, not freely available | |||
| Data Repositories | |||||
| Metabolights | Public data repository for metabolomics data, library capabilities under construction. | Yes, no reanalysis, experimental metadata (13 w/MS2, 131 total) | Aggregator only | 34 | |
| Metabolomics workbench | Public data repository for metabolomics data. | Yes, no reanalysis, extensive metadata required (9 w/open format MS2, 196 total) | Aggregator only | 56 | |
To begin to address this pressing need, GNPS houses both newly-acquired reference spectra (GNPS-Collections) as well as a crowd-sourced library of community-contributed reference spectra (GNPS-Community). GNPS-Collections includes NPs and pharmacologically active compounds totaling 6,629 MS/MS spectra of 4,243 compounds (Fig 2b, Supplementary Table 1, Supplementary Note 1,2, and Supplementary Table 2). The GNPS-Community library has grown to include 2,224 MS/MS spectra of 1,325 compounds from 55 worldwide contributors. While the total number of MS/MS spectra in GNPS libraries is only 4% of the MS/MS spectra collected in third party libraries, GNPS libraries contribute matches of MS/MS spectra at a scale disproportionate to their size (Fig. 2c). The GNPS libraries account for 29% of unique compound matches and 59% of the MS/MS matches in public (88% of public+private) data. This indicates that the GNPS libraries contain compounds that are complementary to the chemical space represented in other libraries (Fig. 2c,d). Moreover, in contrast to third party libraries, spectra submitted to GNPS-Community libraries are immediately searchable by the whole community, such that submissions seamlessly transfer knowledge between laboratories (Fig. 1a) in a process that is akin to the addition of genome annotations to GenBank9.
In order to create a robust library, it is important for submissions to be peer-reviewed and, if necessary, annotations corrected or updated as appropriate. Reference spectra submitted to the GNPS-Community library are categorized by the estimated reliability of the proposed submissions. Gold reference spectra must be derived from structurally characterized synthetic or purified compounds and can only be submitted by approved users. Approval is given to contributors who have undergone training. Training is initiated by contacting the corresponding authors or CCMS administrators. Silver reference spectra need to be supported by an associated publication, while Bronze reference spectra are all remaining putative annotations (Supplementary Table 3). This type of division of spectra is reminiscent of RefSeq/TPA/GenBank9,23 (genomics) and Swiss-Prot/TrEMBL/UniProt24,25 (proteomics), allowing for varying tradeoffs between comprehensiveness and reliability of annotations defined as Gold, Silver, and Bronze (Fig. 2e).
To enable refinements or corrections of annotations, GNPS allows for community-driven, iterative re-annotation of reference MS/MS spectra in a wiki-like fashion, to progressively improve the library and converge towards consensus annotation of all MS/MS spectra of interest. This is a process similar to the iterative annotation of the human genome (e.g., see series of papers on NCBI GenBank9). To date, 563 annotation revisions have been made in GNPS (Supplementary Table 4), most of which added metadata to library spectra or refined compound names. The history of each annotation is retained so that users can discuss the proper annotation and address disagreements via comment threads.
Dereplication using GNPS
High throughput dereplication of NP MS/MS data is implemented in GNPS by querying newly acquired MS/MS spectra against all the accumulated reference spectra in GNPS spectral libraries (Fig. 3a). To date, more than 93 million MS/MS spectra from various instruments (including Orbitrap, Ion Trap, qTof, and FT-ICR) have been searched at GNPS, yielding putative dereplication matches of 7.7 million spectra to 15,477 compounds. In the second stage of dereplication, GNPS goes beyond re-identification by utilizing variable dereplication, which is a modification-tolerant spectral library search that is mediated by a spectral alignment algorithm. Variable dereplication enables the detection of significant matches to either putative analogs of known compounds (e.g., differing by one modification or substitution of a chemical group) or compounds belonging to the same general class of molecules (Fig. 3b).Variable dereplication is not available through any other computational platform. For example, GNPS variable dereplication has detected compounds with different levels of glycosylation on various substrates. As MS/MS fragmentation preferentially results in peaks from glycan fragments, it is possible to detect sets of compounds with related glycans even when the substrates to which the glycans are attached are themselves unrelated26. To date, 3,891 putative analogs have been identified in public data using GNPS variable dereplication (Supplementary Table 5). These 3,891 putative analogs include several unique molecules that could be user-curated and added to GNPS reference libraries (see Molecular Explorer below on accessing and annotating putative analogs).
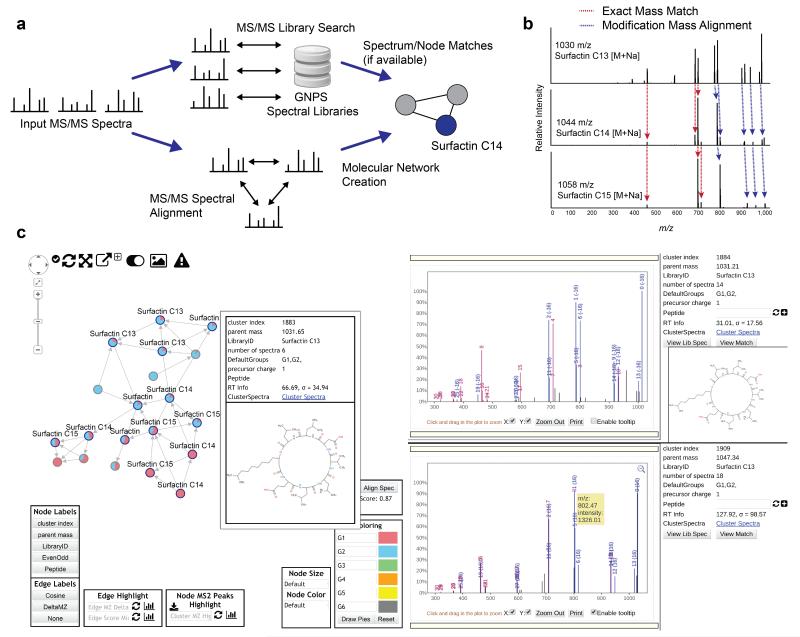
(a) Molecular networks are constructed from the alignment of MS/MS spectra to one another. Edges connecting nodes (MS/MS spectra) are defined by a modified cosine scoring scheme determines the similarity of two MS/MS spectra with scores ranging from 0 (totally dissimilar) to 1 (completely identical). MS/MS spectra are also searched against GNPS Spectral Libraries, seeding putative nodes matches in the molecular networks. Networks are visualized online in-browser or exported for third party visualization software such as Cytoscape31. (b) An example alignment between three MS/MS spectra of compounds with structural modifications that are captured by modification tolerant spectral matching utilized in variable dereplication and molecular networking. (c) In-browser molecular network visualization enables users to interactively explore molecular networks without requiring any external software. To date, more than 11,000 molecular networks have been analyzed using this feature. Within this interface, (i) users are able to define cohorts of input data and correspondingly, nodes within the network are represented as pie charts to visualize spectral count differences for each molecule across cohorts. (ii) Node labels indicate matches made to GNPS spectral libraries, with additional information displayed with mouseovers. These matches provide users a starting point to annotate unidentified MS/MS spectra within the network. (iii) To facilitate identification of unknowns, users can display MS/MS spectra in the right panels by clicking on the nodes in the network, giving direct interactive access to the underlying MS/MS peak data. Furthermore, alignments between spectra are visualized between spectra in the top right and bottom right panels in order to gain insight as to what underlying characteristics of the molecule could elicit fragmentation perturbations.
To assess the reliability of the MS/MS matches found by GNPS dereplication, GNPS users can rate the quality of matches returned by automated GNPS reanalysis (see below). These ratings are 4 star (correct), 3 star (likely correct, e.g. could also be isomers with similar fragmentation patterns), 2 star (unable to confirm the annotation due to limited information) and 1 star (incorrect) (Supplementary Table 6). So far, of the 3,608 matches that have been rated, 139 (3.9%) matches were given 1 or 2 stars (insufficient information (2.9%) or incorrect (1%)) by user ratings. These percentages are consistent with the false discovery rates estimated using spectral library searches of benchmark LC-MS datasets with compound standards (Supplementary Note 3, Supplementary Fig. 1,2 and Supplementary Table 7). Furthermore, these 3,608 match ratings were associated with 2,041 library spectra, therefore the average rating of a library spectrum can offer insight into the reliability of its reference annotation, not unlike Yelp ratings for restaurants. Incorrect matches can arise through either spurious high-scoring matches to library spectra or incorrect annotations for library spectra. Of the 2,041 library spectra with match ratings, 72 (3.5%) spectra had average ratings below 2.5 stars. These percentage ratings were further broken down by spectral library (Fig. 2e). We found that for GNPS-Collection and GNPS-Community libraries, only 29 out of 1746 (1.7%) of the rated library spectra had average ratings below 2.5 stars. These ratings demonstrate that the perceived reliability of GNPS spectral libraries compares favorably with established community resources such as NIST and Massbank, in which 10.5% and 20.1% of the ratings were below 2.5 stars respectively, and provides confidence that the community curation process is robust and that third party libraries integrate well with GNPS. The main advantages of searching using GNPS are the option to run simple or variable dereplication against all publicly accessible reference spectra, and that community-rated matches can be used to improve the quality of the reference libraries and matching algorithms. These dereplication capabilities are not possible with existing published resources.
Molecular Networking
Molecular networks are visual displays of the chemical space present in mass spectrometry experiments. GNPS can be used for molecular networking14–21,27,28, a spectral correlation and visualization approach that can detect sets of spectra from related molecules (so-called spectral networks29) even when the spectra themselves are not matched to any known compounds (Fig. 3a). Spectral alignment15,27 detects similar spectra from structurally related molecules, assuming these molecules fragment in similar ways reflected in their MS/MS patterns (Fig. 3b), analogous to the detection of related protein or nucleotide sequences by sequence alignment. GNPS is currently the only public infrastructure that enables molecular networking. The visualization of molecular networks in GNPS represents each spectrum as a node, and spectrum-to-spectrum alignments as edges (connections) between nodes. Nodes can be supplemented with metadata, including dereplication matches or information that is provided by the user, such as abundance, origin of product, biochemical activity or hydrophobicity which can be reflected in a node’s size or color. It is possible to visualize the map of related molecules as a molecular network21,30–33 (Supplementary Fig. 3) both online at GNPS (Fig. 3c) or exported for analysis in Cytoscape31. Molecular networking analyses of 272 public datasets (Fig. 4a) from a diverse range of samples reveals that on average 35.2% of all unidentified nodes are significantly matched to other spectra of related molecules within a cosine score of 0.8 (increasing to 44.7% of all nodes in more exploratory networks with a cosine score of 0.65 – See Supplementary Table 8). This indicates that a large fraction of all unidentified spectra are identifiable if their or their neighboring nodes’ reference spectra were available in the reference spectral libraries.
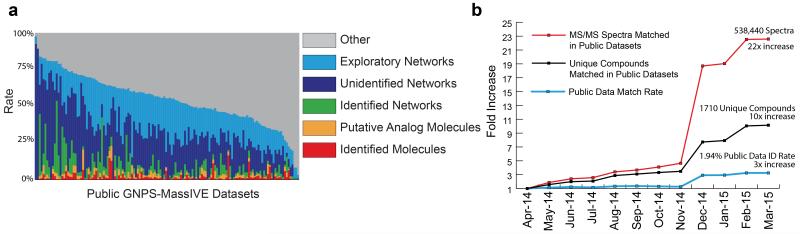
(a) A global snapshot of the state of MS/MS matching of public natural product datasets available in GNPS using molecular networking and library search tools. Identified molecules (1.9% of the data) are MS/MS spectrum matches to library spectra with a cosine greater than 0.7. Putative Analog Molecules (another 1.9% of the data) are MS/MS spectra that are not identified by library search but rather are immediate neighbors of identified MS/MS spectra in molecular networks. Identified Networks (9.9% of the data) are connected components within a molecular network that have at least one spectrum match to library spectra. Unidentified Networks (25.2% of the data) are molecular networks where none of the spectra match to library spectra; these networks potentially represent compound classes that have not yet been characterized. Exploratory Networks (an additional 20.1% of the data) are unidentified connected components in molecular networks with more relaxed parameters (Supplementary Table 8). Thus, 55.3% of the MS/MS spectra at least have one related MS/MS spectrum in spectral networks, with 44.7% having none. In this 44.7% of the data, each MS/MS spectrum has been observed in two separate instances and should not constitute noise. Altogether, this analysis indicates that most of the chemical space captured by mass spectrometry remains unexplored. (b) In the past year, there has been significant growth in the GNPS spectral libraries, driving growth in the match rates of all public data. The number of unique compounds matched in the public data has increased 10x; the number of total spectra matched has increased 22x; and the average match rate has increased 3x. It is expected that identification rates will continue to grow with further contributions from the community to the GNPS-Community spectral library.
Living Data by Continuous Analysis
Funding agencies and publishers have called for raw scientific data, including mass spectrometry data, and analysis methods to be made publically available where possible. Consistent with this aim, GNPS datasets usually comprise the full set of mass spectrometry files produced during a NP research project or the full set of spectra analyzed for a peer-reviewed publication (Supplementary Note 4). While it is potentially advantageous to the community for all data to be made public, GNPS user data can remain private until users explicitly choose to make it public (private data is also analyzable and privately sharable, with >93 million spectra in >250,000 private LC/MS runs already searched using GNPS). GNPS has the largest collection of publicly accessible natural product and metabolomics MS/MS datasets and is the only infrastructure where public data sets can be reanalyzed together and compared with each other (Table 1). To date, GNPS has made 272 public GNPS datasets openly available which are comprised of more than 30,000 mass spectrometry runs with approximately 84 million MS/MS spectra. In common with other public repositories34,35, GNPS datasets can be downloaded. However, data availability on its own does not serve to enable data reuse. GNPS is unique among MS repositories by enabling continuous identification: the periodic and automated re-analysis of all public datasets (Supplementary Note 5,6 and Supplementary Table 9,10). This continuous re-analysis, which incorporates molecular networking and dereplication tools, implements a ‘virtuous cycle’ as illustrated in Figure 1a. Because GNPS spectral libraries are constantly growing due to community contributions and continued generation of reference spectra, the number of matches made by successive re-analyses of public datasets has already grown and is expected to continue to grow over time (Fig. 4b). GNPS users are periodically updated with alerts of new search results.
For example, a Streptomyces roseosporeus project (MSV000078577) was deposited April 8, 2014. At first, only 7 MS/MS spectra were matched. However as of July 14, 2015 36 spectral matches have been made to GNPS libraries. Overall, the total number of compounds matched to GNPS datasets increased more than tenfold, while the number of matched MS/MS spectra in GNPS datasets increased more than twenty-fold in 2015 (Fig. 4b). GNPS users can also subscribe to specific datasets of interest, rather like ‘following’ people on Twitter. When new matches are made, changed, or revoked, all subscribers are notified of new information by an email summarizing changes in identification. From April 2014 to July 2015, 45 updates were initiated by CCMS and automatically sent to subscribers (Supplementary Fig. 4). Update emails have led to substantially more views per dataset, compared to non-GNPS datasets (192 proteomics datasets) deposited in MassIVE. Continuous identification not only keeps a single dataset ‘alive’, it can create connections between datasets and users over time. Similarities between datasets could form the basis of a data-mediated social network of users with potentially related research interests despite seemingly disparate research fields, rather like the “People You May Know” feature on LinkedIn. On average each GNPS user already has 5 suggested collaborators (Supplementary Fig. 5).
Molecular Explorer
Molecular Explorer is a feature that can only be implemented on ‘living data’ repositories and thus exists only in GNPS. Molecular Explorer allows users to find all datasets and putative analogs that have ever been observed for a given molecule of interest. We anticipate that this feature could guide the discovery of previously unknown analogs of existing antibiotics. Public NP data contains more than one hundred unidentified putative analogs of antibiotics such as valinomycin, actinomycin, etamycin, hormaomycin, stendomycin, daptomycin, erythromycin, napsamycin, clindamycin, arylomycin, and rifamycin, highlighting a clear potential to generate leads to discover structurally related antibiotics though the application of GNPS (Supplementary Fig. 6, Supplementary Table 5, and Supplementary Note 7).
To demonstrate this principle we searched for an analog of stenothricin, a broad spectrum antibiotic produced by S. roseosporus with a unique biological response profile36,37 (Supplementary Fig. 7). MS/MS data from S. roseosporus and Streptomyces sp. DSM5940 extracts (MSV000079204) were analyzed by molecular networking and dereplication in GNPS (Supplementary Note 8, Supplementary Fig. 8, and Supplementary Table 11). Nodes corresponding to the stenothricin37 from S. roseosporus were identified in the molecular network. In addition, a small sub-network corresponding to spectra from Streptomyces sp. DSM5940 (Fig. 5a) included 14 nodes that were 41 Da smaller than nodes already known to be stenothricin analogs. This sub-network seemed to indicate that Streptomyces sp. DSM5940 produces a set of 5 abundant analogs of stenothricin which we named stenothricin-GNPS 1-5 (Supplementary Table 12). To our knowledge, a chemical entity that is related to stenothricin with a mass shift of −41 Da has not been described in any database or in the literature. The most abundant analog, stenothricin-GNPS 2 (m/z 1105) was purified and the MS/MS spectra manually compared to MS/MS spectra produced from stenothricin D. This confirmed structural similarity (Fig. 5b,c Supplementary Fig. 9). Differential 2D NMR (Supplementary Fig. 10-14, Supplementary Table 13, and Supplementary Note 9), Marfey’s analysis38 (Supplementary Fig. 15), and genome mining (Supplementary Fig. 16,17, Supplementary Table 14, and Supplementary Note 10) all support that the −41 Da mass shift is due to a lysine to serine substitution.
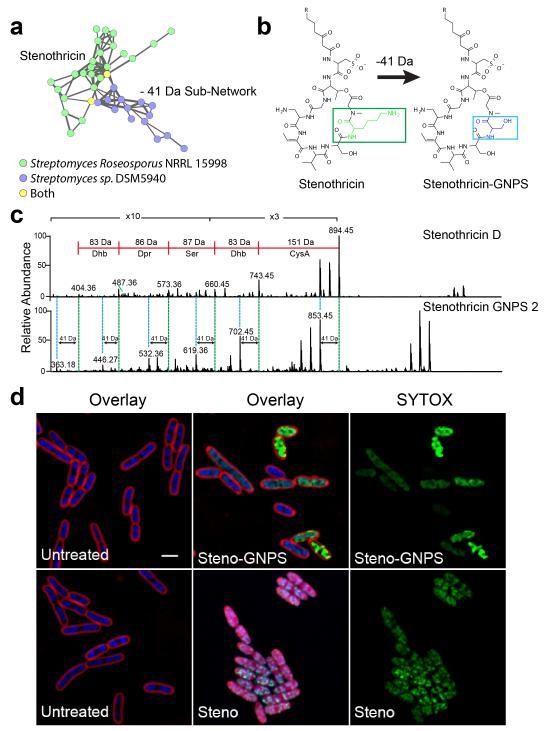
a) The stenothricin molecular family was identified during analysis of a molecular network between chemical extracts of S. roseosporus NRRL 15998 (Green) and Streptomyces sp. DSM5940 (Blue). This analysis indicates that Streptomyces sp. DSM5940 produces a structurally similar compound to stenothricin with a −41 Da m/z difference. An enlarged version of the network can be found in Supplementary. b) Based on preliminary structural analysis, stenothricin-GNPS (41 Da) may contain a Lys to Ser substitution. c) Comparison of the MS/MS of stenothricin D with stenothricin-GNPS 2. d) Although structurally related, stenothricin and stenothricin-GNPS have different effects on E. coli as visualized using fluorescence microscopy. Red is the membrane stain FM4-64, blue is the membrane permeable DNA stain DAPI, green is the membrane impermeable DNA stain SYTOX green. SYTOX green only stains DNA when the cell membrane is damaged. The scale bar represents 2 μm.
The structural comparison between stenothricin D and stenothricin-GNPS has identified a potential role for the lysine residue of stenothricin D in biological function. Stenothricin-GNPS was subjected to fluorescence microscopy based bacterial cytological profiling39,40 (Fig. 5d). Unlike stenothricin D, stenothricin-GNPS is only active against Escherichia coli lptD cells, which are defective in the essential outer membrane protein LptD (Supplementary Fig. 18 and Supplementary Note 11). Although both stenothricin D and stenothricin-GNPS increased membrane permeability of bacterial cells within two hours, stenothricin-GNPS did not have the membrane solubilization function of stenothricin D (Fig. 5d), indicating that the activity of stenothricin D is altered by the presence of a lysine residue that is absent from stenothricin-GNPS. Several published applications of molecular networking and MS/MS based dereplication using GNPS have been reported while the infrastructure has been under development. Specifically, GNPS has enabled the discovery of natural products including colibactin41–45, characterization of biosynthetic pathways46,47, understanding of the chemistry of ecological interactions28,48–52, and development of metabolomics bioinformatics methods53. The application of GNPS workflows to such diverse research areas demonstrates its utility.
Conclusion
GNPS provides a community-led knowledge space in which NP data can be shared, analyzed and annotated by researchers worldwide. It enables a cycle of annotation, in which users curate data, continuous dereplication for product identification, and houses a knowledge base of reference spectral libraries and public datasets. Selected views from community members were sought by nature Biotechnology and are presented, together with author responses, in BOX 1.
The transformation of deposited spectra into living data that is enabled by the GNPS platform could mediate connections between researchers and has the potential to transform data networks into social networks. Of 1,272 compound identifications obtained by continuous identification with the GNPS-Community library, 1,063 (83.6%) were made using reference spectra that were not uploaded by the submitter. In other words, the vast majority of identifications were enabled by other community members. This reuse of knowledge and data is analogous to other community-wide curation efforts including Wikipedia and crowd-sourced dictionaries. Since their initial deposition, 59% of datasets have an increased number of identifications, with the average dataset more than doubling the number of identifications since submission (Supplementary Fig. 19). GNPS enables facile sharing of individual analyses (Supplementary Fig. 20) and uses molecular networks to reveal connections between datasets from different laboratories and biological sources that would otherwise remain disconnected. To date, 3,145 analysis jobs have included files shared between GNPS users, encompassing 548 unique pairs of individuals’ collaborations. GNPS recasts public datasets as “conversation starters” in a data-mediated social network.
Although we have described only one simple application of GNPS in this Perspective ( identification of a stenothricin analog), the community has already begun to utilize GNPS to expedite natural product analysis28,41,43,45,46,50,52. Furthermore, we expect the user base of GNPS to expand to include other communities that use MS/MS data, including those studying metabolomics, exposomes, the chemistry of the human habitat, drug discovery, microbiomes, immunology, food industry, agricultural industry, stratification of patients in clinical trials, clinical adsorption/metabolism and ocean science to name a few, resulting in different GNPS workflows42,44,47,51,53.
As previously shown in genomics9 and protein structure analysis54, the models of global collaboration and social cooperation that are present in GNPS could empower scientific communities to collectively translate big data into shared, reusable knowledge and profoundly influence the way we explore molecules using mass spectrometry.
Online Methods
Spectral Library Searching
Input MS/MS spectra (i.e., query spectra) are considered matched to library spectra if they meet the following criteria: same precursor charge state, precursor m/z is within a user defined Thompson tolerance, share a minimum number of matched peaks, and exceed a user-defined minimum spectral match score. Exact spectral matches between library and query spectra are scored with a normalized dot product57–59. The matching of peaks between two spectra is formulated as a maximum bipartite matching problem15 where peaks from the library and query spectra are represented as nodes with edges connecting library and query peaks. Edges connect peaks that are within a user defined fragment mass tolerance. The bipartite match of library to query peaks that maximizes the normalized dot product is selected. The highest scoring library match for each query spectrum is reported. Estimated false discovery rates of the exact spectral library search are shown in Supplementary Note 3. Parameters of the search can be found in Supplementary Table 8. Source code can be found at the CCMS github page.
Variable Dereplication
Variable dereplication utilizes a modification tolerant spectral library search. Similar to exact spectral matches, except additional edges are added to the bipartite matching between library and query peaks which differ by a δ (as determined by their precursor mass difference δ) +/− the user defined fragment mass tolerance.
Molecular Network Construction
Molecular networks can be constructed from any collection of MS/MS spectra. First, all MS/MS spectra are clustered with MSCluster60 such that MS/MS spectra found to be identical are merged into a consensus spectrum. Consensus spectra are then matched against each other using the modification tolerant spectral matching scheme15. All spectrum-to-spectrum matches that exceed a user defined minimum match score are retained. MS/MS spectra are then represented as nodes in a graph and significant matches between spectra are represented as edges. Further, edges in the graph are only retained if the two nodes, A and B, connected by a given edge satisfy the following properties: i) B must be in the top K highest scoring neighbors of A and ii) A must be in the top K highest scoring neighbors of B. All other edges are removed. Source code can be found at the CCMS github page.
GNPS Collections – Sample Preparation
The NIH Prestwick Phytochemical Library, NIH Natural Product Library, and NIH Small Molecule Pharmacologically Active Library compounds were received as stock solutions of pure compounds (10 mM in DMSO). They were reformatted by 1 μL of each compound into 89 μL of methanol into 96 well plates with 11 distinct compounds in each well. They were further diluted 100-fold for a final 1 μM concentration.
The NIH Clinical Collections and FDA Library part 2 were received as stock solutions of pure compounds (10 mM in DMSO). They were diluted to final concentration of 1 μM in 50:50 methanol:water and formatted onto 96 well plates with 10 compounds per well.
GNPS Collections – LC MS/MS Acquisition
LC-MS/MS acquisition for all in house generated libraries was performed using a Bruker Daltonics Maxis qTOF mass spectrometer equipped with a standard electrospray ionization source (ESI). The mass spectrometer was tuned by infusion of Tuning Mix ES-TOF (Agilent Technologies) at a 3 μL/min flow rate. For accurate mass measurements, lock mass internal calibration used a wick saturated with hexakis (1H,1H,3H-tetrafluoropropoxy) phosphazene ions (Synquest Laboratories, m/z 922.0098) located within the source. Samples were introduced by a Thermo Scientific UltraMate 3000 Dionex UPLC using a 20 μL injection volume. A Phenomenex Kinetex 2.6 μm C18 column (2.1 mm × 50 mm) was used. Compounds from NIH Prestwick Phytochemical Library, NIH Natural Product Library, and NIH Small Molecule Pharmacologically Active Library were separated using a seven minute linear water-acetonitrile gradient (from 98:2 to 2:98 water:acetonitrile) containing 0.1% formic acid. Compounds from NIH Clinical Collections and FDA Library part 2 Library employed a step gradient for chromatographic separation [5% solvent B (2:98 water:acetonitrile) containing 0.1% formic acid for 1.5 min, a step gradient of 5% B-50% B in 0.5 min, held at 50% B for 2 min, a second step of 50% B-100% B in 6 min, held at 100% B for 0.5 min, 100%-5 % B in 0.5 min and kept at 5% B for 0.5 min]. The flow rate was 0.5 mL/min. The mass spectrometer was operated in data dependent positive ion mode; automatically switching between full scan MS and MS/MS acquisitions. Full scan MS spectra (m/z 50 – 1500) were acquired in the TOF and the top ten most intense ions in a particular scan were fragmented using collision induced dissociation (CID) utilizing stepping.
GNPS Collections – Spectral Library Creation
All raw data were centroided and converted to 32-bit uncompressed mzXML file using Bruker Data Analysis. A script was developed to select all possible MS/MS spectra in each LC-MS/MS run that could correspond to a compound present in the sample. For each compound, we calculated the theoretical mass M from its chemical composition and searched for the M+H, M+2H, M+K, and M+Na adducts. Putative identifications included all MS/MS spectra whose precursor m/z had a ppm error <50 compared to the theoretical mass of each possible precursor m/z; all tandem MS/MS spectra with an MS1 precursor intensity of <1E4 were ignored. All candidate identifications were manually inspected and the most abundant representative spectrum for each compound was added to the corresponding library at the gold or bronze level based upon an expert evaluation of the spectrum quality. The best MS/MS spectrum per compound as added to the GNPS-Collections library without filtering or alteration from the mzXML files.
GNPS-Community Contributed Spectral Library Processing and Control
User contributed library spectra are not filtered or altered in any way from the user submission. MS/MS spectra are extracted from the submitted data and are made available in the GNPS libraries. The list and description of metadata fields can be found in GNPS online documentation. To preserve provenance information, the full input file is also retained and made available for download for each library spectrum (e.g. link). Different levels of reference spectra submissions are enforced with access restrictions on a per user basis. The description of each of the quality levels: Gold, Silver and Bronze and be found in Supplementary Table 3. While any MS/MS spectrum can be Bronze quality level in the GNPS libraries, Silver contributions require peer-reviewed publication of the MS/MS spectra, and Gold contributions require MS/MS spectra to be of synthetics or purified compounds with complete structural characterization.
Materials and Strains
Streptomyces sp. DSM5940, obtained from Eberhard-Karls-Universität Tübingen, Germany, was originally isolated from a soil sample collected from the Andaman Islands, India. Streptomyces roseosporus NRRL 15998 was acquired from the Broad Institute, MIT/Harvard, MA, USA, whose parent strain S. roseosporus NRRL 11379 was isolated from soil from Mount Ararat in Turkey. All media components were purchased from Sigma-Aldrich. Organic solvents were purchased from JT Baker at the highest purity.
Streptomyces sp. DSM5940 and S. roseosporus Metabolite Extraction
S. roseosporus and Streptomyces. sp. DSM5940 were inoculated by 4 parallel streaks onto individual ISP2 agar plates61. After incubating for 10 d at 28 °C, the agar was sliced into small pieces and put into a 50 mL centrifuge tube containing 1:1 water:n-butanol and shaken at 225 rpm for 12 h. The n-butanol layer was collected via transfer pipette, centrifuged, and dried with in vacuo.
Streptomyces sp. DSM5940 and S. roseosporus MS/MS Acquisition
MS/MS spectra for crude extracts of S. roseosporus and Streptomyces sp. DSM were collected as previously described37. Briefly, MS/MS spectra were collected using direct infusion using an Advion nanomate-electrospray robot and capillary liquid chromatography using a manually pulled 10 cm silica capillary packed with C18 reverse phase resin. Samples were introduced for capillary LC using a Surveyor system using a 10mL injection (10 ng/μL in 10% ACN). Metabolites were separated using a time variant gradient [(minutes, % of solvent B): (20, 5), (30, 60), (75, 95) where solvent A is water with 0.1% AcOH and B is ACN with 0.1% AcOH] using a 200mL flowrate (1% to instrument source with 1.8kV source voltage). Both methods utilized detection by a Thermo Finnigan LTQ/FT-ICR mass spectrometer. The mass spectrometer was operated in data dependent positive ion mode; automatically switching between full scan high resolution FT MS and low resolution LTQ MS/MS acquisitions. Full scan MS spectra were acquired in the FT and the top six most intense ions in a particular scan were fragmented using collision induced dissociation (CID) at a constant collision energy of 35eV, an activation Q of 0.25, and an activation time of 50 to 80 ms. RAW files were converted to .mzXML using ReAdW.
Molecular Networking Parameters
A molecular network was created at GNPS data from the S. roseosporus and Streptomyces sp. DSM5940 MS/MS data. The specific job is browse-able online (link). Full parameters can be found in Supplementary Table 11.
Stenothricin-GNPS extraction and purification
400 ISP2 agar plates were inoculated with spore suspension of Streptomyces sp. DSM5940 strain and incubated for 10 d at 30 °C. The agar was sliced into small pieces and extracted twice with 1:1 water:n-butanol for 12 h at 28 °C and 225 rpm in two 2.8 L Fernbach flasks. Agar pieces were removed by filtration. The resultant filtrate was centrifuged and the n-butanol layer was collected, dried and resuspended in 1 mL methanol. The extract was fractionated using a Sephadex LH20 column utilizing a methanol mobile phase at a flow rate of 0.5 mL/min. Each fraction was analyzed by dried droplet MALDI-TOF MS for the m/z values corresponding to stenothricin-GNPS. For this analysis, 1 mL of each fraction was mixed 1:1 with a saturated solution of Universal MALDI matrix (Sigma-Aldrich) in 78 % acetonitrile containing 0.1 % TFA and spotted on a Bruker MSP 96 anchor plate. The sample was dried and analyzed by either a Microflex or Autoflex MALDI-TOF MS (Bruker Daltonics). Mass spectra were obtained using the FlexControl software and a single spot acquisition of 80 shots. MALDI-TOF MS data was analyzed by FlexAnalysis software. Fractions containing m/z values putatively assigned to stenothricin-GNPS were combined and further purified by a two-step reversed-phase HPLC procedure (Solvent A: water with 0.1% TFA; Solvent B: ACN with 0.1% TFA). Initial HPLC analysis (SUPELCO C18, 5 μm, 100 Å, 250 × 10.0 mm) utilized a linear gradient from 50% to 75% solvent B in 35 min at flow rate 2 mL/min. Fractions containing target peptide m/z values as detected by MALDI-TOF MS were collected, combined, and evaporated. Subsequent HPLC analysis (Thermo, Syncronis Phenyl HPLC, 5 μm, 150 × 4.6 mm) used an isocratic elution with 35% solvent B. Purified stenothricin-GNPS 2 (m/z 1091) and 3 (m/z 1105) were lyophilized and stored at −80 °C.
Stenothricin-GNPS NMR
50 μg stenothricin-GNPS 2 was dissolved in 30 μL of CD3OD for NMR acquisition. 1H-NMR spectra were recorded on Bruker Avance III 600 MHz NMR with 1.7 mm Micro-CryoProbe at 298 K, with standard pulse sequences provided by Bruker. The NMR spectrum was overlayed with the NMR spectrum from stenothricin D and analyzed using the MestReNova software37.
Genome sequencing and de novo assembly Streptomyces sp. DSM5940
Streptomyces sp. DSM5940 genome was subjected to partial genome sequencing by Ion Torrent and Illumina MiSeq with paired end sequencing. The resulting contigs were assembled by Geneious 5.1.1 using the S. roseosporus 15998 genome sequence as template. Sequences have been deposited in NCBI with accession number assignment pending.
Sequence definition of the gene cluster in Streptomyces sp. DSM5940
To identify the Strenothricin-GNPS gene cluster, the Streptomyces sp. DSM5940 genome was annotated using Artemis62,63. Non-ribosomal peptide synthesis (NRPS) biosynthetic gene clusters were manually assigned using the Artemis Comparison Tool (an “all-against-all” BLAST (NCBI) comparison of proteins within the database)64. The adenylation domains of each NRPS gene cluster were further assessed using NRPSpredictor265,66. The predicted 10 amino acid codes for each A-domain within the NRPS gene clusters was manually compared to those predicted for the putative stenothricin gene cluster from S. roseosporus37. The gene cluster with highest A-domain similarity was putatively identified as the stenothricin-GNPS gene cluster. Full sequence alignment of both the stenothicin-GNPS and stenothricin using ClustalW2 confirmed high sequence identity and similarity67.
Phylogenetic Analysis of C-domains
To determine whether the stenothricin and stenothricin-GNPS gene clusters code for similar amino acid stereochemistry, the condensation domain (C-domain) sequences in the putative stenothricin-GNPS and stenothricin gene clusters were aligned with a subset of C-domain sequences representing the six C-domain families (heterocyclization, epimerization, dual condensation/epimerization (dual), condensation of L amino acids to L amino acids (L to L), and condensation of D amino acids to L amino acids (D to L), and starter) using ClustalW267.
Fluorescence Microscopy
A pre-culture of E. coli lptD cells (NR698) was grown to saturation, then diluted 1:100 into 20 mL LB. Flasks were incubated at 30°C until an OD600 of 0.2 was reached. Cultures were then mixed with the appropriate amount of compound. Compounds were used at the following final concentrations: 1% MeOH, 0.5% DMSO, 20 μg/mL stenothricin D, 40 μg/mL stenothricin-GNPS 2/3. 15 μL of treated cells were transferred into a 1.7 mL tube and incubated at 30°C in a roller. Samples were collected for imaging at 2 hours. 6 μL of cells were added to 1.5 μL of dye mix (30 μg/mL FM 4-64, 2.5 μM SYTOX green and 1.2 μg/mL DAPI) prepared in 1X T-base, and immobilized on an agarose pad (20% LB, 1.2% agarose) prior to microscopy. All microscopy was performed on an Applied Precision Spectris microscope as previous described68 Images were deconvolved using softWoRx V 5.5.1 and the medial focal plane shown. The SYTOX green images were normalized within Figure 5d based on intensity and exposure length relative to the treatment with the highest fluorescence intensity.
Supplementary Material
SI Document
SI Table 5
Acknowledgements
This work was partially supported by National Institution of Health (NIH) Grants 5P41GM103484-07, GM094802, AI095125, GM097509, S10RR029121, UL1RR031980, GM085770, U01TW0007401, U01AI12316-01; NB was also partially supported as an Alfred P Sloan Fellow. In addition, this work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services, under Contract Number HHSN272200800060C. VVP is supported by the NIH Grant K01 GM103809. LMS is supported by National Institutes of Health IRACDA K12 GM068524 award. TLK is supported by the United States - Israel Binational Agricultural Research and Development Fund Vaadia-BARD No. FI-494-13. CP is supported by Science without Borders Program from CNPq. AMCR is supported by São Paulo Research Foundation (FAPESP) grant#2014/01651-8, 2012/18031-7. KK was supported by a fellowship within the Postdoc-Programme of the German Academic Exchange Service (DAAD). MC was supported by a Deutsche Forschungsgemeinschaft (DFG) postdoctoral fellowship. EB is supported by a Marie Curie IOF Fellowship within the 7th European Community Framework Program (FP7-PEOPLE-2011-IOF, grant number 301244-CYANOMIC). CCL was supported by a grant from Ministry of Science and Technology of Taiwan (MOST103-2628-B-110-001-MY3). PC and BOP were supported by the Novo Nordisk Foundation. Lixin Zhang and Xueting Liu are supported by National Program on Key Basic Research Project (2013BC734000) and the National Natural Science Foundation of China (81102369 and 31125002). DP is supported by INSA grant, Rennes. RRS is supported by FAPESP grant#2014/01884-2. DPD is supported by FAPESP grant#2014/18052-0. LMMM is supported by FAPESP grant#2013/16496-5. DBS is supported by FAPESP grant#2012/18031-7. NPL is supported by FAPESP(2014/50265-3), CAPES/PNPD, CNPq-PQ 480 306385/2011-2 and CNPq-INCT_if. EAG is supported by the Notre Dame Chemistry-Biochemistry-Biology Interface (CBBI) program and NIH T32 GM075762. WS and JSM are supported by grants from the National Institutes of Health 1R01DE023810-01 and 1R01GM095373. AE is supported by grant from National Institute of Health K99DE024543. CFM and LJ are supported by the Villum Foundation VKR023113, the Augustinus Foundation 13-4656 and the Aase & Ejnar Danielsens Foundation 10-001120. MSC was supported by UC MEXUS-CONACYT Collaborative Grant CN-12-552. MFT was supported by NIH grant 1F32GM089044. Contributions by BES were supported by NSF grant DEB 1010816 and a Smithsonian Institution Grand Challenges Award. EJNH and JP are supported by the DFG (Forschergruppe 854) and by SNF grant IZLSZ3_149025. KFN and AK are supported by the Danish Council for Independent Research, Technology, and Production Sciences (09-064967) and the Agilent Thought Leader Program. ACS and RSB were supported by NIH/NIAID U19-AI106772. BTM and ME were supported under Department of Defense grant #W81XWH-13-1-0171. Contributions by OBV and KLM were supported by Oregon Sea Grant NA10OAR4170059/R/BT-48, and NIH 5R21AI085540 and U01TW006634-06. EEC, ASM and ARJ were supported by an NSF CAREER Award, a Pew Biomedical Scholar Award (EEC), a Sloan Research Fellow Award (EEC), the Research Corporation for Science Advancement (Cottrell Scholar Award; EEC) and an Indiana University Quantitative Chemical Biology trainee fellowship (ARJ). MM was supported by the Danish Research Council for Technology and Production Science with Sapere Aude (116262). PMA was supported by FNS for fellowship on Subside (200020_146200).
We thank Valerie Paul, Rich Taylor, Lihini Aluwihare, Forest Rohwer, Benjamin Pullman, Jinshu Fang, Martin Overgaard, Michael Katze, Richard D. Smith, Sarkis K Mazmanian, William Fenical, Eduardo Macagno, Xuesong He, and Cajetan Neubauer for feedback and support for their lab personnel to contribute to the work. We thank Bertold Gust and co-workers at the University of Tuebingen for assisting us to obtain Streptomyces sp. DSM5940.
Footnotes
Author contributions:
Design and oversight of the project: PCD and NB
Algorithms: MW and NB
Web-site: MW, JC
In-house library acquisition and analysis: VVP, LMS, NG
User curated library acquisition and analysis: ACS, AE, JSM, WS, WTL, MJM, VVP, LLM, NG, RAQ, AB, CP, TLK, AMCR, AM, MC, KRD, KK, ECO, BSM, EB, EG, DDN, SJM, PDB, XL, LZ, HUH, CFM, LJ, DP, ST, EAG, MSC, CS, KLK, PMA, RGL, RSB, PRJ, MFT, SJ, BES, LMMM, DPD, DBS, NPL, JP, EJNH, AK, RAK, JEK, TOM, PGW, JD, RN, JG, BA, OBV, KLM, EEC, ASM, ARJ, RDK, JJK, KMW, CCH, MM, CCL, YLY
Sample preparation, data generation and web-site beta testing: AE, WTL, MJM, VVP, LMS, NG, RAQ, AB, CP, TLK, AMCR, AM, DF, MC, JC, NB, PCD, ECO, EB, EG, DDN, SJM, PDB, XL, LZ, CZ, CFM, RRS, EAG, MSC, CS, DP, ST, PMA, RGL, BES, LMMM, JP, EJNH, DTM, CABP, ME, BTM, OBV, KLM, EEC, ASM, ARJ, KRD
GNPS Documentation: MW, VVP, LMS, CK, DDN, RRS, LAP
Genome sequencing, assembly and targeted amplification: YP, PC, RG, MG, BOP, LG
Stenothricin GNPS data analysis: WTL, VVP, LMS, YP, PCD
NMR acquisition and analysis: BMD, PDB, LMS
Marfey’s analysis: YP, PDB
Microbiology: YP, ACS, RSB
Peptidogenomics analysis: YP, RDK, PCD
Fluorescence Microscopy: YP, AL, KP
Writing of the paper: MW, VVP, LMS, NG, RK, PCD, and NB
Source code and license is available at the CCMS software tools webpage. Source code is also available with this manuscript as Supplementary Source Code.
Competing Financial Interests
NB has an equity interest in Digital Proteomics, LLC, a company that may potentially benefit from the research results; Digital Proteomics LLC was not involved in any aspects of this research. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. EE, EP, HH, LV, and VM are employees of Sirenas MD. PCD is on the advisory board for Sirenas MD. TA is the Scientific Director of SCiLS GmbH.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nbt.3597
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/nbt.3597.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Longitudinal analyses of infants' microbiome and metabolome reveal microbes and metabolites with seemingly coordinated dynamics.
Commun Biol, 7(1):1506, 14 Nov 2024
Cited by: 0 articles | PMID: 39543263 | PMCID: PMC11564710
Discovery of New Cyclic Lipodepsipeptide Orfamide N via Partnership with Middle School Students from the Boys and Girls Club.
ACS Omega, 9(44):44749-44759, 24 Oct 2024
Cited by: 0 articles | PMID: 39524626
Engineered reduction of S-adenosylmethionine alters lignin in sorghum.
Biotechnol Biofuels Bioprod, 17(1):128, 15 Oct 2024
Cited by: 0 articles | PMID: 39407217 | PMCID: PMC11481400
Goondapyrones A-J: Polyketide α and γ Pyrone Anthelmintics from an Australian Soil-Derived <i>Streptomyces</i> sp.
Antibiotics (Basel), 13(10):989, 18 Oct 2024
Cited by: 0 articles | PMID: 39452255 | PMCID: PMC11505385
Insights into predicting small molecule retention times in liquid chromatography using deep learning.
J Cheminform, 16(1):113, 07 Oct 2024
Cited by: 0 articles | PMID: 39375739 | PMCID: PMC11460055
Review Free full text in Europe PMC
Go to all (1,629) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Implementation of an MS/MS Spectral Library for Monoterpene Indole Alkaloids.
Methods Mol Biol, 2505:87-100, 01 Jan 2022
Cited by: 1 article | PMID: 35732939
Data libraries - the missing element for modeling biological systems.
FEBS J, 287(21):4594-4601, 10 Mar 2020
Cited by: 1 article | PMID: 32100391 | PMCID: PMC7687078
Review Free full text in Europe PMC
Bioactive Natural Products Identification Using Automation of Molecular Networking Software.
J Chem Inf Model, 62(24):6378-6385, 10 Aug 2022
Cited by: 1 article | PMID: 35947427
Funding
Funders who supported this work.
FIC NIH HHS (3)
Grant ID: U19 TW007401
Grant ID: U01 TW006634
Grant ID: U01 TW007401
NCCIH NIH HHS (1)
Grant ID: U41 AT008718
NCRR NIH HHS (2)
Grant ID: UL1 RR031980
Grant ID: S10 RR029121
NIAID NIH HHS (5)
Grant ID: R01 AI095125
Grant ID: R21 AI085540
Grant ID: HHSN272200800060C
Grant ID: U19 AI106772
Grant ID: U01 AI124316
NIDCR NIH HHS (4)
Grant ID: R01 DE023810
Grant ID: R00 DE024543
Grant ID: K99 DE024543
Grant ID: R01 DE020102
NIDDK NIH HHS (1)
Grant ID: T32 DK007202
NIGMS NIH HHS (9)
Grant ID: R01 GM094802
Grant ID: R01 GM095373
Grant ID: K12 GM068524
Grant ID: R01 GM097509
Grant ID: F32 GM089044
Grant ID: K01 GM103809
Grant ID: P41 GM103484
Grant ID: T32 GM075762
Grant ID: R01 GM085770
NNF Center for Biosustainability (1)
Grant ID: Network Reconstruction
Novo Nordisk Fonden (1)
Grant ID: NNF10CC1016517
Swiss National Science Foundation (3)
Grant ID: 146200
Medicines from Marine Microbes
Dr Joern PIEL, ETH Zurich
Grant ID: 149025
Grant ID: 200020
Villum Fonden (1)
Grant ID: 00007262





