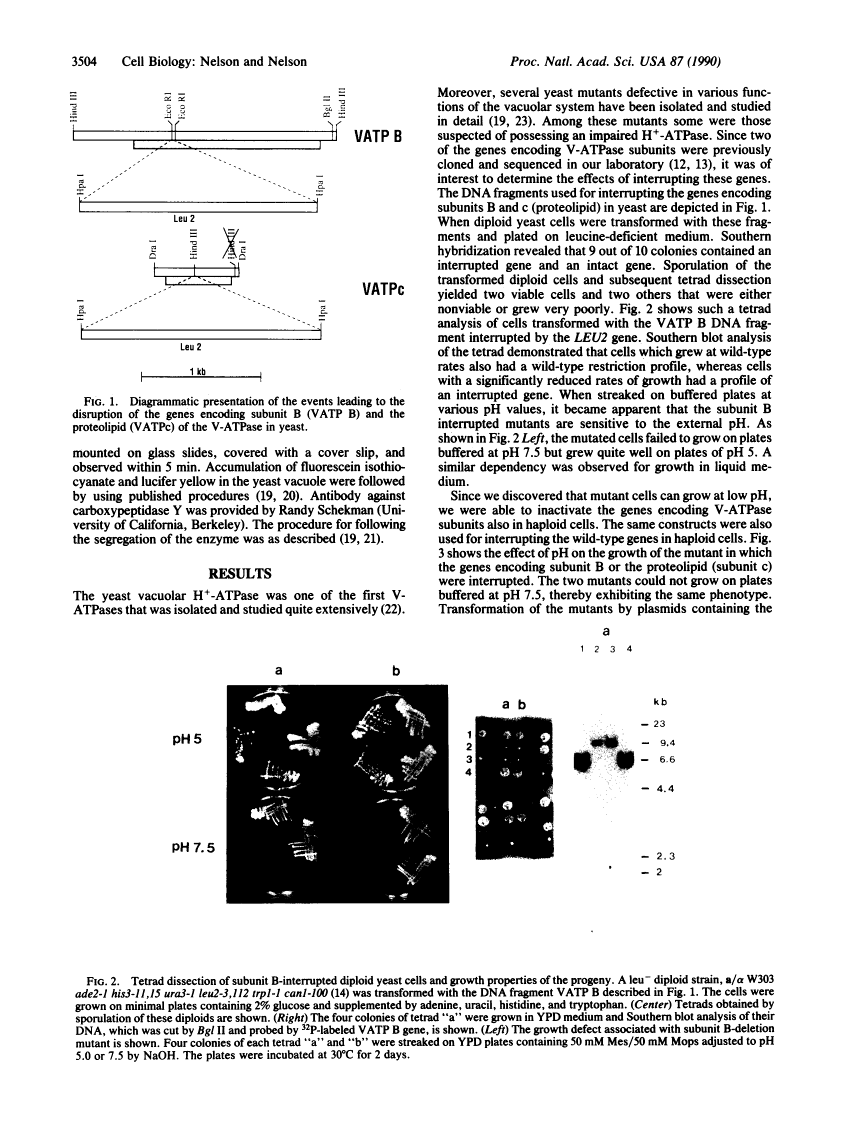
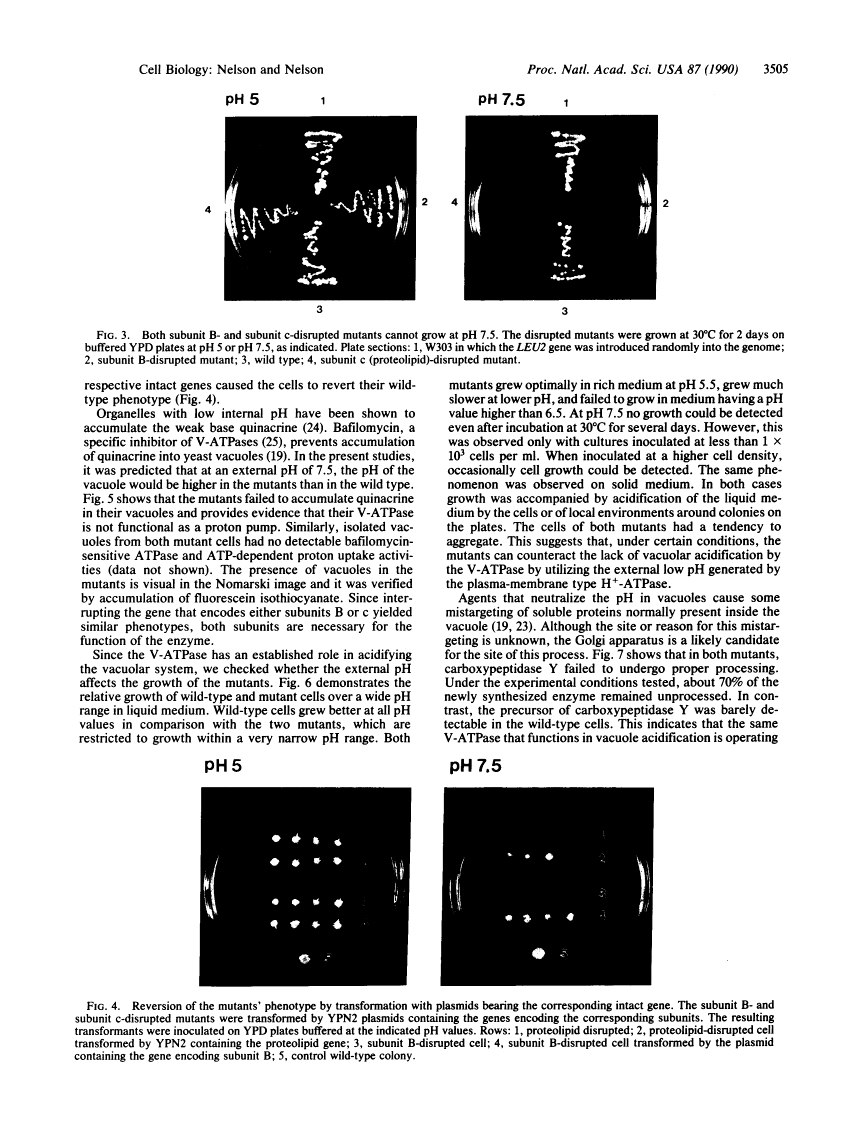
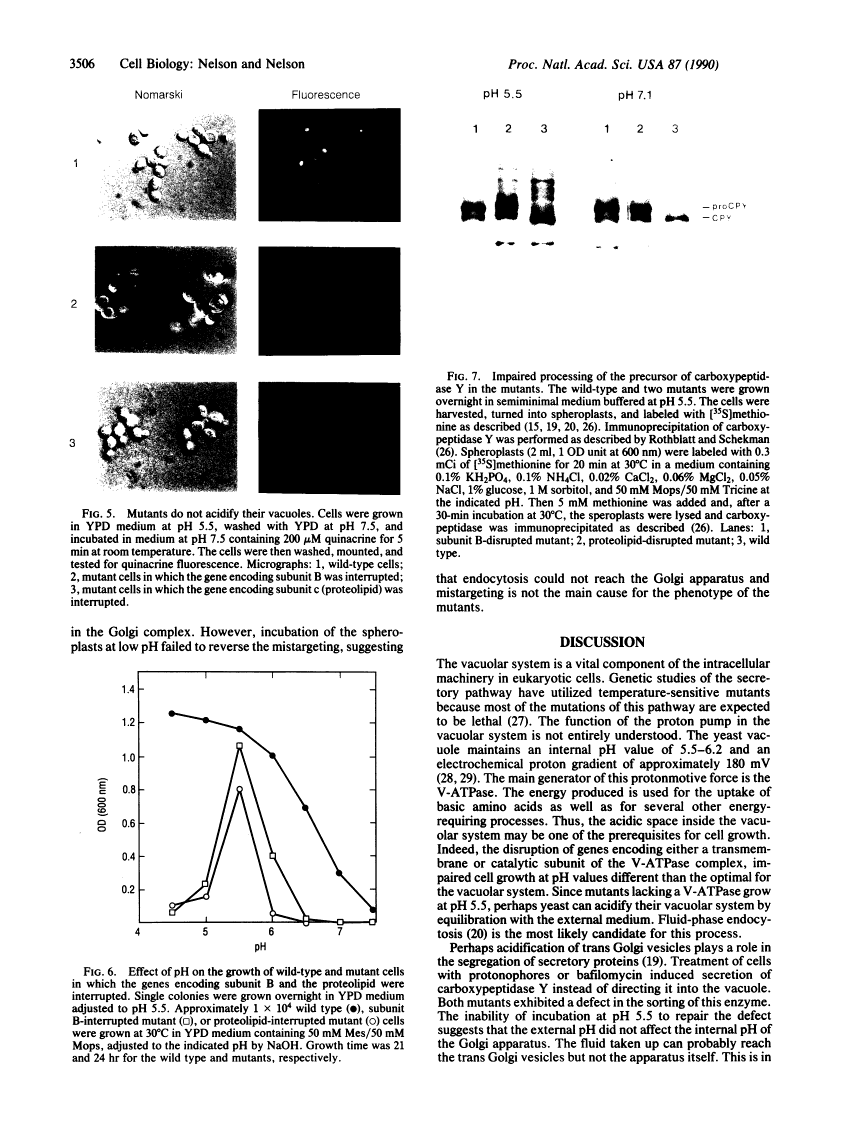
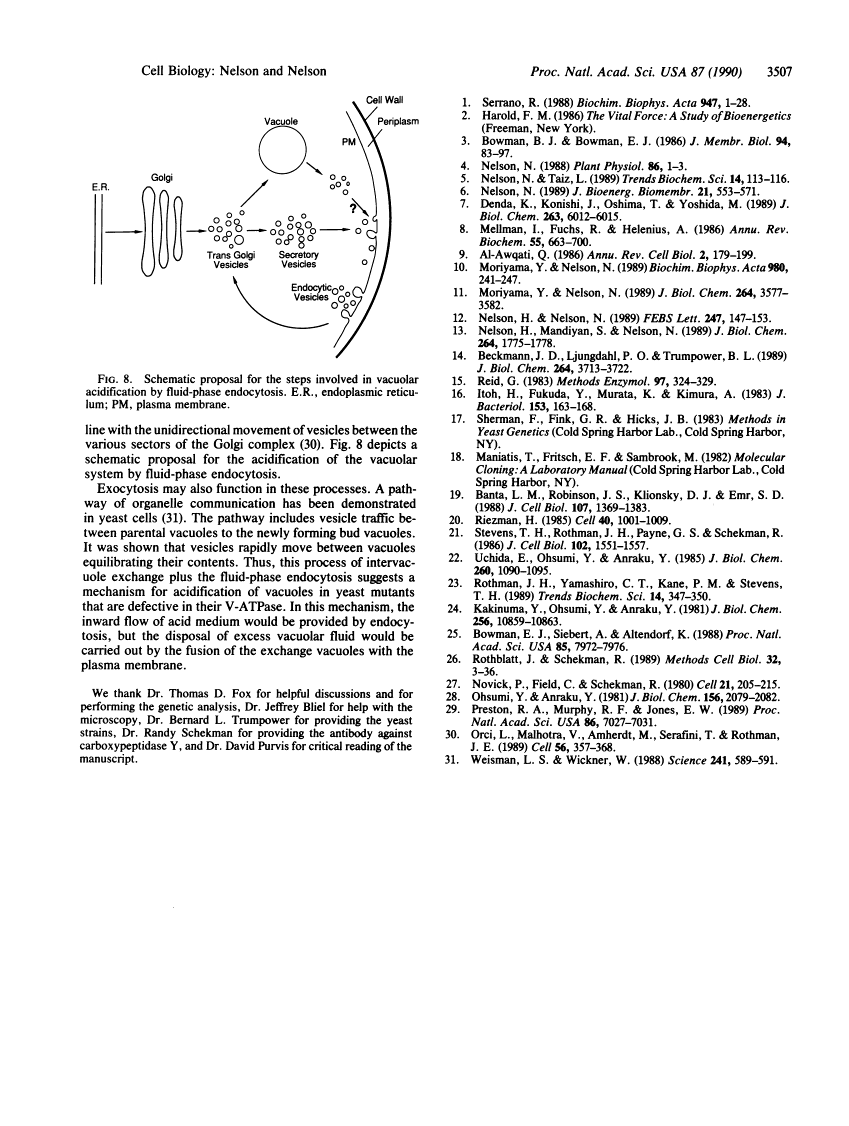
Abstract
Free full text

Disruption of genes encoding subunits of yeast vacuolar H(+)-ATPase causes conditional lethality.
Abstract
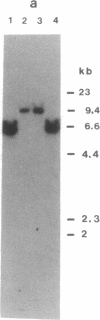
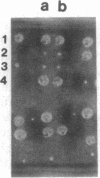
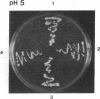
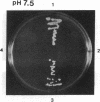
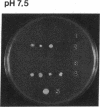
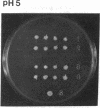
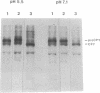
The main function of vacuolar H(+)-ATPases in eukaryotic cells is to generate proton and electrochemical gradients across the membranes of the vacuolar system. The enzyme is composed of a catalytic sector with five subunits (A-E) and a membrane sector containing at least two subunits (a and c). We disrupted two genes of this enzyme, in yeast cells, one encoding a subunit of the membrane sector (subunit c) and another encoding a subunit of the catalytic sector (subunit B). The resulting mutants did not grow in medium with a pH value higher than 6.5 and grew well only within a narrow pH range around 5.5. Transformation of the mutants with plasmids containing the corresponding genes repaired the mutations. Thus failure to lower the pH in the vacuolar system of yeast, and probably other eukaryotic cells, is lethal and the mutants may survive only if a low external pH allows for this acidification by fluid-phase endocytosis.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.4M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Click on the image to see a larger version.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Serrano R. Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim Biophys Acta. 1988 Feb 24;947(1):1–28. [Abstract] [Google Scholar]
- Bowman BJ, Bowman EJ. H+-ATPases from mitochondria, plasma membranes, and vacuoles of fungal cells. J Membr Biol. 1986;94(2):83–97. [Abstract] [Google Scholar]
- Nelson N. Structure, Function, and Evolution of Proton-ATPases. Plant Physiol. 1988 Jan;86(1):1–3. [Abstract] [Google Scholar]
- Nelson N, Taiz L. The evolution of H+-ATPases. Trends Biochem Sci. 1989 Mar;14(3):113–116. [Abstract] [Google Scholar]
- Nelson N. Structure, molecular genetics, and evolution of vacuolar H+-ATPases. J Bioenerg Biomembr. 1989 Oct;21(5):553–571. [Abstract] [Google Scholar]
- Denda K, Konishi J, Oshima T, Date T, Yoshida M. The membrane-associated ATPase from Sulfolobus acidocaldarius is distantly related to F1-ATPase as assessed from the primary structure of its alpha-subunit. J Biol Chem. 1988 May 5;263(13):6012–6015. [Abstract] [Google Scholar]
- Mellman I, Fuchs R, Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. [Abstract] [Google Scholar]
- Al-Awqati Q. Proton-translocating ATPases. Annu Rev Cell Biol. 1986;2:179–199. [Abstract] [Google Scholar]
- Moriyama Y, Nelson N. Lysosomal H+-translocating ATPase has a similar subunit structure to chromaffin granule H+-ATPase complex. Biochim Biophys Acta. 1989 Apr 14;980(2):241–247. [Abstract] [Google Scholar]
- Moriyama Y, Nelson N. Cold inactivation of vacuolar proton-ATPases. J Biol Chem. 1989 Feb 25;264(6):3577–3582. [Abstract] [Google Scholar]
- Nelson H, Nelson N. The progenitor of ATP synthases was closely related to the current vacuolar H+-ATPase. FEBS Lett. 1989 Apr 10;247(1):147–153. [Abstract] [Google Scholar]
- Nelson H, Mandiyan S, Nelson N. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase. J Biol Chem. 1989 Jan 25;264(3):1775–1778. [Abstract] [Google Scholar]
- Beckmann JD, Ljungdahl PO, Trumpower BL. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. I. Construction of a RIP1 deletion strain and isolation of temperature-sensitive mutants. J Biol Chem. 1989 Mar 5;264(7):3713–3722. [Abstract] [Google Scholar]
- Reid GA. Pulse labeling of yeast cells and spheroplasts. Methods Enzymol. 1983;97:324–329. [Abstract] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. [Europe PMC free article] [Abstract] [Google Scholar]
- Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988 Oct;107(4):1369–1383. [Europe PMC free article] [Abstract] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985 Apr;40(4):1001–1009. [Abstract] [Google Scholar]
- Stevens TH, Rothman JH, Payne GS, Schekman R. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol. 1986 May;102(5):1551–1557. [Europe PMC free article] [Abstract] [Google Scholar]
- Uchida E, Ohsumi Y, Anraku Y. Purification and properties of H+-translocating, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1090–1095. [Abstract] [Google Scholar]
- Rothman JH, Yamashiro CT, Kane PM, Stevens TH. Protein targeting to the yeast vacuole. Trends Biochem Sci. 1989 Aug;14(8):347–350. [Abstract] [Google Scholar]
- Kakinuma Y, Ohsumi Y, Anraku Y. Properties of H+-translocating adenosine triphosphatase in vacuolar membranes of SAccharomyces cerevisiae. J Biol Chem. 1981 Nov 10;256(21):10859–10863. [Abstract] [Google Scholar]
- Bowman EJ, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7972–7976. [Europe PMC free article] [Abstract] [Google Scholar]
- Rothblatt J, Schekman R. A hitchhiker's guide to analysis of the secretory pathway in yeast. Methods Cell Biol. 1989;32:3–36. [Abstract] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980 Aug;21(1):205–215. [Abstract] [Google Scholar]
- Ohsumi Y, Anraku Y. Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J Biol Chem. 1981 Mar 10;256(5):2079–2082. [Abstract] [Google Scholar]
- Preston RA, Murphy RF, Jones EW. Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7027–7031. [Europe PMC free article] [Abstract] [Google Scholar]
- Orci L, Malhotra V, Amherdt M, Serafini T, Rothman JE. Dissection of a single round of vesicular transport: sequential intermediates for intercisternal movement in the Golgi stack. Cell. 1989 Feb 10;56(3):357–368. [Abstract] [Google Scholar]
- Weisman LS, Wickner W. Intervacuole exchange in the yeast zygote: a new pathway in organelle communication. Science. 1988 Jul 29;241(4865):589–591. [Abstract] [Google Scholar]
Associated Data
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.87.9.3503
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc53929?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Molecular mechanism of Oxr1p mediated disassembly of yeast V-ATPase.
EMBO Rep, 25(5):2323-2347, 02 Apr 2024
Cited by: 1 article | PMID: 38565737 | PMCID: PMC11094088
Yeast TLDc domain proteins regulate assembly state and subcellular localization of the V-ATPase.
EMBO J, 43(9):1870-1897, 08 Apr 2024
Cited by: 2 articles | PMID: 38589611 | PMCID: PMC11066047
Membrane contact sites regulate vacuolar fission via sphingolipid metabolism.
Elife, 12:RP89938, 27 Mar 2024
Cited by: 1 article | PMID: 38536872 | PMCID: PMC10972560
Structure-based engineering of Tor complexes reveals that two types of yeast TORC1 produce distinct phenotypes.
J Cell Sci, 137(4):jcs261625, 28 Feb 2024
Cited by: 0 articles | PMID: 38415789 | PMCID: PMC10941655
Tender love and disassembly: How a TLDc domain protein breaks the V-ATPase.
Bioessays, 45(7):e2200251, 15 May 2023
Cited by: 4 articles | PMID: 37183929
Go to all (165) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Isolation of vacuolar membrane H(+)-ATPase-deficient yeast mutants; the VMA5 and VMA4 genes are essential for assembly and activity of the vacuolar H(+)-ATPase.
J Biol Chem, 268(1):221-227, 01 Jan 1993
Cited by: 63 articles | PMID: 8416931
The Saccharomyces cerevisiae VMA6 gene encodes the 36-kDa subunit of the vacuolar H(+)-ATPase membrane sector.
J Biol Chem, 268(17):12749-12757, 01 Jun 1993
Cited by: 60 articles | PMID: 8509410
The Saccharomyces cerevisiae VMA7 gene encodes a 14-kDa subunit of the vacuolar H(+)-ATPase catalytic sector.
J Biol Chem, 269(39):24150-24155, 01 Sep 1994
Cited by: 23 articles | PMID: 7929071
Structure and function of the yeast vacuolar membrane proton ATPase.
J Bioenerg Biomembr, 21(5):589-603, 01 Oct 1989
Cited by: 26 articles | PMID: 2531738
Review