Abstract
Free full text

Complex Evolutionary History of the Mammalian Histone H1.1–H1.5 Gene Family
Abstract
H1 is involved in chromatin higher-order structure and gene regulation. H1 has a tripartite structure. The central domain is stably folded in solution, while the N- and C-terminal domains are intrinsically disordered. The terminal domains are encoded by DNA of low sequence complexity, and are thus prone to short insertions/deletions (indels). We have examined the evolution of the H1.1–H1.5 gene family from 27 mammalian species. Multiple sequence alignment has revealed a strong preferential conservation of the number and position of basic residues among paralogs, suggesting that overall H1 basicity is under a strong purifying selection. The presence of a conserved pattern of indels, ancestral to the splitting of mammalian orders, in the N- and C-terminal domains of the paralogs, suggests that slippage may have favored the rapid divergence of the subtypes and that purifying selection has maintained this pattern because it is associated with function. Evolutionary analyses have found evidences of positive selection events in H1.1, both before and after the radiation of mammalian orders. Positive selection ancestral to mammalian radiation involved changes at specific sites that may have contributed to the low relative affinity of H1.1 for chromatin. More recent episodes of positive selection were detected at codon positions encoding amino acids of the C-terminal domain of H1.1, which may modulate the folding of the CTD. The detection of putative recombination points in H1.1–H1.5 subtypes suggests that this process may has been involved in the acquisition of the tripartite H1 structure.
Introduction
Histone H1 is a main component of eukaryotic chromatin. It binds to nucleosomes and linker-DNA in the chromatin fiber, playing a key role in the folding of the nucleofilament. H1 may contribute to transcriptional regulation through several mechanisms, including nucleosome positioning (Pennings et al. 1994), binding to scaffold-associated regions (Izaurralde et al. 1989; Roque et al. 2004), effects on DNA methylation (Fan et al. 2005), modulation of chromatin higher-order structure (Thomas 1999), and binding to nuclear proteins (Halle et al. 2006).
Histone H1 is the most divergent and heterogeneous group of histones. H1 is encoded by a multigene family that has evolved faster than have the core histone families. H1 has multiple isoforms (Parseghian et al. 1994; Talbert et al. 2012). In mammalians, the somatic H1s include H1.1 to H1.5, H1x, and H1.0. Germ-line-specific H1s have been designated as H1t, H1T2, Hils1 (all testis-specific), and H1oo (oocyte-specific).
Histone H1 has three structural domains: a short amino-terminal domain (NTD) (20–35 amino acids), a central globular domain (GD) (~80 amino acids) and a long carboxy-terminal domain (CTD) (~100 amino acids) (Hartman et al. 1977). The central domain is globular and stably folded in solution, and its composition is dominated by hydrophobic amino acids. The NTD contains two distinct sub-regions. The distal half is devoid of basic residues, whereas the half immediately adjacent to the globular domain is highly basic (47%) (Böhm and Mitchell 1985). The CTD is also highly basic (~40%), with basic residues, mostly Lys, uniformly distributed (Subirana 1990). Both the basic subdomain of the NTD and the CTD are intrinsically disordered, but they become extensively folded upon interaction with DNA (Vila, Ponte, Collado, Arrondo, Jiménez, et al. 2001; Vila et al. 2002; Roque et al. 2005).
The H1.1–H1.5 subtypes have evolved mainly in their N- and C-terminal domains. The composition of the C-terminal domain and, to a lesser extent, that of the N-terminal domain, is dominated by the amino acids Lys, Ala, and Pro, residues well-known for promoting protein disorder (Lu et al. 2009). Lysine is positively charged and hydrophilic, alanine has a very small side-chain and proline is a breaker of α-helix. These residues are often arranged in simple repeats, such as SPKK, PKK, PKKA, AAKK, etc. (Churchill and Travers 1991). The terminal domains are thus of low-sequence complexity. The DNA coding for the N- and C-terminal domains is also of low-sequence complexity. Low complexity DNA sequences are prone to slippage events during replication that can cause short insertions/deletions (indels) (Tautz et al. 1986; Ponte et al. 2003). In contrast, the globular domain has a sequence complexity equivalent to that of common globular proteins. It is very similar in all subtypes (93–100% sequence identity) in both interspecies and intraspecies comparisons, and it has been suggested that it may constitute a footprint of this class of subtypes (Eirín-López et al. 2004).
The origin of histone H1 can be traced back to eubacteria, where H1-related lysine-rich DNA-binding proteins have been found, long before the addition of the globular domain (Kasinsky et al. 2001). These proteins have an amino acid composition similar to the CTD and to the basic sub-region of the NTD. H1-like proteins, completely lacking the GD are present in some unicellular eukaryotes, such as Euglenozoa and Alveolata. The appearance of the “winged-helix” motif, present in the globular domain of metazoan H1s, occurred much later in protists, independently of the appearance of the H1 basic-rich regions and core histones.
The analysis of histone H1 sequences is difficult because of the lack of conserved orthologs across even moderately distant classes or phyla. Mammals present some exceptions to the lack of detectable orthology. The somatic H1 subtypes H1.1–H1.5 form a clade, and the individual subtypes have orthologs in mammalian species, which can be clearly identified by their gene organization as well as by their sequences. They are known as replication-dependent subtypes because their synthesis rates increase during the S phase. The H1.1–H1.5 genes and the H1t gene are located together with core histone genes in two large clusters on the short arm of chromosome 6 in humans (Albig, Kioschis, Poutska et al. 1997) or on chromosome 13 in mouse (Drabent et al. 1995; Wang et al. 1997). All H1 genes outside these clusters are solitary (orphon) genes, located on other chromosomes.
It has been shown that H1 subtypes can be knocked out singly and in pairs without noticeable effects on large-scale genome structure and organization, and that the other subtypes compensate the knocked out subtypes to maintain total H1 stoichiometry. However, when three subtypes are inactivated, mice are not able to complete gestation (Fan et al. 2003, 2005).
Biochemical, cytological, and developmental observations suggest that the H1.1–H1.5 subtypes are functionally differentiated (Happel and Doenecke 2009; Parseghian 2015; Millán-Ariño et al. 2016). Evolutionary evidence also supports the functional differentiation of the subtypes of the H1.1–H1.5 gene family. In vertebrates, the rates of nonsynonymous nucleotide substitution differ significantly among subtypes. Furthermore, the synonymous substitution rates greatly exceed the nonsynonymous rates, indicating the presence of strong purifying (negative) selection (Ponte et al. 1998; Eirín-López et al. 2004). In addition, the divergence of H1.1–H1.5 subtypes occurred much earlier than did the mammalian radiation (Ponte et al. 1998; Graur and Li 2005). A phylogenetic analysis of a large group of H1 subtypes showed that they cluster by type in the topologies (Eirín-López et al. 2004), confirming that they are more closely related between than within species (Ponte et al. 1998; Albig, Meergans, Doenecke 1997). Taken together, these results support the view that H1 histones have not been subject to concerted evolution. The generation and diversification of H1 isoforms is better explained by the evolutionary process of birth-and-death with strong purifying selection at the protein level, as first proposed by Nei and Hughes (1992). In this model, new genes are created by gene duplication. Some duplicated genes stay in the genome for a long time, while others are inactivated or deleted from the genome. Protein homogeneity is maintained by the effect of strong purifying selection. This model has been proposed as the primary mode of evolution for numerous multigene families (Nei and Rooney 2005), including histone H1 (Eirín-López et al. 2004).
In the present work, we have examined the evolution of the members of the mammalian histone H1.1–H1.5 gene family. Our results have revealed a high degree of preferential conservation of the basic amino acids and of some subtype-specific post-translational modification (PTM) sites. A conserved pattern of indels in the N- and C-terminal domains, ancestral to the splitting of mammalian orders, was also observed. We have found evidences of positive selection events in certain residues of H1.1. Positive selection episodes appeared to have taken place, both previous and after mammalian radiation. Previous episodes may have contributed to the differentiation of H1.1, while later episodes may reflect the adaptive potential of this subtype. Putative recombination points were found flanking the GD, suggesting that a process of genetic exchange was involved in the acquisition of the tripartite structure, typical of H1.
Material and Methods
Sequence Data
There are several nomenclatures for the H1 subtypes (Parseghian et al. 1994; Talbert et al. 2012). We have used the numerical nomenclature (Albig, Kioschis, Poutska et al. 1997). The equivalence with the nomenclature that uses Roman letters is given in parentheses: H1.1 (H1a), H1.2 (H1c), H1.3 (H1d), H1.4 (H1e), and H1.5 (H1b) (Lennox and Cohen 1983). We have analyzed the sequence data of 27 mammalian species, including representatives of several mammalian orders (i.e., Primates, Artiodactyla, Rodentia, Carnivora, and Cetacea). The full set of the H1.1–H1.5 sequences was available in 24 species, while only some of the subtypes could be used in the other three species. The nucleotide sequences were obtained from the RefSeq database of The National Center for Biotechnology Information (NCBI). The accession number, subtype and species are listed in supplementary table S1, Supplementary Material online.
Sequence Multiple Alignment
The alignment of the nucleotide sequences was made on the basis of the alignment of the translated amino acid sequences. An initial alignment was obtained using the ClustalW/X program (Thomson et al. 1997) running under MEGA v.6 (Tamura et al. 2013). The alignment was then manually curated, which improved the quality of the alignment as analyzed by the calculation of the sum of pairs score (SP-score) at http://www.mtt.fi/AlignmentQuality/ (supplementary table S2, Supplementary Material online) (Ahola et al. 2008).
Insertions/Deletions Frequencies
The number of indels in each sequence was estimated on the basis of the alignment of 130 sequences. For each sequence insertions or deletions were taken into account only when present in the minority of the sequences in the global alignment. Indels present in paralogous and orthologous sequences were counted separately, but every indel was counted only once. The indel frequency was calculated by dividing the number of indels by the number of residues of the protein. The average of the frequencies of indels present in paralogous and orthologous alignments for each subtype was compared using a Mann–Whitney's U-test, the non-parametric version of a t-test.
Phylogenetic Analyses
A maximum likelihood phylogenetic tree was reconstructed with the PhyML 3.0 package (Guindon et al. 2010), on the basis of the sequence multiple alignment of 130 sequences using the best fitted nucleotide substitution model obtained with jModelTest (Posada 2008). The best fitted model was selected by the lowest BIC score and its parameters are shown in supplementary table S3, Supplementary Material online. The confidence level of each branch was estimated by using 1000 bootstrap replications. This tree was used for both the branch and branch-site analyses of selective pressure.
Branch Analysis of Selective Pressure
The estimation of the omega values (ω) for the different branches of the paralog tree, corresponding to the individual H1 subtypes, was performed by running Codeml of the Phylogenetic Analysis by Maximum Likelihood (PAML) v.4.9 software package (Yang 2007). Several hypotheses were built to detect positive selection or the shift in the selective pressure in the H1 subtypes. A Likelihood Ratio Test (LRT) was performed to accept or reject the hypothesis of different ω values among the tree branches. The significance of the LRT was calculated as assuming that twice the difference in the log of maximum-likelihoods was distributed as a χ2 distribution, with the degrees of freedom (df) given by the difference in the number of parameters in the models (Beliawski and Yang 2005).
Branch-Site Analyses of Positive Selection
Branch-site models allow the ω ratio to vary both among sites and among lineages. We have used two different approaches, PAML and BUSTED (Bayesian Unrestricted Test for Episodic Diversification) (Murrell et al. 2015). Both analyses allowed the selection of specific tree branches to test for positive selection, called foreground branches. The branches leading to the cluster of sequences of each subtype of the paralog tree were used as foreground branches. We applied the branch-site test (Zhang et al. 2005) from Codeml of the PAML suite (Yang, 2007). The test is based on the comparison between two nested models: a model (MA) that allows positive selection on one or more branches and a model (MA1) that does not allow positive selection. Model MA estimates the probability of one specific site to fit to four different site classes. The first and the second classes assume that either both background or foreground can be under purifying or under neutral selection. In the third class, foreground branch is under positive selection in comparison to the background that is under negative selection. Finally, the forth class assumes that the foreground is under positive selection, while the background is neutral. As previously mentioned, a likelihood ratio test was used to accept one of the models, but, in this case the P-value obtained for the χ2 distribution of 2LRT was divided by two (df =
= 1). When the LRT suggested the action of positive selection, the Bayes Empirical Bayes (BEB) analysis was used to evaluate the posterior probability that each codon belongs to the site class of positive selection on the foreground branch. We also ran BUSTED, available as a web-application of the HyPhy package at http://www.datamonkey.org/busted. This method considers three ω categories (ω1 ≤
1). When the LRT suggested the action of positive selection, the Bayes Empirical Bayes (BEB) analysis was used to evaluate the posterior probability that each codon belongs to the site class of positive selection on the foreground branch. We also ran BUSTED, available as a web-application of the HyPhy package at http://www.datamonkey.org/busted. This method considers three ω categories (ω1 ≤ ω2 ≤
ω2 ≤ 1 ≤
1 ≤ ω3) shared by all branches and sites, which are calculated separately for the background and foreground branches. The alternative model allows for ω3
ω3) shared by all branches and sites, which are calculated separately for the background and foreground branches. The alternative model allows for ω3 >
> 1 on the foreground branch, while the null model considers ω3
1 on the foreground branch, while the null model considers ω3 =
= 1 on this branch. The LRT was compared with a χ2 distribution (df
1 on this branch. The LRT was compared with a χ2 distribution (df =
= 2) and the null model was rejected if the P-value was lower than 0.05. When positive selection was suggested, a second LRT was done, where twice the difference of the likelihood for the alternative and the null model at each site was compared with a χ2 distribution (df
2) and the null model was rejected if the P-value was lower than 0.05. When positive selection was suggested, a second LRT was done, where twice the difference of the likelihood for the alternative and the null model at each site was compared with a χ2 distribution (df =
= 1). As recombination is not taken into account in the branch-site models, only the branches with P-value lower than 0.001 were further analyzed. Conservatively, we report only the sites with P-value
1). As recombination is not taken into account in the branch-site models, only the branches with P-value lower than 0.001 were further analyzed. Conservatively, we report only the sites with P-value ≤
≤ 0.05 in BUSTED or a posterior probability
0.05 in BUSTED or a posterior probability ≥
≥ 0.95 in the BEB analysis.
0.95 in the BEB analysis.
Site-Specific Analyses of Positive Selection
The presence of residues under positive selection within each of the H1.1–H1.5 subtypes was examined using the Codeml program of the PAML v.4.9 software package (Yang 2007). From the multiple alignment of the 130 sequences (supplementary fig. S1, Supplementary Material online), five ortholog alignments were obtained (supplementary fig. S2, Supplementary Material online). To obtain accurate results for detecting positive selection at amino acid sites, the existence of putative recombination breakpoints within each ortholog alignment was first analyzed (Anisimova et al. 2003) using the GARD package (Kosakovsky-Pond et al. 2006), available at http://www.datamonkey.org/. The significance of the recombination breakpoints was verified with the KH post-test. When recombination breakpoints were detected the site-by-site analysis was performed with the partitioned sequences. New maximum-likelihood phylogenetic trees were reconstructed with PhyML for the individual partitions of H1.1–H1.5 based on best fitted nucleotide substitution model as previously described (supplementary table S3, Supplementary Material online). Three pairs of opposing models were compared. M0 allows for a single ω value across the whole phylogenetic tree at all sites. M0 was compared with M3, assuming three ω values (discrete). The second pair of models compared included M1 (neutral), which assumes two site classes with ω0 <
< 1 and ω1
1 and ω1 =
= 1 fixed, and M2 (positive selection), which adds a third class with ω
1 fixed, and M2 (positive selection), which adds a third class with ω >
> 1 estimated from the data. The third pair compared was M7 and M8. M7 assumes that ω ratios are distributed among sites according to a beta distribution allowing codons to evolve neutrally or under negative purifying selection, while M8 includes an extra class of sites with the ω ratio freely estimated from the data, allowing for positive selection. All model pairs were compared by means of a Likelihood Ratio Test (LRT) using the χ2 distribution to accept or reject the hypothesis of the null model. Sites with Bayes Empirical Bayes posterior probabilities
1 estimated from the data. The third pair compared was M7 and M8. M7 assumes that ω ratios are distributed among sites according to a beta distribution allowing codons to evolve neutrally or under negative purifying selection, while M8 includes an extra class of sites with the ω ratio freely estimated from the data, allowing for positive selection. All model pairs were compared by means of a Likelihood Ratio Test (LRT) using the χ2 distribution to accept or reject the hypothesis of the null model. Sites with Bayes Empirical Bayes posterior probabilities >
> 0.5 were considered as positive. PAML site-analyses were also performed independently for codon alignments of the three domains (NTD, GD, and CTD) of each subtype.
0.5 were considered as positive. PAML site-analyses were also performed independently for codon alignments of the three domains (NTD, GD, and CTD) of each subtype.
When positively selected sites were suggested by PAML, a Random Effects Likelihood (REL) approach (Kosakovsky-Pond and Frost 2005) and a parsimony approach, ADAPTSITE (Suzuki et al. 2001), were used to confirm the results. REL was run as implemented at www.datamonkey.org/, using the partitioned sequences. Sites with a Bayes factor >
> 50 were identified as negatively or positively selected. Adaptsite-p and adaptsite-t programs in the ADAPTSITE package were run to estimate the dN/dS rate ratio and the radical/conservative [(cr/sr)/(cc/sc)] amino acid substitution rate ratio at each codon of the partitions with putative positively selected residues and to determine the probability of those values under selective neutrality (Suzuki et al. 2001). Following the program requirements, all codons in the alignment with gaps were removed and a neighbor-joining tree, constructed by the program Njtree was supplied. Substitution matrices were estimated under the Tamura–Nei model (Tamura et al. 2004) in MEGA 6.0 (Tamura et al. 2013).
50 were identified as negatively or positively selected. Adaptsite-p and adaptsite-t programs in the ADAPTSITE package were run to estimate the dN/dS rate ratio and the radical/conservative [(cr/sr)/(cc/sc)] amino acid substitution rate ratio at each codon of the partitions with putative positively selected residues and to determine the probability of those values under selective neutrality (Suzuki et al. 2001). Following the program requirements, all codons in the alignment with gaps were removed and a neighbor-joining tree, constructed by the program Njtree was supplied. Substitution matrices were estimated under the Tamura–Nei model (Tamura et al. 2004) in MEGA 6.0 (Tamura et al. 2013).
Secondary Structure Predictions
Secondary structure predictions were carried out using the Network Protein sequence analysis server available at http://npsa-pbil.ibcp.fr/ (Combet et al. 2000). Four different prediction methods were used to obtain a consensus prediction: HNN (Hierarchical Neural Network) (Guermeur et al, 1999), MLRC (Multivariate Linear Regression Combination) (Guermeur et al. 1999), PHD (Rost 1996), and SOPM (Self-Optimized Prediction Method) (Geourjon and Deléage 1994).
Intrinsic Disorder Prediction
IUPred, available at http://iupred.enzim.hu/, was used to predict intrinsic disorder at residue level (Dosztányi et al. 2005). This approach is based on the different potential of folded proteins and intrinsically disordered proteins to form stabilizing interactions. It estimates the contribution of an amino acid to order/disorder depending on its chemical type, its sequential environment, and, its potential interaction partners.
Tertiary Structure Analysis
Three-dimensional models of the globular domain were generated with I-TASSER (Yang et al. 2015) using the consensus sequence of H1.1 and that of H1.2–H1.5. All images were modified and represented using PyMOL Molecular Graphics System (version 0_99rc6, 2010). Changes in stability of helix-I of the GD due to point mutations were estimated at the INPS-MD (Impact of Non-synonymous mutations on Protein Stability—Multi Dimension) web server, http://inpsmd.biocomp.unibo.it/inpsSuite/default/index (Savojardo et al. 2016).
Results
Conserved Features in H1 Subtypes
The alignment of 130 sequences (also referred as global H1.1–H1.5 alignment) belonging to the H1.1–H1.5 gene family from species representing different mammalian orders showed several highly conserved features (fig. 1; supplementary fig. S1, Supplementary Material online). It can be observed that while both terminal domains have large sequence variability, the sequence of the GD is remarkably conserved, with more than 70% of identical residues in the 130 sequences. It is also striking that more than 90% of the basic residues are conserved throughout all the protein in all subtypes, including the residues of the GD involved in DNA-binding (Goytisolo et al. 1996). The conserved pattern of basic residues is especially apparent in the CTD, where low sequence identity in the non-basic residues can be observed among subtypes (fig. 1, supplementary fig. S1, Supplementary Material online). The overall conservation of the amount and position of basic residues is presumably associated with their role in the binding of H1 to DNA in chromatin.
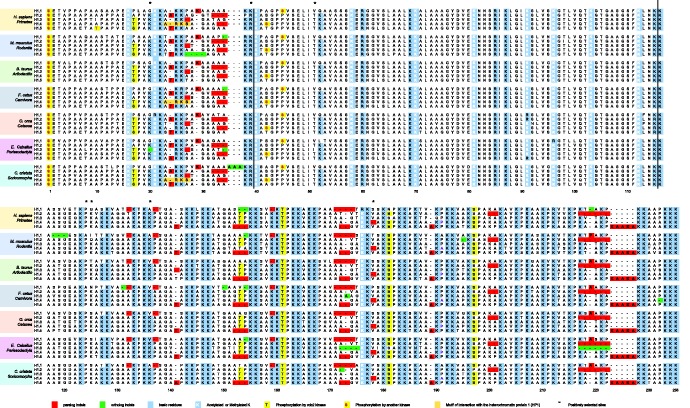
Multiple sequence alignment of histone H1.1–H1.5 subtypes. Selected sequences from the complete alignment (supplementary fig. S2, Supplementary Material online) representing different mammalian orders clustered by species. The limits of the domains are indicated by vertical lines. Insertions/deletions present in paralogous comparisons are highlighted in red; insertions/deletions present in orthologous comparisons are highlighted in green. The conserved positions of basic residues among paralogous sequences are highlighted in light-blue. In white, lysines post-translationally modified; in yellow, phosphorylated residues; in orange, the ARKS peptide, containing the methyl-phos switch that controls H1.4 interaction with the heterochromatin protein (HP1); asterisks indicate positively selected sites in H1.1.
Close examination of the alignment showed conservation of residues that have been found post-translationally modified in mammalian species (fig. 1, supplementary fig. S1, Supplementary Material online) (Sarg et al. 2015; Izzo and Schneider, 2016). This subset comprises nine basic residues: two in the NTD, one in the CTD and six in the GD. Some subtype-specific phosphorylation sites, including those of cyclin-dependent kinases, are conserved as well. In addition, the HP1-binding motif (ARKS) is also conserved in H1.4 of primates and carnivora.
Conserved Pattern of Indels in the H1.1–H1.5 Gene Family
The analysis of the global H1.1–H1.5 alignment revealed the presence of short indels, between 1 and 11 amino acids long, in the N- and C-terminal domains. In the NTD, indels were restricted to a short basic region spanning 11 positions, adjacent to the GD. In the CTD, indels were spread rather uniformly along the sequence, with the longest located at the C-terminal end of the domain.
The indels found in the global alignment were classified according to their presence in paralog or ortholog sequences and counted separately. Then, the frequency of indels per residue was calculated for each sequence (supplementary table S4, Supplementary Material online). The average for each subtype is shown in Table 1. In all subtypes, the frequency of indels in paralog sequences was significantly higher than was the frequency of indels in ortholog sequences, confirming that individual subtypes are more closely related among species than within a species. H1.1 had the highest indel frequency (9.7 × 10−3 indels per residue), while H1.4 had the lowest frequency (5.5 × 10−4 indels per residue). Considering that H1.1 is the most variable of the H1.1–H1.5 subtypes and H1.4 the most conserved (Ponte et al. 1998), the more variable subtypes thus appear to be more tolerant to insertions/deletions.
Table 1
Frequency of Indels Averaged by Subtype.
| Subtype | Average Frequency of Indels in Paralog Sequences (E-02) | Average Frequency of Indels in Ortholog Sequences (E-02) | P-value |
|---|---|---|---|
| H1.1 | 2.97 | 0.97 | 0.0001*** |
| H1.2 | 1.87 | 0.19 | 0.0001*** |
| H1.3 | 0.46 | 0.25 | 0.001** |
| H1.4 | 0.92 | 0.05 | 0.0001*** |
| H1.5 | 2.63 | 0.23 | 0.0001*** |
Note.—The frequencies were calculated by dividing the total number of indels in paralog and ortholog comparisons by the number of residues of each sequence, and afterward were averaged by subtype. P-value corresponds to the probability that the frequency of indels among paralogs is the same as the frequency of indels among orthologs estimated by a Mann–Whitney U-test.
When the paralogs of the different lineages were compared, a pattern of indels common to all lineages became apparent (shown in red, in fig. 1). The conservation of the indel pattern in the different lineages indicates that this pattern is ancestral to the radiation of mammalian orders.
H1 Subtypes Are under Different Degrees of Purifying Selection
As expected, the phylogenetic tree with the 130 sequences shows that H1.1–H1.5 subtypes are clustered together in different branches of the tree (fig. 2). Estimation of omega (ω), defined as the ratio between nonsynonymous and synonymous substitution rates (ω =
= dN/dS), is a measure of selective pressure. Maximum-likelihood estimation of ω for the tree branches would allow the detection of positive selection for the functional divergence, following a gene duplication event and also of a long-term shift in the selective pressure among the different branches. The null hypothesis, H0, of a general average ω value in the entire tree was contrasted with different alternative hypotheses: H1 and H2, propose that subtypes H1.1 and H1.5, respectively, have different ω values than do the rest of the tree. H3 proposes that subtypes H1.2, H1.3, and H1.4 have significantly different ω values among each other and also from the rest of the subtypes (fig. 3). The Likelihood Ratio Test (LRT) allowed to reject the null hypothesis in all cases, suggesting that H1.1–H1.5 subtypes are under different degrees of purifying selection (table 2).
dN/dS), is a measure of selective pressure. Maximum-likelihood estimation of ω for the tree branches would allow the detection of positive selection for the functional divergence, following a gene duplication event and also of a long-term shift in the selective pressure among the different branches. The null hypothesis, H0, of a general average ω value in the entire tree was contrasted with different alternative hypotheses: H1 and H2, propose that subtypes H1.1 and H1.5, respectively, have different ω values than do the rest of the tree. H3 proposes that subtypes H1.2, H1.3, and H1.4 have significantly different ω values among each other and also from the rest of the subtypes (fig. 3). The Likelihood Ratio Test (LRT) allowed to reject the null hypothesis in all cases, suggesting that H1.1–H1.5 subtypes are under different degrees of purifying selection (table 2).
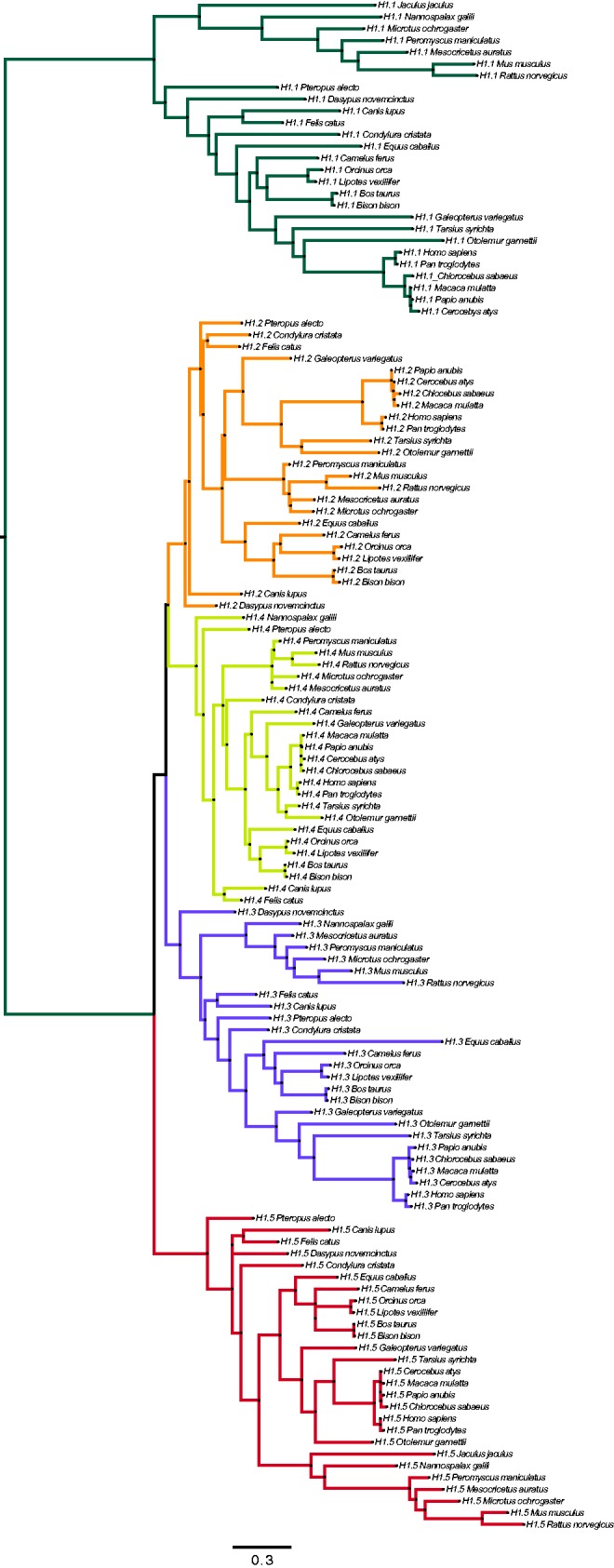
Phylogenetic tree of the 130 sequences of the mammalian H1.1–H1.5 subtypes. The maximum likelihood phylogenetic tree was reconstructed with the PhyML 3.0 package, based on the best fitted nucleotide substitution model obtained with jModelTest (T92 +
+ G
G +
+ I), as described in the Materials and Methods section. For each sequence, the subtype and species are indicated. Scale bar shows nucleotide substitutions per codon.
I), as described in the Materials and Methods section. For each sequence, the subtype and species are indicated. Scale bar shows nucleotide substitutions per codon.
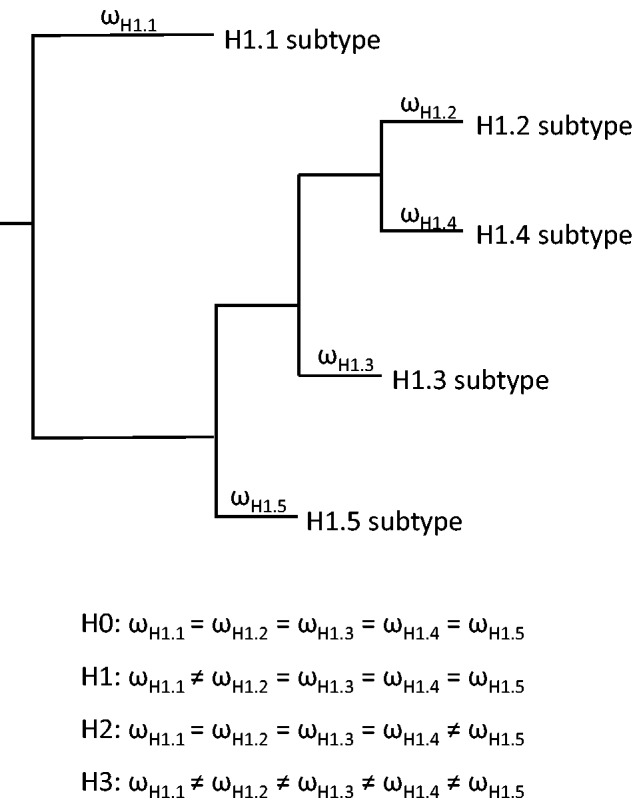
Analysis of the selective pressure in the different branches of the phylogenetic tree. (A) Schematic representation of the topology of the tree and the branch-specific ω ratios. H0, null hypothesis, all the subtypes are under the same selective pressure; H1–H3 alternative hypothesis for different selective pressures in the individual subtypes.
Table 2
Averaged ω in Branches of Phylogenetic Tree of Mammalian H1.1–H1.5 Gene Family.
| Hypothesis | lnL | Branches | Omega (ω) | LRT | P-value |
|---|---|---|---|---|---|
| H0 | −22708.4 | All the branches | 0.14116 | ||
| H1 | −22692.1 | H1.1 | 0.18982 | 32.592414 | 1.137E−08 |
| Rest of the branches | 0.12314 | ||||
| H2 | −22690.59 | H1.5 | 0.08853 | 35.622658 | 2.395E−09 |
| Rest of the branches | 0.15575 | ||||
| H3 | −22694.48 | H1.2 | 0.1097 | 27.834734 | 0.00001347 |
| H1.3 | 0.1741 | ||||
| H1.4 | 0.0952 | ||||
| Rest of the branches | 0.1497 |
Note.—H0, null hypothesis, which considers that all the subtypes are under the same selective pressure; H1–H3 alternative hypothesis considering different selective pressures for the individual subtypes. In italics, p-values lower than 0.05, which allowed to reject the null hypothesis. LRT, likelihood ratio test, calculated as 2(lnLalternative hypothesis − lnL null hypothesis).
No positive selection was detected acting on any of the five subtypes. They had ω values lower than 0.2, indicating the presence of a global strong purifying selection in all the family (table 2). H1.5 is the subtype under the strongest negative selection, followed closely by H1.4, while H1.1 is the subtype with higher average ω value. It should be noted that if functional divergence of H1 subtypes had evolved by positive selection of certain amino acid would not affect significantly the ω ratios among branches (Beliawski and Yang 2005).
Detection of Positive Selection in H1 Subtypes
Lineage-specific methods, including the previous branch analysis, detect positive selection for a lineage only if the average dN over all sites is higher than the average dS. If adaptive evolution occurs at a few time points and affects a few amino acids these models might lack power in detecting positive selection (Yang and Nielsen 2002). Branch-site models allow the ω ratio to vary both among sites and among lineages and, therefore, are capable of detecting positive selection when the averaged ω value for a specific branch is lower than one. We used two different methods: the maximum-likelihood models (MA/MA1) implemented in the PAML suite (Yang 2007) and the branch-site unrestricted statistical test for episodic diversification (BUSTED) (Murrell et al. 2015). These two approaches rely on different assumptions of ω (nonsynonymous/synonymous rate ratio) variation among branches (Murrell et al. 2015).
Branches leading to the five H1 subtypes were tested for positive selection (foreground branches) separately (fig. 3, supplementary table S5, Supplementary Material online). The strongest evidence of positive selection was detected in the branch leading to H1.1, which has P-values lower than 0.001 in both methods (supplementary table S5, Supplementary Material online, table 3). PAML detected six sites (positions 20, 39, 51, 125, 136, and 178) with posterior probabilities calculated with Bayes Empirical Bayes (BEB) >
> 0.95. All sites detected by PAML were also selected by BUSTED with P-values
0.95. All sites detected by PAML were also selected by BUSTED with P-values <
< 0.05, together with three additional sites (table 3).
0.05, together with three additional sites (table 3).
Table 3
Positively Selected Sites Detected in H1.1.
| Type of Analysis | Program | Positively Selected Sites (PSS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Branch-site | PAML | 20, 39, 51, 125, 136, 178 | ||||||||
| BUSTED | 20, | 39, | 41, | 51, | 125, | 131, | 136, | 178, | 214 | |
| Site-specific | PAML | 124, | 125, | 127, | 136, | 157, | 187 | |||
| REL | 124, 125 | |||||||||
| Adaptsite | 125 | |||||||||
Note.—The positions of the positively selected sites are referred to the multiple sequence alignment in figure 1. PSS detected for more than one method are gray shaded; in italics, PSS located at the N-terminal domain; underlined, PSS located at the globular domain. The rest of the PSS are located at the C-terminal domain.
One of the limitations of the two branch-site models implemented in PAML and BUSTED is that recombination is not taken into account. Therefore, a site-by-site analysis with PAML v 4.9 (Yang 2007) to detect individual residues under positive selection (adaptive or diversifying selection) was also performed. This software package uses maximum-likelihood methods that provide a powerful framework for detecting positive selection when sites undergoing positive selection are interspersed among sites dominated by negative selection, as is the case of histone H1 (Wong et al. 2004). In the site-specific analysis, the subtypes were screened separately. Prior to the estimation of the parameters for the different models, the presence of recombination breakpoints in the ortholog alignments (supplementary fig. S2, Supplementary Material online) was analyzed. The GARD analyses revealed the presence of putative recombination breakpoints in all subtypes, validated by the KH post-test. One recombination breakpoint was detected in the CTD of H1.4, relatively close to the GD. Two recombination breakpoints were detected in the rest of the subtypes, approximately located at the boundaries of the GD. Taking this information into account, the site-by-site analysis was performed with the partitioned sequence alignments.
In H1.2–H1.5 subtypes, no positive selected sites were detected, as all the comparisons between M1–M2 and M7–M8 model pairs were not significant (supplementary table S6, Supplementary Material online). However, all comparisons between M0 (one ratio) and M3 (discrete) were significant in the partitions formed mostly by residues of the terminal domains, suggesting variability in the ω ratio among sites (Yang and Nielsen 2002) (supplementary fig. S3, Supplementary Material online).
The comparison of the averaged ω values estimated by the accepted model of the M7–M8 pair for each domain showed that the GD had the lowest averaged ω values, followed by the NTD and the highest values corresponded to the CTD. In addition, significant differences between the averaged ω values of the three domains were found in subtypes H1.1–H1.3, while in H1.4 and H1.5, the more conserved subtypes, only the CTD had significant differences with the rest of the domains (supplementary table S7, Supplementary Material online). Considering these results and that H1 has one highly ordered domain (GD) flanked by two intrinsically disordered domains, which may have different patterns of evolution, the site-by-site analysis with PAML was repeated with the sequence divided by domains with very similar results (supplementary table S8, Supplementary Material online).
The use of the site-specific models revealed positions under positive selection in the CTD of H1.1 (table 3). The discrete model (M3) fits the data significantly better than does the one ratio model (M0). Model M3 suggests that 14% of the CTD sites (15 amino acids) are under positive selection and identifies nine positions under positive selection with posterior probabilities higher than 0.5, as calculated with Naive Empirical Bayes (NEB) approach. Model 8 also fits the data significantly better than model 7 and suggests 10% of the sites are under positive selection. This model detects six positions under positive selection with posterior probabilities calculated with Bayes Empirical Bayes (BEB) >
> 0.5 (positions 124, 125, 127, 136, 157, and 187). The positions detected with model 8 were among the nine positions detected with M3, and one of them, position 125 (table 3, supplementary table S6, Supplementary Material online), had a BEB>
0.5 (positions 124, 125, 127, 136, 157, and 187). The positions detected with model 8 were among the nine positions detected with M3, and one of them, position 125 (table 3, supplementary table S6, Supplementary Material online), had a BEB> 0.95. Unlike M3 and M8, model M2 does not suggest positive selection.
0.95. Unlike M3 and M8, model M2 does not suggest positive selection.
We also used two different site-specific methods: Random Effects Likelihood (REL) and ADAPTSITE to confirm the detection of positive selection in H1.1 CTD. REL allows for tests of selection at a single codon site, while taking into consideration rate variation across synonymous sites. It has high power to detect positive selection in intermediate size datasets (16–32 sequences), such as our 27 H1.1 sequences (Kosakovsky-Pond and Frost 2005). Two sites, 124 and 125, with Bayes factor >
> 50 were detected as evolving under positive selection by REL and confirmed the PAML results (table 3 and supplementary table 6, Supplementary Material online). In addition, 45 sites were detected as negatively selected, confirming the overall negative selection (supplementary table S9, Supplementary Material online).
50 were detected as evolving under positive selection by REL and confirmed the PAML results (table 3 and supplementary table 6, Supplementary Material online). In addition, 45 sites were detected as negatively selected, confirming the overall negative selection (supplementary table S9, Supplementary Material online).
In ADAPTSITE-p, substitutions are inferred using parsimony reconstruction of ancestral sequences, and an excess of non-synonymous substitutions is tested for each site (Suzuki et al. 2001). In this method, amino acid substitutions are classified as conservative or radical, according to whether they retain their charge. It is a parsimony method very conservative in detecting positive selection (Wong et al. 2004). Statistic support was found by ADAPTSITE-t for conservative positive selection at position 125 (P-value 0.037) and for negative selection in 46 sites (supplementary table S9, Supplementary Material online).
Possible Biological Implications of Positive Selection in H1.1
Taking into account the results of the analysis of positive selection, only sites that were identified by more than one method were considered reliable. Using this criteria, seven sites appeared to be under positive selection in H1.1 (table 3). Of the seven sites, two are located in the NTD, one in the GD, and four in the CTD.
The positions located in the NTD were detected by both branch-site methods and represent substitutions in H1.1 of conserved amino acids in the rest of the subtypes that could affect H1.1 affinity for DNA (fig. 1). The first site, at position 20 (referred to the sequence alignment in fig. 1), is a substitution of a lysine in H1.2-H1.5 by an uncharged residue. In the second site (position 39), an arginine is substituted by a lysine, which is positively charged like arginine, but has lower affinity for DNA.
The PSS located at the beginning of the GD (position 51) belongs to the first α-helix of the winged-helix motif (helix-I). In this position, H1.2–H1.5 have a threonine residue that has been substituted by valine in H1.1 (fig. 1). Two 3D-structural models were generated using the consensus sequence of H1.1 and that of H1.2–H1.5 (fig. 4A). The overlap of both models showed great structural similarity, except for helix-I that was shorter in H1.1 model (fig. 4A and B). Stability calculations considering the mutations in helix-I in H1.1 consensus sequence revealed that the T→V mutation is unfavorable for structure stability as the change in ΔG was above 0 [ddG(change) =
= dG(mutant) − dG(wild-type)] (fig. 4C).
dG(mutant) − dG(wild-type)] (fig. 4C).
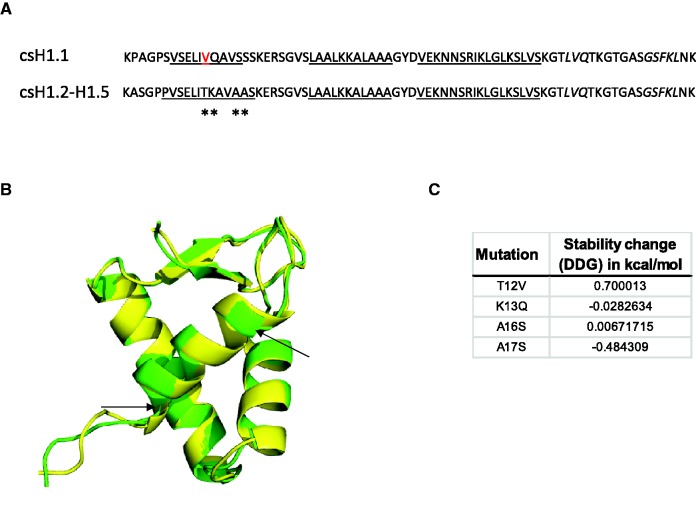
Potential structural changes in the GD of H1.1. (A) Consensus sequences for H1.1 and H1.2–H1.5 subtypes including the secondary structure prediction used in the 3D-model generated with I-TASSER. In red, the positively selected sites (position 51); underlined, residues predicted to form α-helix; in italics, residues predicted in strand conformation; asterisks, changes in helix-I of H1.1. (B) Overlap of the structural models of H1.1, in green and H1.2–H1.5, in yellow, using PyMOL. Arrows point to the limits of helix-I in the H1.1 model. (C) Stability calculations for mutations in helix-I of H1.1 performed by the INPS-MD prediction server.
Four PSS were detected in the CTD by more than one method, positions 124, 125, 136, and 178. Despite the similarity of the percentages of basic residues (~38%) in all H1.1 sequences, the percentages of other amino acids types are variable among the different species. In particular, percentages of non-polar residues varied from 39% to 54%, basically due to changes in the amount of alanine and valine, while the percentage of polar amino acids varied from 8% to 22%, depending on the amounts of serine and threonine present in the sequences (supplementary table S10, Supplementary Material online). Three of the positively-selected sites, 124, 125, and 136 (fig. 5A) are located in the first 25 residues of the CTD, proximal to the GD. This region may adopt both α-helix and extended strand conformations, depending on the specific amino acid composition (fig. 5B). In addition, disorder predictions in the region with the three positively selected sites (PSS) revealed changes in protein disorder. In particular, the region analyzed appeared to be more disorder-prone in primates, but more order-prone in rodentia, artiodactyla, and cetacea (fig. 5C). It is worth noting that basic residues occupy positions 124 and 136 in H1.2–H1.5. The last PSS of the CTD, position 178, is located between two CDK consensus sequences, within a region predicted to be in random coil conformation (supplementary fig. S4, Supplementary Material online). In this position, the valine present in H1.2–H1.5 subtypes was substituted by less hydrophobic or polar residues (S, T, A, and P) in H1.1, as detected by both branch-site methods (table 3).
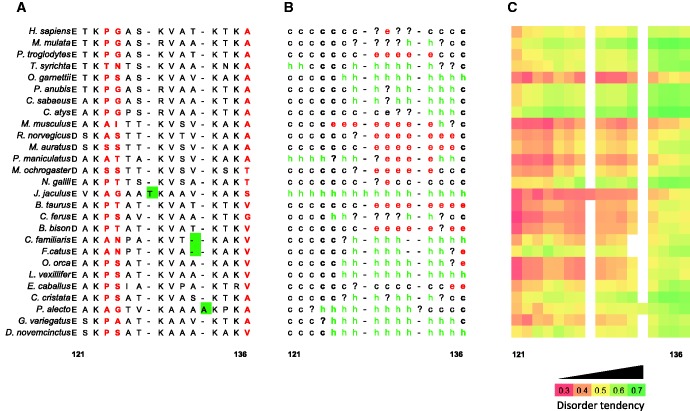
Putative structural effects of positive selection in the CTD of H1.1. (A) Multiple sequence alignment of the 27 H1.1 sequences in the region proximal to the GD, clustering positively selected sites. In red, positively selected sites; in green, indels present in H1.1 sequences. The numbers correspond to the positions in the sequence alignment in figure 1. (B) Consensus secondary structure prediction for each sequence. The analysis was performed at the Network Protein sequence analysis server available at http://npsa-pbil.ibcp.fr/. The different secondary structure motifs are labeled as follows: h, α-helix; e, extended strand; c, random coil;?, ambiguous states; -, indel in the sequence alignment. (C) Heat map of disorder tendency by residue as predicted by IUPred.
Discussion
In mammals, histone H1 comprises eleven subtypes. Subtypes H1.1–H1.5 are evolutionarily close, forming a clade (Talbert et al. 2012). H1.1–H1.5 genes are clustered with core histones, and they are expressed in a replication-dependent manner through the cell-cycle. We have examined the evolution of the mammalian H1.1–H1.5 gene family using 130 sequences, belonging to 27 different species, finding some conserved features and more importantly, evidences of positive selection in certain residues of H1.1.
Overall, basic residues are strongly conserved throughout the protein, even in the terminal domains, which have poor sequence identity between subtypes. Basic amino acids may be preferentially conserved in the terminal domains because of their major role in the induction and stabilization of chromatin higher-order structure. Therefore, a degree of functional equivalence of H1 subtypes could be achieved, helping to the interpretation of knockout experiments with the H1.1–H1.5 group of subtypes (Fan et al. 2003, 2005). The apparent functional overlap of H1 subtypes can be made compatible with their functional differentiation, assuming that specific functions optimally performed by particular subtypes can, to some extent, be fulfilled sub-optimally by other subtypes without compromising survival. Experimental evidences showing differences between H1 subtypes in their genomic distribution, expression patterns, chromatin binding affinities, PMTs and protein–protein interactions are in favor of subtype specificity (Millán-Ariño et al. 2016).
Examination of the multiple sequence alignment showed the conservation of several subtype-specific post-translational modification sites. The position and number of cyclin-dependent kinase (CDK) phosphorylation sites among subtypes are conserved both in the NTD and the CTD, indicating the importance of this PTM in the regulation of H1 function throughout the cell-cycle (Liao and Mizzen 2016). Even the specific phosphorylated residue, serine or threonine is conserved, as it is supposed to be related to the phase of the cell-cycle in which the phosphorylation occurs (Sarg et al. 2006). Some of the mitotic specific phosphorylation sites are conserved as well, including T10 in H1.5 and S35 in H1.4 (Happel et al. 2009; Chu et al. 2011). Interestingly, although S35 phosphorylation has been characterized in H1.4, consensus sequences for phosphorylation by Aurora B kinase, responsible for this modification, are detected at equivalent positions in subtypes H1.2, H1.3 and H1.5. In addition, the tetrapeptide ARKS responsible for the interaction with HP1, which is regulated by a methyl-phos switch, is conserved in primates and carnivora, but only appears sporadically in the rest of species (Daujat et al. 2005; Hergeth et al. 2011).
We also identified the presence of an indel pattern in the terminal domains conserved among paralogs, and thus, ancestral to the radiation of mammalian orders. Indels in protein coding genes are mostly small, spanning 1–5 amino acids. They occur almost exclusively in loops linking structural elements at the solvent-exposed surfaces of protein structures and are likely to be involved in intermolecular interactions and species-specific adaptations (Ajawatanawong and Baldauf 2013). Moreover, indel surveys have also been used to identify regions of the human genome under positive selection (Chen et al. 2009). Terminal domains of H1 are coded by low-complexity DNA, which is prone to slippage events during replication, originating short insertions/deletions, favoring the rapid divergence of the subtypes (Tautz et al. 1986; Ponte et al. 2003). The conservation of the indel pattern in H1 subtypes, suggest that indels might be the footprints of positive selection events associated with subtype differentiation, previous to the radiation of mammalian orders. The virtual absence of indels in ortholog sequences confirms that strong purifying selection has maintained the pattern of indels among paralogs because it is associated with function.
One the most compelling evidence of subtype functional specificity comes from the fact that sequences of gene members are more closely related between than within species, suggesting a birth-and-death mode of evolution (Eirín-López et al. 2004). Under this model of evolution new genes are created by repeated gene duplication and protein homogeneity is maintained by a strong purifying selection (Nei and Hughes 1992). In our analysis, mammalian orthologs also clustered together, indicating the higher resemblance of individual subtypes from different species in comparison with the subtypes present in a single species. In addition, the analysis of the averaged ω values in the tree branches revealed that even though all the subtypes are under strong purifying selection, the ω values have significant differences among branches, indicating that H1 subtypes are under different degrees of purifying selection. However, substitutions fixed by positive selection on a background of purifying selection were detected in H1.1.
Signatures of positive selection, ancestral to mammalian radiation and related to subtype differentiation were provided by the branch-site analyses carried out with all the sequences under study. PAML and BUSTED found strong signs of episodic positive selection in the branch clustering H1.1 sequences. This branch is longer than the counterparts to other genes, supporting the larger divergence of this subtype. In addition, H1.1 presents the highest number of ortholog indels, resulting in a variable number of amino acids. H1.1 is present in actively proliferating cells and is the predominant H1 variant in prepachytene spermatocytes, comprising approximately 70% of the total H1 (Pan and Fan 2016). H1.1 has relatively low chromatin binding affinity and is a weak chromatin condenser (Th'ng et al. 2005; Orrego et al. 2007; Clausell et al. 2009), which presumably contributes to the adoption of loosely compacted chromatin states necessary for replication and in spermatogenesis. In the latter, open chromatin is thought to facilitate genetic recombination in pachytene spermatocytes and/or the replacement of histones with transition proteins in early spermatids. Genomic mapping of H1.1 in human fibroblasts showed a distinct binding profile, suggesting a special role of this subtype in chromatin function in those cells. In contrast with subtypes H1.2–H1.5, H1.1 is present at promoters and CpG islands, enriched in intergenic regions and associated with polycomb-type chromatin (Izzo et al. 2013; Millán-Ariño et al. 2016).
The analysis at residue level revealed several positively selected sites, which may have a role in the phenotypic features of H1.1. In particular, the loss of basic residues in three of the positive selected sites located in the terminal domains (20, 125, and 136) coupled to the substitution of arginine by lysine in position 39, contribute to the decrease in the overall basicity of this subtype. In addition, the substitution of a threonine residue by valine in the helix-I of the globular domain is unfavorable for the stability of this domain. These changes, especially those in the CTD, may explain the lower affinity of H1.1 for chromatin, and therefore underscore the role of positive selection in subtype differentiation. The role of charged amino acids of the CTD in subtype affinity is evidenced in recent FRAP studies that suggest that the increased proximity of an acidic residue to the GD may be associated with the lower affinity for chromatin of mouse H1.1 (Flanagan et al. 2016).
Evidences of positive selection after mammalian radiation has been found in the CTD of H1.1 using site-specific methods. It is important to highlight that in this case the 27 sequences of H1.1 were analyzed independently and divided in three partitions due to the presence of putative recombination points. Two of the identified positions of the CTD, 125 and 136, were previously mentioned in the branch-site analysis due to the loss of a positive charge in H1.1 CTD when compared with H1.2–H1.5 subtypes. These positions were also detected to be under positive selection in H1.1 sequences, highlighting their relevance for this subtype specific functions. Interestingly, positions 125 and 136, plus another PSS (position 124) are found in a region of 25 residues of the CTD, adjacent to the GD. The equivalent region of mouse H1.0 has been shown to fold in α-helix and is associated with altering linker DNA conformation, stabilizing the folding and facilitating self-association of the chromatin fiber (Vila, Ponte, Collado, Arrondo, Suau 2001; Lu and Hansen 2004). Changes in secondary structure and intrinsic disorder propensities among H1.1 orthologs suggest that positive selection may be associated with the structural flexibility of the CTD. In fact, it has been shown that the CTD has great conformational flexibility, depending on the ionic conditions, post-translational modifications and the linker DNA conformation (Roque et al. 2005; Roque et al. 2008; Fang et al. 2012; Lopez et al., 2015; Fang et al. 2016). It seems possible that positive selection may somehow promote the folding of the CTD in a conformation or conformations optimal for H1.1 physiological role in different species.
The results show that evolutionary footprints in histone H1 subtypes are associated with both intrinsically disordered domains (NTD and CTD), indicating the importance of intrinsic disorder in the production and maintenance of genetic variation with adaptive potential (Brown et al. 2011). The more abundant amino acid in both terminal domains is lysine, which is associated with highly conserved low-complexity regions (Radó-Trilla and Albá 2012). This fact is consistent with our results, where more than half of the negatively selected sites found by REL and ADAPTSITE in the CTD are lysine residues. The conservation of the number and position of basic amino acids, as well as, the indel pattern in paralog sequences also provides a solid argument in favor of the accuracy of our results.
Low complexity regions are also proposed to promote genetic diversity by favoring recombination (DePristo et al. 2006; Lenz et al. 2014). As previously mentioned, recombination breakpoints, located close to the boundaries of the GD were detected in H1 subtypes. This finding suggests the possibility that recombination, or any other event of genetic exchange, may have been involved in the origin of H1 tripartite structure (fig. 6). A plausible model for metazoan H1 origin is a recombination event between genes encoding lysine-rich DNA binding proteins, evolutionary related to those found in eubacteria and genes encoding the “winged-helix” motif present in the GD, which appeared much later in protists, and is also present in some transcription factors (Kasinsky et al. 2001). This hypothesis would also explain that the NTD is found after the acquisition of the GD (Kasinsky et al. 2001). Later on, the combined action of gene duplication, slippage and positive selection in a birth-and-death mode of evolution may have originated the actual mammalian subtypes, which are conserved by strong purifying selection.
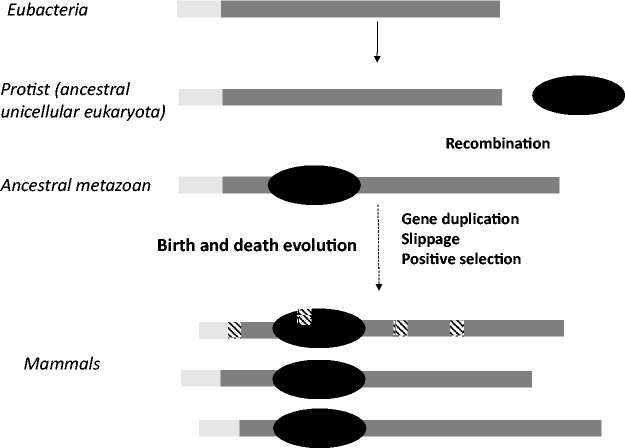
Hypothetical model of evolution of mammalian subtypes. Lysine-rich DNA binding proteins, related to H1, have been detected in eubacteria. The acquisition of the globular domain in protist may have involved a recombination event, originating the typical tripartite structure of metazoan H1. From protists to mammals, H1 has evolved under a birth-and-death mode of evolution. Subtype differentiation may have been determined by slippage and positive selection after gene duplication events. The mammalian subtypes are represented by the different lengths of the terminal domains. In dark-gray, highly basic regions; in light-gray, hydrophobic regions of the NTD; in black, the globular domain; in black line pattern, the sites under positive selection.
In summary, we have found conservation and variation between the members of mammalian H1.1–H1.5 gene family. Conserved features include overall basicity and some PTM sites that may explain to some extent the apparent functional overlap between H1 subtypes. Two major distinct features were found, ancestral to the radiation of mammalian orders, which may be associated with subtype differentiation. In the first place, a conserved pattern of indels was identified in paralogs. This pattern may constitute a footprint of slippage events after gene duplication, associated with subtype specificity. In the second place, strong evidence of positive selection was found in the branch of the phylogenetic tree clustering H1.1 sequences. Positive selection seems to has acted on specific positions of H1.1 causing a reduction in the basicity of the terminal domains and affecting the stability of the GD, and therefore, favoring the lower affinity and chromatin condensing capacity characteristic of this subtype. Apparently, more recent events of positive selection have occurred in the CTD of H1.1 sequences. Most of the positions under positive selection are clustered in the region of the CTD adjacent to the GD, where they may modulate the folding of this domain when bound to chromatin. Additionally, the presence of putative recombination points at both sides of the GD in most of the analyzed subtypes suggests the importance of this process in the origin of metazoan H1.
Acknowledgments
We thank Oscar Conchillo-Solé, Noel Hernández, David Mas and Irantzu Pallarès for their technical support. This work was supported by the Ministerio de Ciencia e Innovación (BFU2008-00460). Spain.
References
- Ahola V, Aittokallio T, Vihinen M, Uusipaikka E. 2008. Model-based prediction of sequence alignment quality. Bioinformatics 24:2165–2171. [Abstract] [Google Scholar]
- Ajawatanawong P, Baldauf SL. 2013. Evolution of protein indels in plants, animals and fungi. BMC Evol Biol. 13:140.. [Europe PMC free article] [Abstract] [Google Scholar]
- Albig W, Meergans T, Doenecke D. 1997. Characterization of the H1.5 gene completes the set of human H1 subtype genes. Gene 184:141–148. [Abstract] [Google Scholar]
- Albig W, Kioschis P, Poutska A, Meergans K, Doenecke D. 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40:314–322. [Abstract] [Google Scholar]
- Anisimova M, Nielsen R, Yang Z. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164:1229–1236. [Europe PMC free article] [Abstract] [Google Scholar]
- Beliawski JP, Yang Z. 2005. Maximum Likelihood Methods for Detecting Adaptive Protein Evolution In: Nielsen R, editor. , editor. Statistics for biology and health, Part II Springer Science, New York: p 103–124. [Google Scholar]
- Böhm L, Mitchell TC. 1985. Sequence conservation in the N-terminal domain of histone H1. FEBS Lett. 193:1–4. [Abstract] [Google Scholar]
- Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. 2011. Evolution and disorder. Curr Opin Struct Biol. 21:441–446. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen CH, Chuang TJ, Liao BY, Chen FC. 2009. Scanning for the signatures of positive selection for human-specific insertions and deletions. Genome Biol Evol. 1:415–419. [Europe PMC free article] [Abstract] [Google Scholar]
- Chu CS, Hsu PH, Lo PW, Scheer E, Tora L, Tsai HJ, Tsai MD, Juan LJ. 2011. Protein kinase A-mediated serine 35 phosphorylation dissociates histone H1.4 from mitotic chromosome. J Biol Chem. 286:35843–35851. [Europe PMC free article] [Abstract] [Google Scholar]
- Churchill MEA, Travers AA. 1991. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 16:92–97. [Abstract] [Google Scholar]
- Clausell J, Happel N, Hale TK, Doenecke D, Beato M. 2009. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One 4:e0007243.. [Europe PMC free article] [Abstract] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deléage G. 2000. NPS@: network protein sequence analysis. Trends Biochem Sci. 25:147–150. [Abstract] [Google Scholar]
- Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. 2005. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 280:38090–38095. [Abstract] [Google Scholar]
- DePristo MA, Zilversmit MM, Hartl DL. 2006. On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene 378:19–30. [Abstract] [Google Scholar]
- Drabent B, Franke K, Bode C, Kosciessa U, Bourterfa H, Hameister H, Doenecke D. 1995. Isolation of two murine H1 histone genes and chromosomal mapping of the H1 gene complement. Mamm Genome 6:505–511. [Abstract] [Google Scholar]
- Dosztányi Z, Csizmók V, Tompa P, Simon I. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. [Abstract] [Google Scholar]
- Eirín-López JM, González-Tizón AM, Martínez A, Méndez J. 2004. Birth-and-death evolution with strong purifying selection in the histone H1 multigene family and the origin of orphon H1 genes. Mol Biol Evol. 21:1992–2003. [Abstract] [Google Scholar]
- Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. 2003. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 23:4559–4572. [Europe PMC free article] [Abstract] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123:1199–1212. [Abstract] [Google Scholar]
- Fang H, Clark DJ, Hayes JJ. 2012. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 40:1475–1484. [Europe PMC free article] [Abstract] [Google Scholar]
- Fang H, Wei S, Lee TH, Hayes JJ. 2016. Chromatin structure-dependent conformations of the H1 CTD. Nucleic Acids Res. 44:9131–9141. [Europe PMC free article] [Abstract] [Google Scholar]
- Flanagan TW, Files JK, Casano KR, George EM, Brown DT. 2016. Photobleaching studies reveal that a single amino acid polymorphism is responsible for the differential binding affinities of linker histone subtypes H1.1 and H1.5. Biol Open. 5:372–380. [Europe PMC free article] [Abstract] [Google Scholar]
- Geourjon C, Deléage G. 1994. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 7:157–164. [Abstract] [Google Scholar]
- Goytisolo FA, Gerchman SE, Yu X, Rees C, Graziano V, Ramakrishnan V, Thomas JO. 1996. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 15:3421–3429. [Europe PMC free article] [Abstract] [Google Scholar]
- Graur D, Li WH. 2005. Fundamentals of molecular evolution. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Guermeur Y, Geourjon C, Gallinari P, Deléage G. 1999. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15:413–421. [Abstract] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [Abstract] [Google Scholar]
- Halle TK, Contreras A, Morrison AJ, Herrera RE. 2006. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1α. Mol Cell 22:693–699. [Abstract] [Google Scholar]
- Happel N, Doenecke D. 2009. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431:1–12. [Abstract] [Google Scholar]
- Happel N, Stoldt S, Schmidt B, Doenecke D. 2009. M phase-specific phosphorylation of histone H1.5 at threonine 10 by GSK-3. J Mol Biol. 386:339–350. [Abstract] [Google Scholar]
- Hartman PG, Chapman GE, Moss T, Bradbury EM. 1977. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur J Biochem. 77:45–51. [Abstract] [Google Scholar]
- Hergeth SP, Dundr M, Tropberger P, Zee BM, Garcia BA, Daujat S, Schneider R. 2011. Isoform-specific phosphorylation of human linker histone H1.4 in mitosis by the kinase Aurora B. J Cell Sci. 124:1623–1628. [Abstract] [Google Scholar]
- Izaurralde E, Käs E, Laemmli UK. 1989. Highly preferential nucleation of histone H1 assembly on scaffold associated regions. J Mol Biol. 210:573–585. [Abstract] [Google Scholar]
- Izzo A, Kamieniarz-Gdula K, Ramirez F, Noureen N, Kind J, Manke T, van Steensel B, Schneider R. 2013. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 3:2142–2154. [Abstract] [Google Scholar]
- Izzo A, Schneider R. 2016. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta. 1859:486–495. [Abstract] [Google Scholar]
- Kasinsky HE, Lewis JD, Dacks JB, Ausió J. 2001. Origin of H1 linker histones. FASEB J. 15:34–42. [Abstract] [Google Scholar]
- Kosakovsky-Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 22:1208–1222. [Abstract] [Google Scholar]
- Kosakovsky-Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. [Abstract] [Google Scholar]
- Lennox RW, Cohen LH. 1983. The human H1 complement of dividing and nondividing cells of the mouse. J Biol Chem. 258:262–268. [Abstract] [Google Scholar]
- Lenz C, Haerty W, Golding GB. 2014. Increased substitution rates surrounding low-complexity regions within primate proteins. Genome Biol Evol. 6:655–665. [Europe PMC free article] [Abstract] [Google Scholar]
- Liao R, Mizzen CA. 2016. Interphase H1 phosphorylation: regulation and functions in chromatin. Biochim Biophys Acta 1859:476–485. [Abstract] [Google Scholar]
- Lopez R, Sarg B, Lindner H, Bartolomé S, Ponte I, Suau P, Roque A. 2015. Linker histone partial phosphorylation: effects on secondary structure and chromatin condensation. Nucleic Acids Res. 43:4463–4476. [Europe PMC free article] [Abstract] [Google Scholar]
- Lu X, Hamkalo B, Parseghian MH, Hansen JC. 2009. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry 48:164–172. [Europe PMC free article] [Abstract] [Google Scholar]
- Lu X, Hansen JC. 2004. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J Biol Chem. 279:8701–8707. [Abstract] [Google Scholar]
- Millán-Ariño L, Izquierdo-Bouldstridge A, Jordan A. 2016. Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochim Biophys Acta. 1859:510–519. [Abstract] [Google Scholar]
- Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, et al. 2015. Gene-wide identification of episodic selection. Mol Biol Evol. 32:1365–1371. [Europe PMC free article] [Abstract] [Google Scholar]
- Nei M, Hughes AL. 1992. Balanced polymorphism and evolution by the birth-and-death process in the MHC loci In: Aizawa TM, Sasazuki T., editors. editors. Eleventh histocompatibility workshop and conference. Oxford, (England: ): Oxford University Press; p. 27–38. [Google Scholar]
- Nei M, Rooney AP. 2005. Concerted and Birth-and-death evolution of multigene families. Annu. Rev Genet. 39:121–152. [Europe PMC free article] [Abstract] [Google Scholar]
- Orrego M, Ponte I, Roque A, Buschati N, Mora X, Suau P. 2007. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 5:22.. [Europe PMC free article] [Abstract] [Google Scholar]
- Pan C, Fan Y. 2016. Role of H1 linker histones in mammalian development and stem cell differentiation. Biochim Biophys Acta. 1859:496–509. [Europe PMC free article] [Abstract] [Google Scholar]
- Parseghian MH. 2015. What is the role of histone H1 heterogeneity? A functional model emerges from a 50-year mystery. AIMS Biophys. 2:724–772. [Europe PMC free article] [Abstract] [Google Scholar]
- Parseghian MH, Henschen AH, Krieglstein KG, Hamkalo BA. 1994. A proposal for a coherent mammalian histone H1 nomenclature correlated with amino acid sequences. Protein Sci. 3: 575–587. [Europe PMC free article] [Abstract] [Google Scholar]
- Pennings S, Meersseman G, Bradbury EM. 1994. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci U S A. 91:10275–10279. [Europe PMC free article] [Abstract] [Google Scholar]
- Ponte I, Vidal-Taboada JM, Suau P. 1998. Evolution of the vertebrate histone class: evidence for the functional differentiation of the subtypes. Mol Biol Evol. 15:702–708. [Abstract] [Google Scholar]
- Ponte I, Vila R, Suau P. 2003. Sequence complexity of histone H1subtypes. Mol Biol Evol. 20:371–380. [Abstract] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [Abstract] [Google Scholar]
- Radó-Trilla N, Albà M. 2012. Dissecting the role of low-complexity regions in the evolution of vertebrate proteins. BMC Evol Biol. 12:155.. [Europe PMC free article] [Abstract] [Google Scholar]
- Roque A, Iloro I, Ponte I, Arrondo JLR, Suau P. 2005. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J Biol Chem. 280:32141–32147. [Abstract] [Google Scholar]
- Roque A, Orrego M, Ponte I, Suau P. 2004. The preferential binding of histone H1 to DNA scaffold-associated regions is determined by its C-terminal domain. Nucleic Acids Res. 32:6111–6119. [Europe PMC free article] [Abstract] [Google Scholar]
- Roque A, Ponte I, Arrondo JL, Suau P. 2008. Phosphorylation of the carboxy-terminal domain of histone H1: effects on secondary structure and DNA condensation. Nucleic Acids Res. 36: 4719–4726. [Europe PMC free article] [Abstract] [Google Scholar]
- Rost B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266: 525–539. [Abstract] [Google Scholar]
- Sarg B, Lopez R, Lindner H, Ponte I, Suau P, Roque A. 2015. Identification of novel post-translational modifications in linker histones from chicken erythrocytes. J Proteomics. 113:162–177. [Abstract] [Google Scholar]
- Sarg B, Helliger W, Talasz H, Förg B, Lindner HH. 2006. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J Biol Chem. 281:6573–6580. [Abstract] [Google Scholar]
- Savojardo C, Fariselli P, Martelli PL, Casadio R. 2016. INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics 32:2542–2544. [Abstract] [Google Scholar]
- Subirana JA. 1990. Analysis of the charge distribution of the C-terminal region of Histone H1 as related to its interaction with DNA. Biopolymers 29:1351–1357. [Abstract] [Google Scholar]
- Suzuki Y, Gojobori T, Nei M. 2001. ADAPTSITE: detecting natural selection at single amino acid sites. Bioinformatics 17:660–661. [Abstract] [Google Scholar]
- Talbert PB, Ahmad K, Almouzni G, Ausió J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, et al. 2012. A unified phylogeny-based nomenclature for histone variants. Epigenet Chromat. 5:7. [Europe PMC free article] [Abstract] [Google Scholar]
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 101:11030–11035. [Europe PMC free article] [Abstract] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [Europe PMC free article] [Abstract] [Google Scholar]
- Tautz D, Trick M, Dover GA. 1986. Cryptic simplicity in DNA is a major source of genetic variation. Nature 322:652–656. [Abstract] [Google Scholar]
- Th'ng JP, Sung R, Ye M, Hendzel MJ. 2005. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J Biol Chem. 280:27809–27814. [Abstract] [Google Scholar]
- Thomas JO. 1999. Histone H1: location and role. Curr Opin Cell Biol. 11:312–317. [Abstract] [Google Scholar]
- Thomson JD, Gibson TJ, Piewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. [Europe PMC free article] [Abstract] [Google Scholar]
- Vila R, Ponte I, Collado M, Arrondo JL, Jiménez MA, Rico M, Suau P. 2001. DNA-induced alpha-helical structure in the NH2-terminal domain of histone H1. J Biol Chem. 276:46429–46435. [Abstract] [Google Scholar]
- Vila R, Ponte I, Collado M, Arrondo JL, Suau P. 2001. Induction of secondary structure in a COOH-terminal peptide of histone H1 by interaction with the DNA: an infrared spectroscopy study. J Biol Chem. 276:30898–30903. [Abstract] [Google Scholar]
- Vila R, Ponte I, Jimenez MA, Rico M, Suau P. 2002. An inducible ´ helix-Gly-Gly-helix motif in the N-terminal domain of histone H1e: a CD and NMR study. Protein Sci. 11:214–220. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang ZF, Sirotkin AM, Buchold GM, Skoultchi AI, Marzluff WF. 1997. The mouse histone H1 genes: gene organization and differential regulation. J Mol Biol. 271:124–138. [Abstract] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041–1051. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods. 12:7–8. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 19:908–917. [Abstract] [Google Scholar]
- Yang Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [Abstract] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [Abstract] [Google Scholar]
Articles from Molecular Biology and Evolution are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/molbev/msw241
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/mbe/article-pdf/34/3/545/24366773/msw241.pdf
Citations & impact
Impact metrics
Article citations
Unraveling Histone Loss in Aging and Senescence.
Cells, 13(4):320, 09 Feb 2024
Cited by: 1 article | PMID: 38391933 | PMCID: PMC10886805
Review Free full text in Europe PMC
Casting histone variants during mammalian reproduction.
Chromosoma, 132(3):153-165, 22 Jun 2023
Cited by: 3 articles | PMID: 37347315 | PMCID: PMC10356639
Review Free full text in Europe PMC
A rapidly evolving single copy histone H1 variant is associated with male fertility in a parasitoid wasp.
Front Cell Dev Biol, 11:1166517, 31 May 2023
Cited by: 0 articles | PMID: 37325562 | PMCID: PMC10264595
Adaptive Selection in the Evolution of Aquaglyceroporins in Mammals.
J Mol Evol, 91(4):441-457, 07 May 2023
Cited by: 0 articles | PMID: 37149832
Previously Unidentified Histone H1-Like Protein Is Involved in Cell Division and Ribosome Biosynthesis in Toxoplasma gondii.
mSphere, 7(6):e0040322, 05 Dec 2022
Cited by: 4 articles | PMID: 36468865 | PMCID: PMC9769792
Go to all (15) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Birth-and-death evolution with strong purifying selection in the histone H1 multigene family and the origin of orphon H1 genes.
Mol Biol Evol, 21(10):1992-2003, 14 Jul 2004
Cited by: 42 articles | PMID: 15254261
Post-translational modifications of the intrinsically disordered terminal domains of histone H1: effects on secondary structure and chromatin dynamics.
Chromosoma, 126(1):83-91, 21 Apr 2016
Cited by: 11 articles | PMID: 27098855
Review
Histone H1 and its isoforms: contribution to chromatin structure and function.
Gene, 431(1-2):1-12, 14 Nov 2008
Cited by: 250 articles | PMID: 19059319
Review
Evolution of the vertebrate H1 histone class: evidence for the functional differentiation of the subtypes.
Mol Biol Evol, 15(6):702-708, 01 Jun 1998
Cited by: 53 articles | PMID: 9615451





