Abstract
Free full text

VBASE2, an integrative V gene database
Abstract
The database VBASE2 provides germ-line sequences of human and mouse immunoglobulin variable (V) genes. It acts as an interconnecting platform between several existing self-contained data systems: VBASE2 integrates genome sequence data and links to the V genes in the Ensembl Genome Browser. For a single V gene sequence, all references to the EMBL nucleotide sequence database are provided, including references for V(D)J rearrangements. Furthermore, cross-references to the VBASE database, the IMGT database and the Kabat database are available. A DAS server allows the display of VBASE2 V genes within the Ensembl Genome Browser. VBASE2 can be accessed either by a web-based text query or by a sequence similarity search with the DNAPLOT software. VBASE2 is available at http://www.vbase2.org, and the DAS server is located at http://www.dnaplot.com/das.
INTRODUCTION
Immunogenetics is dependent on a reliable and comprehensive database of variable gene segments in order to analyse the immune repertoire. Various approaches have been made to generate databases containing variable gene segments. The first and original database in this context is the Kabat database (1), which is a very valuable collection of sequences that are not necessarily included in the nucleotide sequence databases EMBL-Bank/GenBank/DDBJ. The Kabat database is the first database to classify the variable gene segments into families that are dependent on small sequence motifs. It also provides statistics on the variability of individual positions within the gene segments. The database has recently been commercialized. The next milestone was the establishment of the IMGT/LIGM database (2,3). This database collects all entries containing V gene notification from the EMBL-Bank/GenBank/DDBJ databases (4) and provides useful additional sequence annotation and classification. Furthermore, a systematic V gene nomenclature and a unique numbering system have been introduced. However, the IMGT/LIGM database does not sort the EMBL entries by their V gene sequences. In a heroic approach, the database VBASE (http://www.mrc-cpe.cam.ac.uk/vbase-ok) was compiled manually by analysing all human immunoglobulin variable gene segments known at the time. Rearrangements were assigned to a certain germ-line V gene and somatic mutations were excluded. The VBASE database is of great value although it was not updated after its first and final release in 1997.
Here we present the VBASE2 database. It follows the rationale of VBASE in sorting the EMBL entries by their V gene sequences. In contrast to VBASE, VBASE2 is generated automatically, and it provides new information and sequences as it implements the current knowledge derived from the genome sequencing projects by linking to the Ensembl Genome Browser (5). VBASE2 also connects the existing immunoglobulin sequence databases, thereby integrating the distinct knowledge resources.
THE VBASE2 DATASET
The current VBASE2 dataset contains immunoglobulin germ-line V genes from the heavy chain and lambda and kappa light chain loci of human and mouse. The current release holds 498 human and 554 mouse V gene sequences.
Automatic generation
The sequence data and database cross-references provided by VBASE2 are generated automatically so that manual annotation is not required. An overview about the procedure is given in Figure Figure1.1. By a BLAST search (6) of known germ-line V genes all potential V gene sequences are extracted from the EMBL-Bank, including the high throughput genomic (HTG) and whole genome shotgun (WGS) sections (4). Potential V gene sequences from Ensembl are extracted by a BLAST search against the Ensembl chromosome sequences. The DNAPLOT software is used to align, sort and compare the V gene sequences, identify J elements, RSS elements and pseudogenes. Synthetic sequences are detected and removed. All germ-line configured V genes are matched to the rearranged sequences. To assign a rearrangement to a germ-line sequence a 100% match in the V gene region is required. Thus, the sequence comparison is restricted to the FR1–FR3 region, excluding potential N nucleotides in CDR3. The current procedure assigns V gene alleles to different V gene entries, and allele assignment is not yet included in the database. V gene families are assigned using family consensus sequences. In addition, DNAPLOT is used to compare the VBASE2 dataset with the LIGM dataset from the IMGT database, the VBASE database and the last freely available version of the Kabat database (ftp://ftp.ebi.ac.uk/pub/databases/kabat/). Owing to the changes in the source sequence databases, Ensembl and EMBL-Bank/GenBank/DDBJ, the VBASE2 dataset is updated regularly.
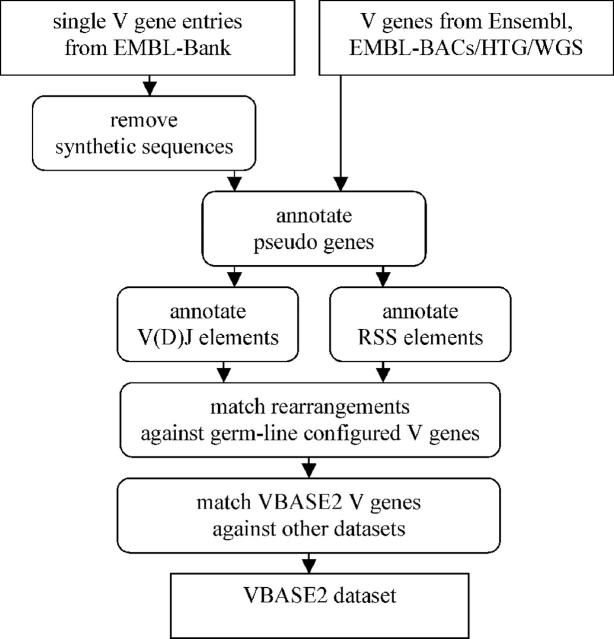
The data generation procedure. The procedure analysing the V gene sequences retrieved by the BLAST search is performed using the DNAPLOT program and interconnecting Perl scripts. EMBL-Bank entries containing a single V gene are filtered for synthetic sequences. All V gene sequences are checked for stop codons to detect pseudo genes. Rearrangements are detected by an alignment against J elements, RSS element detection allows the detection of germ-line configured V gene entries. In a multiple alignment step, all rearranged V gene sequences are matched to the germ-line configured V genes. All germ-line V genes are matched against the VBASE, IMGT/LIGM and KABAT database.
Sequence class assignment
Depending on their sequence sources, the V genes are grouped into three classes (Table (Table1).1). Class 1 holds sequences for which a genomic sequence and a rearranged sequence are known. Class 2 contains sequences that have not been found in a rearrangement, thus lacking evidence of functionality. This class includes pseudogenes and orphans, but it might also contain V genes of rare usage or V genes for which rearrangements are known only in a somatic mutated version. Class 3 contains sequences, which have been observed in different V(D)J rearrangements that give strong evidence of the absence of mutations, but lack a genomic reference.
Table 1.
| Class 1 | Class 2 | Class 3 | Total | |
|---|---|---|---|---|
| Human IGHV | 59 | 204 | 3 | 266 |
| Human IGKV | 46 | 100 | 2 | 148 |
| Human IGLV | 38 | 46 | 0 | 84 |
| Mus IGHV | 121 | 212 | 11 | 344 |
| Mus IGKV | 75 | 123 | 7 | 205 |
| Mus IGLV | 3 | 2 | 0 | 5 |
The number of V genes from the three immunoglobulin loci in human and mouse are shown. Class 1 sequences are supported by a genomic sequence and a rearrangement. Class 2 contains sequences with genomic evidence only and Class 3 holds sequences, which have been found in rearrangements only.
Cross-references, V gene annotation and features
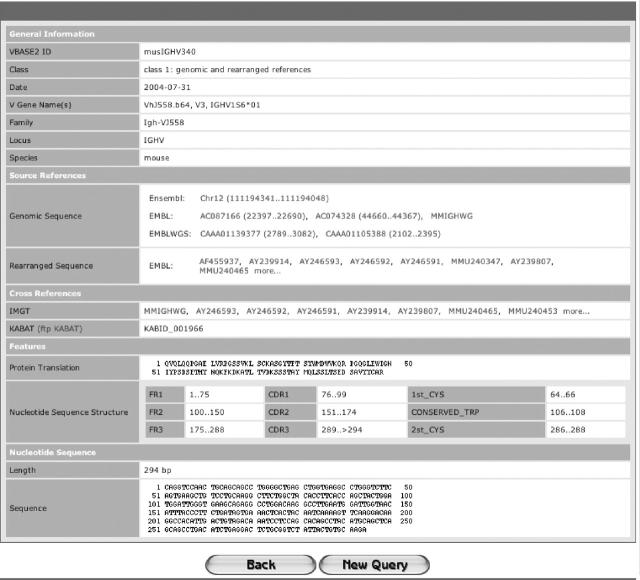
Each V gene entry holds a list of source references linking to EMBL-Bank and/or Ensembl (Figure (Figure2).2). If the EMBL-Bank reference is a BAC sequence, the V gene position within the BAC is given, as many BAC sequences have not yet been annotated. Sequences containing stop codons are labelled as pseudogenes, V genes allocated to another chromosomal locus are marked as orphans. As several names may have been assigned to the same V gene all known names for each V gene are listed. Furthermore, hits in the IMGT-, KABAT- and VBASE-databases are shown. These cross-references allow access to manually annotated data available in these databases. Also, the protein translation and the positions of the complementary determining regions (CDRs) are indicated.
ACCESSING THE VBASE2 DATABASE
The VBASE2 database can be accessed at http://www.vbase2.org. V gene entries can be requested either by a text-based query or a sequence similarity search with the DNAPLOT tool.
The Direct Query form
For a text-based query the VBASE2 website provides the selection of species, V gene locus and V gene family. Text fields allow the search for V gene names, VBASE2 sequence IDs and V gene reference IDs from the EMBL, IMGT, VBASE and Kabat database. By choosing a class the search can be restricted to a certain sequence quality. By pasting a nucleotide or protein sequence into the sequence input field the user can search for a matching VBASE2 sequence. However, as this query will only report a 100% identity match this field is more useful to search for the appearance of certain sequence fragments rather than to compare a complete V gene sequence with the VBASE2 dataset.
The DNAPLOT query
To compare a complete V gene sequence or rearrangement with the VBASE2 dataset, the DNAPLOT query provides a sequence similarity search tool. The query returns a V gene alignment referring to the IMGT unique numbering (3), containing the query sequence and the best VBASE2 matches. Queries containing a V gene rearrangement return the name of the D- and J-element and also the automatically assigned V gene family is given (Figure (Figure33).
Ensembl DAS server
Those VBASE2 V genes that can be mapped onto a chromosome in Ensembl have a link to the gene location in the Ensembl Genome Browser. The VBASE2 V genes can also be viewed within the browsers' Contig View by selecting the DAS server at http://www.dnaplot.com/das, and clicking on the V gene links to the corresponding VBASE2 database entry.
IMPLEMENTATION
VBASE2 is implemented in a relational database structure using PostgreSQL DBMS. The web interface uses PHP scripts for dynamic web pages. The website requires a HTML 4.0-compliant browser with JavaScript enabled. The automatic generation procedure uses the NCBI BLASTALL program, the DNAPLOT program and Perl scripts.
CONCLUSIONS
VBASE2 connects several separated data collections and thereby combines all V gene annotation and classification data from the distinct resources. Furthermore, it shows the chromosomal location of a V gene in Ensembl, and a DAS server enables the display of the V genes in the Ensembl Genome Browser. During the automatic data generation process, sequences are sorted and evaluated only on the basis of their sequence information. Classification and cross-references allow the user to validate the sequence quality. Currently, the VBASE2 database contains germ-line V gene sequences of the immunoglobulin loci of human and mouse. A forthcoming challenge in the future development of the database is the assignment of haplotypes and V gene alleles. Another important step is the extension of the stored V gene sequence to the end of the RSS element. Furthermore, the scope of the database will be extended; as the process of sequence extraction and evaluation only requires the extension of the computer programs and the underlying sequence tables, the database can be expanded to T-cell receptor sequences and to other species.
ACKNOWLEDGEMENTS
We are grateful to Rolf Hühne who set up the NGFN-BLAST service, supplying the base for the VBASE2 dataset generation procedure. We also thank Miguel Nunes for continuous improvements of the DNAPLOT program and Andreas Kahari for support with the DAS server. We thank Ian Tomlinson for allowing us to call our database ‘VBASE2’ and for his helpful discussion. This work was funded by the German Bundesministerium für Bildung und Forschung (BMBF) for the bioinformatics competence center ‘Intergenomics’ (grant no. 031U110A/031U210A).
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gki088
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/33/suppl_1/D671/7622126/gki088.pdf
Citations & impact
Impact metrics
Article citations
AIRR-C IG Reference Sets: curated sets of immunoglobulin heavy and light chain germline genes.
Front Immunol, 14:1330153, 09 Feb 2024
Cited by: 5 articles | PMID: 38406579 | PMCID: PMC10884231
HSP60 mimetic peptides from Mycobacterium leprae as new antigens for immunodiagnosis of Leprosy.
AMB Express, 13(1):120, 27 Oct 2023
Cited by: 0 articles | PMID: 37891336 | PMCID: PMC10611693
Human-murine chimeric autoantibodies with high affinity and specificity for systemic sclerosis.
Front Immunol, 14:1127849, 16 Jun 2023
Cited by: 0 articles | PMID: 37398644 | PMCID: PMC10311643
Role of the mechanisms for antibody repertoire diversification in monoclonal light chain deposition disorders: when a friend becomes foe.
Front Immunol, 14:1203425, 13 Jul 2023
Cited by: 4 articles | PMID: 37520549 | PMCID: PMC10374031
Review Free full text in Europe PMC
AIRR community curation and standardised representation for immunoglobulin and T cell receptor germline sets.
Immunoinformatics (Amst), 10:100025, 19 Feb 2023
Cited by: 3 articles | PMID: 37388275 | PMCID: PMC10310305
Go to all (112) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome Information Broker for Viruses (GIB-V): database for comparative analysis of virus genomes.
Nucleic Acids Res, 35(database issue):D339-42, 07 Dec 2006
Cited by: 14 articles | PMID: 17158166 | PMCID: PMC1781101
The UCSC Genome Browser Database: update 2006.
Nucleic Acids Res, 34(database issue):D590-8, 01 Jan 2006
Cited by: 838 articles | PMID: 16381938 | PMCID: PMC1347506
The EMBL Nucleotide Sequence Database.
Nucleic Acids Res, 32(database issue):D27-30, 01 Jan 2004
Cited by: 83 articles | PMID: 14681351 | PMCID: PMC308854
[Introduction to Go! Poly, a human genome polymorphism database].
Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 18(6):482-485, 01 Dec 2001
Cited by: 0 articles | PMID: 11774222
Review