Abstract
Free full text

Local Inflammatory Cues Regulate Differentiation and Persistence of CD8+ Tissue-Resident Memory T Cells
Associated Data
SUMMARY
Many pathogens initiate infection at mucosal surfaces and tissue-resident memory T (Trm) cells play an important role in protective immunity, yet the tissue-specific signals that regulate Trm differentiation are poorly defined. During Yersinia infection, CD8+ T cell recruitment to areas of inflammation within the intestine is required for differentiation of the CD103−CD69+ Trm subset. Intestinal proinflammatory microenvironments have elevated IFN-β and IL-12, which regulated Trm markers including CD103. Type I interferon receptor- or IL-12 receptor-deficient T cells functioned similarly to WT cells during infection; however, the inability T cells to respond to inflammation resulted in defective differentiation of CD103−CD69+ Trm cells and reduced Trm persistence. Intestinal macrophages were the main producers of IFN-β and IL-12 during infection and deletion of CCR2+ IL-12-producing cells reduced the size of the CD103− Trm population. These data indicate intestinal inflammation drives phenotypic diversity and abundance of Trm cells for optimal tissue-specific immunity.
Graphical abstract
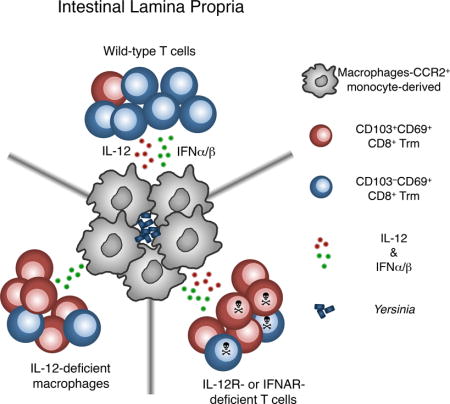
INTRODUCTION
During infection, effector CD4+ and CD8+ T cells enter a variety of peripheral tissues and differentiate into a population of tissue-resident memory T cells (Trm) that persist locally for extended periods and are unable to re-enter the circulation (Mueller and Mackay, 2016; Schenkel and Masopust, 2014). Pathogen-specific Trm cells outnumber those in the lymphoid organs (Steinert et al., 2015) and play important roles in protecting against secondary infection (Mueller and Mackay, 2016; Schenkel and Masopust, 2014). Trm cells can serve as a potent alarm by producing IFNγ, leading to the recruitment of additional innate and adaptive immune cells into the tissue (Schenkel et al., 2014; 2013) and priming the tissue to enhance pathogen resistance (Ariotti et al., 2014). Therefore, it is of significant interest to identify the signals that drive the differentiation and maintenance of Trm populations in response to infection and immunization.
Clustering of CD4+ and CD8+ Trm cells with other immune cells around areas of microbial invasion is commonly observed during tissue infection, and these immune cell aggregates constitute distinct microenvironments that support Trm responses. These structures have been identified in the brain (Wakim et al., 2010), female reproductive tract (Iijima and Iwasaki, 2014), skin (Collins et al., 2016; Natsuaki et al., 2014), lung (Anderson et al., 2014), and intestine (Bergsbaken and Bevan, 2015). In addition to T cells, immune cell clusters are comprised of macrophages and dendritic cells, but lack blood/lymphatic vasculature and B cells, distinguishing them from tertiary lymphoid structures (Bergsbaken and Bevan, 2015; Iijima and Iwasaki, 2014; Wakim et al., 2010). The production of chemokines by macrophages in these clusters is required for the recruitment and differentiation of Trm cells (Bergsbaken and Bevan, 2015) and their long term retention within the tissue (Collins et al., 2016; Iijima and Iwasaki, 2014). Presentation of microbial antigens in these structures can affect Trm differentiation and survival in some tissues (Wakim et al., 2010; Khan et al., 2016). T cell clustering may also limit the spread of infection, as they form around areas of pathogen invasion and replication (Bergsbaken and Bevan, 2015; Hickman et al., 2015; Wakim et al., 2010). We are only beginning to understand the signals that are required for the formation and maintenance of these structures and the signals they provide to Trm cells.
Trm cells are phenotypically distinct from memory T cells in lymphoid tissues, and expression of CD69 and the integrin CD103 are often used to define T cells as tissue-resident. In some tissues, CD103+CD69+ cells make up the majority of the Trm population and CD103− T cells are only transiently present and quickly re-enter the circulation (Gebhardt et al., 2011). Some tissues, including the intestine, contain subsets of T cells that lack CD103 and/or CD69 expression, but are nonetheless capable of maintaining tissue residence (Bergsbaken and Bevan, 2015; Hondowicz et al., 2015; Steinert et al., 2015). This phenotypic heterogeneity among Trm populations often depends on the tissue of residence and whether Trm cells are generated by local tissue infection. Currently, it is unclear whether these distinct Trm populations differentially contribute to pathogen control. CD103 has been shown to alter migration of T cells within the intestinal epithelium (Edelblum et al., 2012) and enhance killing of E-cadherin-expressing antigen presenting cells (Le Floc’h et al., 2007), suggesting differential expression of CD103 may regulate Trm function. Cytokine and chemokine receptors are also differentially regulated in CD103+ and CD103− Trm subsets (Mackay et al., 2013; Wakim et al., 2012), suggesting CD103 expression may divide Trm cells into functionally distinct subsets.
During local infection with the bacterial pathogen Yersinia pseudotuberculosis, two populations of CD8+ Trm cells are present in the lamina propria (LP), a CD103+CD69+ and a CD103−CD69+ subset. Immune cell clustering in areas of bacterial infection leads to the differentiation of the CD103−CD69+ CD8+ Trm subset (Bergsbaken and Bevan, 2015), but the inflammatory signals regulating this process are unknown. Here we identify IFN-β and IL-12 as important regulators of CD103−CD69+ Trm differentiation in proinflammatory microenvironments. Absence of IFNAR1 or IL-12Rβ2 on T cells allowed proper expansion and trafficking of T cells, but once in the intestine, cytokine receptor-deficient cells failed to differentiate into the CD103−CD69+ Trm population and did not persist after infection. Intestinal macrophage populations produced IFN-β and IL-12 during infection, and deletion of IL-12-producing CCR2+ cells also reduced CD103−CD69+ Trm differentiation. These studies identify type I IFN and IL-12 in intestinal proinflammatory microenvironments as key regulators of the differentiation and persistence of Trm cells. Maximizing the number and function of Trm cells within the tissue during immunization may be critical for resistance to mucosal infection and a determinant of mucosal vaccine efficacy.
RESULTS
IFN-β and IL-12 suppress TGF-β-mediated CD103 expression
Simultaneous exposure to TGF-β and certain inflammatory cytokines ex vivo can inhibit CD103 expression by LCMV primed CD8+ T cells (Casey et al., 2012). These data suggest inflammatory signals sensed by T cells entering the intestine could override the developmental program initiated by TGF-β and lead to differentiation of the CD103−CD69+ Trm subset. The ability of inflammatory cytokines to control CD103 expression in Yersinia-primed effector CD8+ T cells was examined. OT-I T cells were isolated from mice 7 days after Yptb-OVA infection and cultured in the presence of cytokines for 20 hours. As expected, in vivo primed cells cultured in the presence of TGF-β alone increased expression of CD103 compared to untreated T cells (Figure 1A, ,1B).1B). Addition of IFN-β or IL-12 to these cultures prevented CD103 expression; however, IL-18 did not significantly alter the TGF-β-mediated upregulation of CD103 (Figure 1A, ,1B).1B). Other cytokines were tested, and only IFN-β and IL-12 were able to completely prevent the expression of CD103 (data not shown). CD69 expression was also examined under these conditions, and expression of CD103 and CD69 were not mutually exclusive. Culture of in vivo primed T cells with TGF-β and IFN–β, IL-12, or IL-18 all resulted in a significant increase in the expression of CD69 compared to TGF-β alone (Figure 1C).
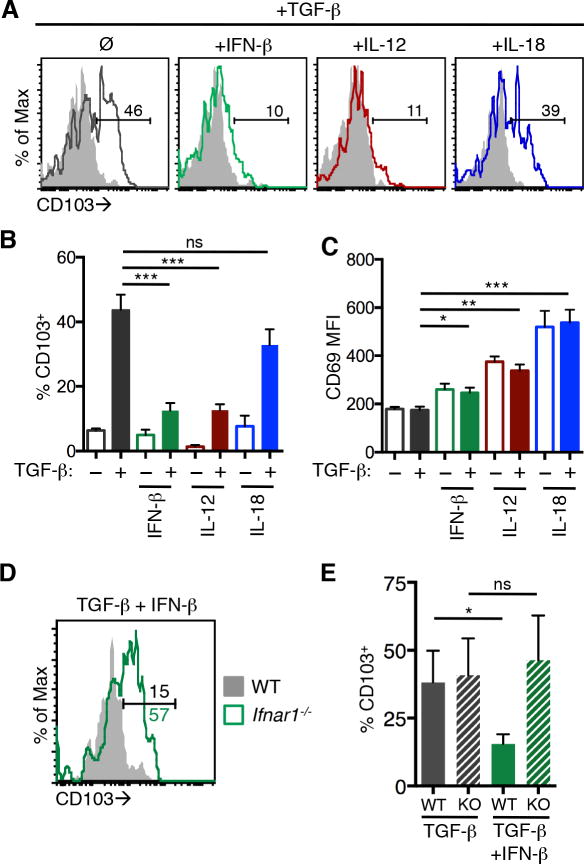
Mice received WT OT-I T cells (A–C) or an equal number of WT and Ifnar1−/− OT-I T cells (D–E) and were infected with Yptb-OVA. Seven days post infection, cells from the MLNs and spleen were cultured with the indicated cytokines for 20 hours and CD103 and CD69 expression were analyzed. (A) Representative histograms of CD103 expression on OT-I T cells stimulated with the indicated cytokines in the absence (shaded histograms) or presence of TGF-β (open histograms). Numbers represent the percent of cells within the CD103+ gate. Mean percent CD103+ cells (B) and CD69 MFI (C) from technical replicates and representative of 3 experiments. Error bars represent SD. (D) Representative histograms of CD103 expression on in vivo primed WT (shaded histogram) and Ifnar1−/− (open histogram) OT-I T cells cultured with TGF-β and IFN-β. Numbers represent the percent of cells within the CD103+ gate. (E) Mean percent CD103+ cells from triplicate samples from one representative experiment of 2. Error bars represent SD. *p≤0.05, **p≤0.005, ***p≤0.0005 by unpaired t-test. See also Figure S1.
To determine whether the regulation of CD103 and CD69 expression by cytokine stimulation was T cell-intrinsic, wild-type (WT) and Ifnar−/− OT-I T cells were primed in vivo with Yptb-OVA and then cultured with IFN-β and TGF-β. WT OT-I T cells did not express CD103 after culture with TGF-β and IFN-β, but Ifnar−/− OT-I T cells maintained CD103 expression under these conditions (Figure 1D, ,1E).1E). A similar percent of WT and Ifnar−/− cells upregulated CD103 when cultured with TGF-β alone (Figure 1E). IFN-β also directly regulated CD69 levels on effector T cells (Figure S1). These data indicate IFN-β and IL-12 can negatively regulate expression of the Trm marker CD103 in response to TGF-β, while positively affecting the expression of CD69 on effector CD8+ T cells.
IFN-β and IL-12 are produced in intestinal proinflammatory microenvironments
IFN-β-YFP and IL12p40-YFP reporter mice were utilized to track cytokine production within the intestinal tissue during Yersinia infection. We observed small numbers of YFP+ cells in the intestine of uninfected control mice (Figure 2A, left panels, 2B). Seven days after Yptb-OVA infection, we observed large clusters of YFP+ cells in the LP near the crypts in both IFN-β- and IL-12-YFP reporter mice (Figure 2A, right panels, 2B). Fewer YFP+ cells were seen in the villi, consistent with proinflammatory microenvironments previously observed during Yersinia colonization of the intestinal tissue (Bergsbaken and Bevan, 2015). At this timepoint, antigen-specific T cells are beginning to enter the intestine, and CD8β+ cells were enriched in areas that contain cytokine producing cells (Figure 2C), indicating local IFN-β and IL-12 production could influence Trm differentiation.
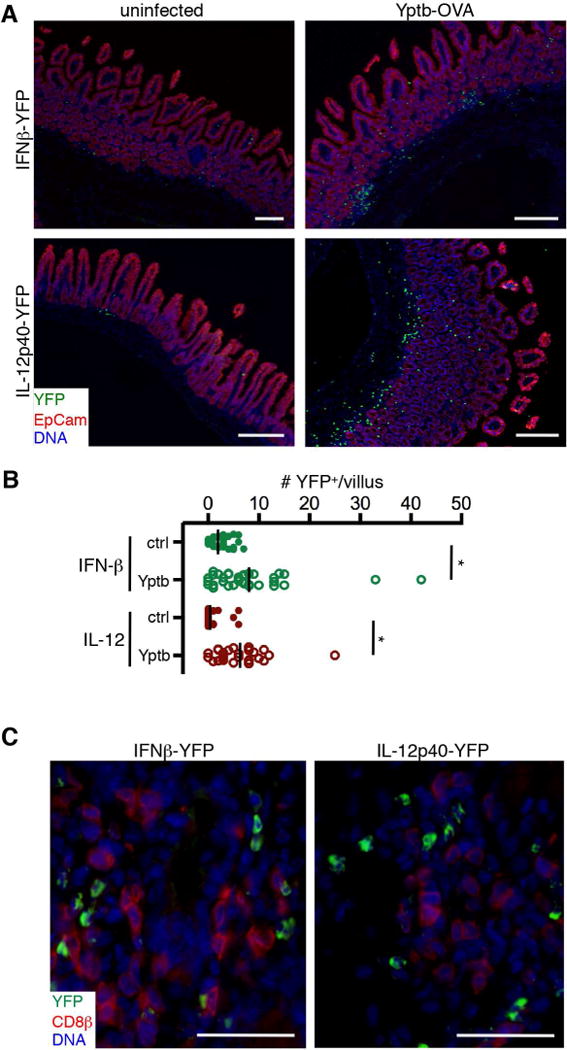
IFNβ-YFP and IL12p40-YFP mice were infected with Yptb-OVA and tissues were isolated 7 days post infection. (A) Expression of YFP (green), Epcam (red), and DNA (blue) in control uninfected intestine (left panels) and Yptb-OVA infected intestine (right panels). Scale bars, 250μm. (B) The number of YFP+ cells/villus in uninfected control and Yptb-infected IFN-β-YFP and IL-12p40-YFP mice. *p<0.0001 by the Mann-Whitney test. (C) Infiltrating CD8β+ T cells (red) are found in areas of cytokine expression (green). Scale bars, 40μm. All images are representative of 2–4 mice for each condition. See also Figure S2.
To determine whether CD103−CD69+ CD8+ Trm cells are exposed to lower levels of TGF-β as a consequence of their localization, Tgfb expression was examined in intestinal microenvironments using laser capture microdissection. LP regions containing CD8+ T cell clusters were compared to areas lacking T cell clusters, and Tgfb mRNA was present at higher levels in areas containing CD8+ T cell clusters compared to other areas of the LP (Figure S2A). In addition, at 9 days post infection, CD103+ and CD103− LP T cells express similar levels of Tgfbr2 (Figure S2B). These data indicate T cells entering these intestinal microenvironments are exposed to TGF-β but that high local concentrations of inflammatory cytokines would prevent CD103+CD69+ Trm differentiation.
CD103+ and CD103− CD8+ Trm populations adopt and maintain unique gene expression profiles
Regulation of T-box transcription factor expression is critical for the differentiation of CD8+ Trm cells in both the lung and skin (Laidlaw et al., 2014; Mackay et al., 2015). T-bet has been shown to negatively regulate CD103 expression in lung Trm cells via direct binding of T-bet to the Itgae (CD103) promoter (Laidlaw et al., 2014). Exposure of CD8+ T cells to IFN-β and IL-12 could lead to elevated T-bet expression, preventing the upregulation of CD103 in this Trm subset. The expression of Tbx21 (T-bet) was analyzed in LP Trm cells by qRT-PCR, and Tbx21 expression was similar in both CD103+ and CD103− populations at days 9 and 32 post infection (Figure 3A). This was confirmed by intracellular staining for T-bet protein (Figure 3B); about 40% of Trm cells in both CD103+ and CD103− Trm populations expressed elevated levels of T-bet at >30 days post infection (Figure 3C). Thus, altered expression of T-bet alone is not sufficient to regulate the differentiation of CD103−CD69+ intestinal Trm population.
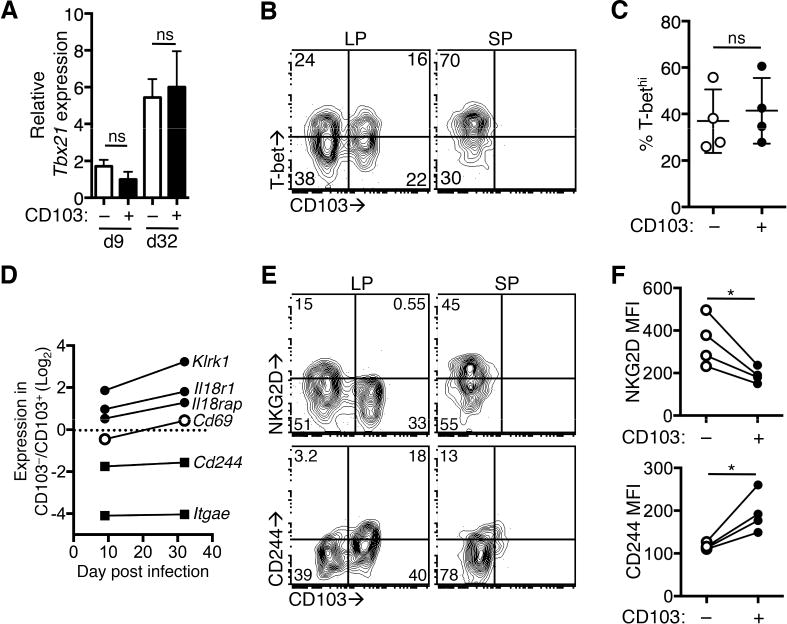
Mice received OT-I T cells and were infected with Yptb-OVA and T cells were analyzed at 9 and >30 days post infection. (A) CD103+ and CD103− OT-I T cells were sorted from the LP at the indicated timepoints and expression of Tbet was determined by qRT-PCR. Values are expressed relative to actb expression. Mean and SD represent technical replicates, representative of 2 experiments. (B) T-bet expression in CD103+ and CD103− LP and splenic (SP) OT-I T cells at 30 days post infection. Numbers are the percent of cells in each quadrant. (C) Percent T-bethi of CD103+ and CD103− LP Trm populations. Symbols represent individual mice from one experiment and are representative of 2 experiments. (D) CD103+ and CD103− T cells were sorted from the LP at the indicated timepoints and gene expression was determined by qRT-PCR. Values are expressed relative to actb expression and the fold change in gene expression in CD103−/CD103+ populations is shown. The dashed line indicates equivalent expression by both populations. (E) OT-I T cells from the LP and spleen were analyzed for expression of CD103 and NKG2D (top panel) or CD244 (bottom panel) at >30 days post infection. Numbers represent percent of cells in each quadrant. Representative plots of at least 6 mice from 2 or more experiments. (F) MFI of NKG2D (top panel) and CD244 (bottom panel) on CD103+ and CD103− LP OT-I T cells at >30 days post infection. Lines connect populations from individual mice. *p<0.05 by paired t-test. See also Figure S3.
IL-12 and type I IFN result in phosphorylation of STAT4 in CD8+ T cells (Nguyen et al., 2002), and we examined the expression of several genes that are regulated by IL-12/IFN-β/pSTAT4 (Good et al., 2009; Wei et al., 2010) in CD103+ and CD103− LP Trm subsets at 9 and 34 days after Yptb infection. This included genes regulating IL-18 responsiveness (Il18r1, Il18rap), Klrk1 (encoding NKG2D), and Cd244, a marker of Trm cells from a variety of tissues (Mackay et al., 2013) that is often associated with T cell dysfunction (Schietinger and Greenberg, 2014). We determined these genes were differentially expressed by qRT-PCR, while CD69 is expressed equivalently by CD103+ and CD103− LP Trm populations (Figure 3D). We also observed differential surface expression of NKG2D and CD244 in LP Trm populations at >30 days post infection. CD103− LP T cells expressed elevated levels of NKG2D; in contrast, the CD103+ subset had increased expression of CD244 (Figure 3E, left panels, 3F). NKG2D and CD244 were both expressed on a subset of splenic OT-I T cells (Figure 3E, right panels). Taken together, these data indicate that Yptb-OVA-primed CD8+ T cells are exposed to IFN-β and IL-12 and adopt and maintain a transcriptional signature that suggests differential exposure to these cytokines.
Both CD103+ and CD103− Trm populations persist long after infection is cleared (Bergsbaken and Bevan, 2015); however, to confirm the stability of these populations, CD103+CD69+ and CD103−CD69+ LP Trm cells were sorted from the intestine at >30 days post infection and stimulated for 24 hours with anti-CD3/CD28 beads in combination with IL-12 or TGF-β. These stimuli failed to alter the expression of CD103 on LP Trm cells (Figure S3A,B), indicating that delivery of early, transient signals to T cells during infection results in stable Trm populations.
Cytokine receptor-deficient T cells enter the intestine and display enhanced expression of Trm markers
To analyze the role of inflammatory cytokines in the differentiation of Trm cells in vivo, Il18r1−/−, Ifnar1−/−, and Il12rb2−/− OT-I T cells were transferred into mice at a 1:1 ratio with WT T cells. Mice were infected with Yptb-OVA and the expansion and trafficking of T cells into the LP were analyzed at 9 days post infection. Cytokine receptor-deficient and WT OT-I T cells expanded to a similar degree in the spleen (Figure 4A, left) and both entered the intestinal tissue in response to infection (Figure 4A, right). After local infection, CD8+ T cells that enter the intestine quickly acquire a Trm phenotype (Bergsbaken and Bevan, 2015; Sheridan et al., 2014), and 25–60% of WT cells in the LP were CD103+ at 9 days post infection (Figure 4B, top panel, panel,4C).4C). There was no significant difference in the expression of CD103 on Il18r1−/− and WT OT-I T cells, and Ifnar1−/− T cells had slightly elevated CD103 expression, although this was not statistically significant (Figure 4B, top panel, panel,4C).4C). However, CD103 was significantly elevated on Il12rb2−/− LP T cells, with 55–85% of cells expressing CD103 (Figure 4B, top panel, panel,4C).4C). CD69 expression was similar between all groups (Figure 4B, bottom panel, panel,4D).4D). We analyzed other markers differentially regulated in CD103+ and CD103− LP Trm subsets to determine if they were similarly altered in the absence of IL-12Rβ2. We found that CD244 expression was slightly increased in CD103+ WT cells at 9 days post infection (Figure 4E), and an increased percentage of Il12rb2−/− CD103+ LP T cells had already upregulated CD244 at 9 days post infection compared to WT cells (31% vs. 14%, Figure 4E). Cytokine signaling is not required for expansion or intestinal trafficking of CD8+ T cells, but in the absence of IL-12 signaling, LP T cells show enhanced differentiation into the CD103+ subset and early expression of other Trm markers, including CD244.
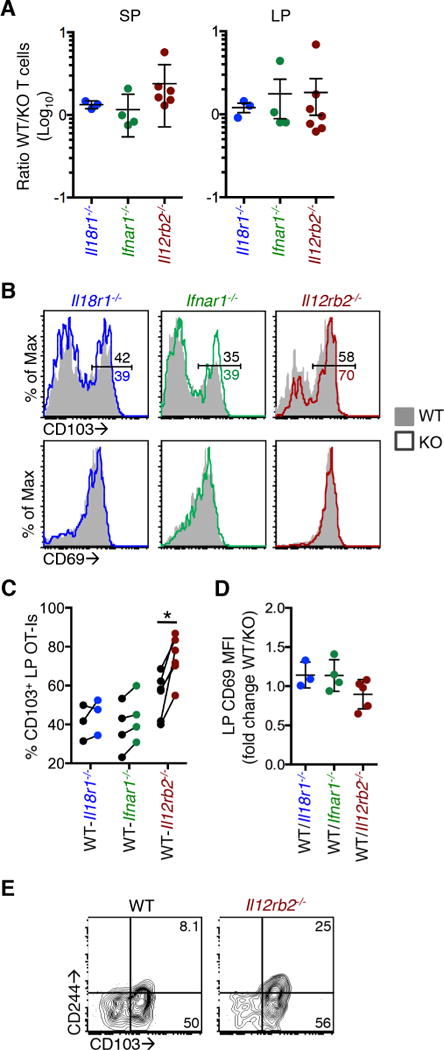
Mice received 5×103 WT and 5×103 Il18r1−/−, Ifnar1−/−, or Il12rb2−/− OT-I T cells and were infected with Yptb-OVA by oral gavage. T cells were analyzed at 9 days post infection. (A) Ratio of WT to Il18r1−/−, Ifnar1−/−, or Il12rb2−/− OT-I T cells in the spleen (left panel) and LP (right panel). Ratios do not significantly differ from 1.0 using a one sample t-test. (B) Representative histograms of CD103 (top panels) and CD69 (bottom panels) expression on LP T cells. Numbers (top panels) are the percent in the CD103+ gate. (C) Percent CD103+ of WT and cytokine receptor-deficient OT-I T cells isolated from the LP. Lines connect data points from individual mice. (D) CD69 expression on LP cells expressed as the ratio of CD69 MFI on WT/cytokine receptor-deficient T cells. (E) Representative plot of CD103 and CD244 expression on WT and Il12rb2−/− LP OT-I T cells. Data are pooled from 2 or more experiments (A,C,D) or from one experiment and representative of at least two experiments (B,E). *p<0.005 by paired t-test.
Il12rb2−/− T cells enter proinflammatory microenvironments and exhibit normal effector function
We hypothesized that cytokine receptor-deficient T cells were localizing to proinflammatory microenvironments during Yptb-OVA infection, but their inability to respond to cytokines led to aberrant differentiation into the CD103+CD69+ Trm subset. We have previously demonstrated that CXCR3 mediates recruitment of CD8+ T cells to proinflammatory microenvironments and this localization is required for differentiation of the CD103− Trm population (Bergsbaken and Bevan, 2015). We confirmed similar expression of CXCR3 on splenic WT and Il12rb2−/− OT-I T cells after Yptb-OVA infection (Figure 5A). Localization of T cells was analyzed directly using immunohistochemistry; CD8+ T cell clusters in the ileum of infected mice contained similar numbers of WT and Il12rb2−/− OT-I T cells (Figure 5B). There was no significant difference in the ratio of WT to Il12rb2−/− OT-I T cells in clustered populations compared to non-clustered populations (Figure 5C). A greater percentage of Il12rb2−/− OT-I T cells within proinflammatory microenvironments expressed CD103 when compared to WT cells (Figure S4), confirming that cytokine receptor-deficient T cells enter proinflammatory microenvironments, but their failure to respond to IL-12 in these areas leads to elevated CD103 expression.

Mice received 5×103 WT and 5×103 Il12rb2−/− OT-I T cells and were infected with Yptb-OVA by oral gavage. T cells were analyzed at 9 days post infection (A–E). (A) Representative histogram of CXCR3 expression on splenic WT and Il12rb2−/− OT-I T cells. (B) Localization of GFP+ CD45.2+ WT (green and red) and CD45.2+ Il12rb2−/− (red) OT-I T cells in the ileum. Representative image from one of 4 mice. (C) Ratio of WT to Il12rb2−/− LP OT-I T cells within individual T cell clusters containing at least 10 cells and non-clustered T cells from individual fields. Data are pooled from 4 mice. p=0.14 by Mann-Whitney. (D) LP cells were restimulated with SIINFEKL peptide in the presence of Brefeldin A for 4 hours and IFNγ production was analyzed. IFNγ+ gates were drawn using unstimulated controls. Representative plots of 5 mice from 3 experiments. (E) Granzyme B expression in LP T cells, representative of 5 mice from 3 experiments. (F) Day 7 effector CD8+ T cells from WT and Il12rb2−/− mice were transferred into WT mice that were infected with Yptb 3 days earlier. Two days after T cell transfer, CFU were enumerated in the ileum (left panel) and spleen (right panel) and compared to infected mice receiving no T cells. Data are pooled from 4 experiments and geometric mean is shown. *p<0.05, **p<0.005 by the Mann-Whitney test. See also Figure S4.
Clustered CD103− LP T cells mediate control of bacterial replication during primary infection (Bergsbaken and Bevan, 2015), and both cytolytic molecules and cytokines contribute to control of Yersinia replication by CD8+ T cells (Szaba et al., 2014). The production of effector molecules by WT and Il12rb2−/− OT-I T cells from the LP was analyzed; approximately 50% of each population produced IFN-γ after peptide restimulation (Figure 5D). Granzyme B expression was also similar between WT and cytokine receptor-deficient populations in the LP (Figure 5E). This allowed us to determine if localization per se or localization coupled with proper Trm differentiation is required for control of bacterial replication. WT and Il12rb2−/− mice were infected with Yptb, and 7 days later, effector CD8+ T cells were isolated from the mesenteric lymph nodes (MLNs) and spleen. WT and Il12rb2−/− CD8+ T cells were transferred into separate mice that had been infected with Yptb 3 days prior. Two days after transfer, the spleen and ileum were isolated and the number of Yptb colony forming units (CFU) was determined. Transfer of either WT or Il12rb2−/− CD8+ T cells reduced bacterial CFU in both the spleen and ileum compared to mice receiving no T cells (Figure 5F). Thus, localization T cells and not IL-12 signaling is critical for control of bacterial replication in the intestine, and early expression of markers (CD103, CD244) restricted to cells outside of CD8+ T cell clusters does not inhibit their antimicrobial function during primary infection.
Cytokine receptor-deficient Trm cells show reduced persistence
To analyze the persistence of Trm populations in the absence of cytokine signaling, the ratio of WT to cytokine receptor-deficient T cells in the LP on days 9 and >30 post infection was compared. There was no significant difference in the ratio of WT to Il18r1−/− or Ifnar1−/− OT-I T cells at 9 and >30 days post infection (Figure 6A); however, there was a significant loss of Il12rb2−/− T cells from the LP over time (Figure 6A). The increased expression of CD103 by Il12rb2−/− LP T cells compare to WT cells was even more pronounced at >30 days post infection (Figure 6B, ,6C).6C). At this timepoint, we also observed elevated CD103 expression in Ifnar1−/− T cells compared to WT, while the expression of CD103 by Il18r1−/− OT-I T cells remained similar to that by WT cells (Figure 6B, ,6C).6C). Cytokine receptor-deficient and WT T cells all expressed similar levels of CD69 (Figure 6D). Other surface markers that are differentially expressed in CD103+ and CD103− LP Trm populations were also regulated by IL-12. WT T cells expressed NKG2D on the CD103− population, and Il12rb2−/− LP Trm cells failed to upregulate NKG2D on either the CD103+ or CD103− population (Figure 6E, top). There was also an increase in the proportion of CD244-expressing Il12rb2−/− T cells compared to WT cells (Figure 6E, bottom). These data identify IL-12 and type I IFN as critical regulators of CD103−CD69+ Trm differentiation, with Il12rb2−/− T cells in particular showing an almost complete loss of the CD103−CD69+ Trm population after infection. There was a statistically significant difference in the number of WT and Il12rb2−/− CD103−CD69+ LP Trm cells but not in the CD103+CD69+ LP Trm population (Figure 6F). The reduced survival of the Il12rb2−/− CD103−CD69+ LP T cells correlated with lower expression of the antiapoptotic factor Bcl-2 when compared to WT CD103−CD69+ T cells (Figure 6G). No significant difference in Bcl-2 expression was observed in WT and Il12rb2−/− CD103+CD69+ LP T cells (Figure 6G).

Mice received 5×103 WT and 5×103 Il18r1−/−, Ifnar−/−, or Il12rb2−/− OT-I T cells and were infected with Yptb-OVA by oral gavage. (A) Ratio of WT to Il18r1−/−, Ifnar−/−, or Il12rb2−/− OT-I T cells in the LP on 9 and >30 days post infection. (B) Representative CD103 expression on LP T cells. (C) Percent CD103+ of LP OT-I T cells at >30 days post infection. Lines connect data points from individual mice. (D) CD69 expression on LP cells expressed as the ratio of CD69 MFI on WT/cytokine receptor-deficient T cells. (E) WT and Il12rb2−/− OT-I T cells were analyzed for expression of NKG2D (top panels) and CD244 (bottom panels) on CD103+ and CD103−subsets. Numbers are percent of cells in each quadrant. Plots are representative of at least 5 mice from 2 or more experiments. (F) Absolute number of OT-I T cells in the CD103−CD69+ (left) and CD103+CD69+ (right) subsets at 9 and >30 days post infection. Numbers represent the fold difference in the number of WT and Il12rb2−/− OT-I T cells at each timepoint. (G) MFI of Bcl-2 in the CD103−CD69+ and CD103+CD69+ subsets at 9 days post infection. *p<0.05, **p<0.005 by the Mann-Whitney (A), paired t-test (C,E,G), or ratio paired t-test (F). Data are pooled from 2 or more experiments (A,D,F) or from one experiment and representative of at least two experiments (B,C,E,G). See also Figure S5.
It has been hypothesized that T cells are programmed to differentiate into CD103+ Trm cells prior to tissue entry. KLRG1− T cells preferentially form CD103+ Trm cells in the skin and intestine (Mackay et al., 2013; Sheridan et al., 2014); therefore, one could predict that the reduced expression of KLRG1 on Il12rb2−/− T cells in the lymphoid tissue during Yptb-OVA infection (Figure S5A) would result in increased CD103 expression independently of local cytokines. To more definitively address the role of local versus systemic cytokine exposure in Trm differentiation, we transferred WT and Ifnar1−/− OT-I T cells into mice and infected with VSV-OVA, which does not lead to intestinal infection or LP T cell clustering (data not shown). Ifnar1−/− T cells have reduced KLRG1 expression when compared to WT cells during infection (Figure S5B); however, the percent of CD103+ LP T cells was not significantly different between the two groups (Figure S5C). These data indicate that reducing the number of terminally differentiated KLRG1+ cells in the lymphoid organs does not result in a predictable increase in CD103 expression in the tissue. In addition, we do not observe any difference the persistence of Ifnar1−/− and WT LP Trm cells after VSV-OVA infection (Figure S5D). These data indicate that cytokine signaling in lymphoid organs is less critical for driving Trm differentiation and persistence than is the local cytokine milieu within the tissue.
IL-12 producing inflammatory monocytes drive CD103− Trm differentiation
Inflammatory monocytes are recruited into the intestine in response to infection and monocytes that enter the intestine in this context differentiate into IL-12-, TNFα-, and IL-6-producing macrophages/dendritic cells, while resident macrophages retain an anti-inflammatory signature (Zigmond et al., 2012; Bergsbaken and Bevan, 2015). We hypothesized that this population of newly recruited cells produced IFN-β and IL-12 in proinflammatory microenvironments. We examined this by evaluating expression of markers associated with intestinal macrophage populations (CD11b and CD11c) on cytokine-producing cells identified using reporter mice as described in Figure 2A. IFN-β-and IL-12-producing cells had characteristics of recently recruited inflammatory monocytes; they were clustered in the LP near the crypts at sites of infection and expressed both CD11b and CD11c (Figure 7A).
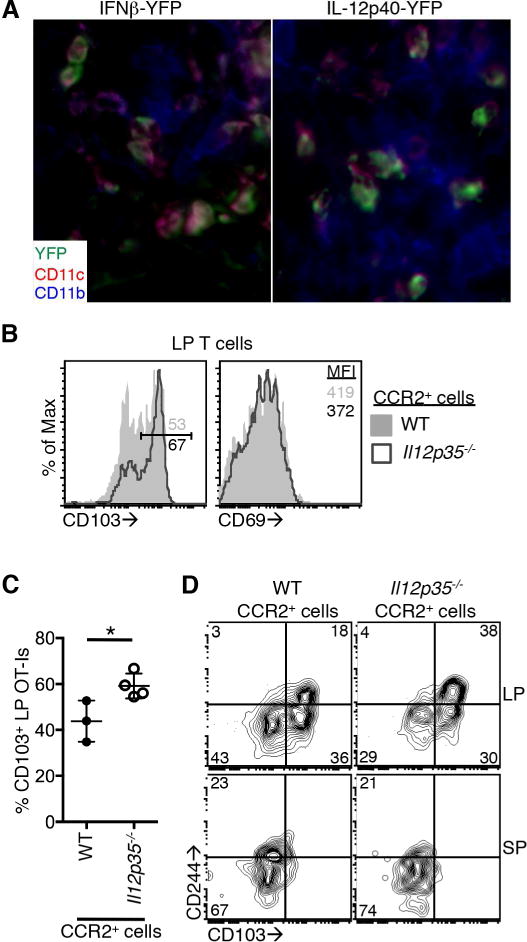
(A) IFNβ-YFP and IL12p40-YFP mice were infected with Yptb-OVA and tissues were isolated 7 days post infection. Expression of YFP (green) in CD11b+ (blue) and CD11c+ (red) cells in the intestinal LP. Images are representative of 3 mice. (B–D) WT:CCR2-DTR or Il12p35−/−:CCR2-DTR mixed bone marrow chimeric mice received 1×104 WT OT-I T cells and were infected with Yptb-OVA. Mice were given daily injections of DT beginning at 5 days post infection. Twelve days after infection, OT-I T cells in the LP and spleen were analyzed. (B) Representative histograms of CD103 (left) and CD69 (right) expression on OT-I T cells from mice containing WT (filled histogram) or Il12p35−/− (open histogram) monocytes/monocyte-derived cells. Numbers are the percent of the population in the CD103+ gate or the CD69 MFI. (C) Percent of LP OT-I T cells expressing CD103. Data are pooled from 2 experiments. (D) CD103 and CD244 expression on OT-I T cells from the LP (top panels) and spleen (bottom panels). Numbers are percent of cells in each quadrant. Representative of 3 or more mice per group pooled from 2 experiments. *p<0.05 by unpaired t-test. See also Figure S6.
To more definitively assess the role of monocytes/monocyte-derived cells in the differentiation of Trm cells in the intestine, we generated mixed bone marrow chimeras reconstituted with a 1:1 ratio of WT or Il12p35−/− to CCR2-DTR (Hohl et al., 2009) bone marrow. This allowed deletion of IL-12-producing CCR2+ cells at defined timepoints after infection. These mice received OT-I T cells, were infected with Yptb-OVA, and treated with diphtheria toxin (DT) from 5 to 12 days post infection. At 12 days post infection, the expansion and intestinal trafficking of OT-I T cells did not require IL-12 production by CCR2+ cells using this experimental system (Figure S6A). The expression of CD103 on LP T cells was increased in the absence of IL-12 production by CCR2+ cells when compared to T cells exposed to IL-12 (Figure 7B, left, left,7C).7C). As expected, the expression of CD69 was not significantly different between the two groups (Figure 7B, right, Figure S6B). In addition, expression of CD244 was enhanced on LP T cells generated in the absence of IL-12-producing CCR2+ cells when compared to those from mice that had WT CCR2+ cells (Figure 7D, top). Expression of CD244 on splenic OT-I T cells was similar in both groups (Figure 7D, bottom). These data highlight the importance of recently recruited inflammatory monocytes in producing IL-12 and driving differentiation of CD103− Trm cells in the context of intestinal infection.
DISCUSSION
Work presented here identifies IL-12 and type I IFN as critical regulators of Trm differentiation. These cytokines act on effector T cells to prevent CD103 expression in response to TGF-β, which is abundantly expressed in the intestinal tissue. The ability of CD8+ T cells to respond to these cytokines in vivo was not required for localization or effector function of CD8+ Trm cells during acute infection; however, cytokine receptor-deficient T cells showed impaired differentiation into the CD103−CD69+ Trm subset and failed to persistent long term. Type I IFN and IL-12 were produced by intestinal macrophage populations and depletion of these cells also resulted in reduced CD103− CD69+ Trm differentiation. These data indicate intestinal inflammation during infection is required for the differentiation and persistence of the full complement of Trm cells, and this should be an important consideration in the design of immunization strategies to combat mucosal pathogens.
IL-12 and type I IFN both result in phosphorylation of STAT4 in CD8+ T cells (Nguyen et al., 2002) and we hypothesize pSTAT4 may contribute to the regulation of intestinal Trm differentiation. Upon their entry into the tissue, CD8+ T cells express transcription factors that are Trm-specific (Hobit) while maintaining factors associated with both effector memory (Blimp-1, T-bet) and central memory (Eomes, KLF2) differentiation (Mackay et al., 2015; 2016; Wakim et al., 2012). The balance of these factors regulates the differentiation of intestinal Trm subsets, and our data indicate this is also influenced by exposure to local inflammatory cues. Itgae (the gene that encodes CD103) and other genes differentially regulated in CD103+ and CD103− Trm subsets are bound and/or regulated by pSTAT4 (Good et al., 2009; Wei et al., 2010). Expression of CD103 and other markers is maintained after inflammation and T cell clustering is resolved (Bergsbaken and Bevan, 2015) and CD103+ and CD103− populations are remarkably stable ex vivo. CD103 expression is negatively regulated by DNA methylation at the promoter in naïve and effector T cells (Scharer et al., 2013), suggesting a potential mechanism for regulation of Trm differentiation and maintenance. The stability of Trm populations in vivo in response to secondary infection by the same pathogen or in response to inflammation induced by other pathogens is currently unclear.
CD103+ and CD103− Trm subsets localize differently and therefore carry out distinct functions during acute infection; however, CD103 itself does not seem to play a role in Trm retention (Sheridan et al., 2014) and questions still remain about the unique roles played by these Trm subsets during subsequent infections. However; many additional genes are differentially regulated in these populations. CD244 is upregulated in CD103+ Trm cells from a variety of tissues (Mackay et al., 2013; Wakim et al., 2012), and while it is often associated with T cell dysfunction (Schietinger and Greenberg, 2014), the increased expression of CD244 by Il12rb2−/− LP T cells did not adversely affect their function. Expression of NKG2D, which can act as a costimulatory molecule and mediate TCR-independent cytotoxicity (Meresse et al., 2004; Chu et al., 2013), was upregulated along with that of IL-18 receptor in CD103− Trm cells. These expression patterns suggest that CD103− Trm populations could display innate function during infection by unrelated pathogens. Identification of strategies to specifically deplete CD103+ and CD103− Trm subsets in the intestine will be required to address this further.
Infection of the intestinal tissue is not required for trafficking or long term maintenance of Trm cells (Masopust et al., 2001); however, local immunization has proven more effective than systemic immunization in driving protective immunity against mucosal pathogens in a variety of settings (Mueller and Mackay, 2016; Schenkel and Masopust, 2014; Sheridan et al., 2014). Several aspects of tissue infection have been implicated in maximizing Trm-mediated immunity: tissue inflammation leads to increased recruitment of Trm precursors into the tissue (Mackay et al., 2012), local antigen presentation enhances the differentiation of Trm populations (Wakim et al., 2010; Khan et al., 2016), and clusters of lymphoid cells that are formed during local infection support the persistence of Trm cells by enhancing their survival and retention within the tissue (Collins et al., 2016; Iijima and Iwasaki, 2014). In the context of intesintal infection, antigen does not lead to preferential differentiation of either intestinal Trm subset. In addition, CD8+ T cell clustering is not required for maintenance of intestinal CD8+ Trm populations; intestinal lymphoid cell clusters do not persist indefinitely and loss of clustering does not result in a concomitant loss of CD103− Trm populations (Bergsbaken and Bevan, 2015). Here we show that although T cell clustering is not required indefinitely, early localization to these areas and exposure to inflammatory cytokines is important for differentiation and survival of intestinal Trm cells. It is becoming increasingly clear that although local infection is not absolutely required for the differentiation of Trm cells, the inflammation, antigen presentation, and tissue microenvironment established during tissue infection drives superior Trm responses. Although mucosal inflammation is not thought of as a desirable side effect during immunization, our data suggest that some degree of inflammation may be necessary to achieve optimal protective immunity. Local tissue inflammation, specifically recruitment of inflammatory monocytes and induction of IL-12 or type I IFN, may be required for differentiation and maintenance of the full complement of CD8+ Trm cells in the intestine and possibly other mucosal sites.
EXPERIMENTAL PROCEDURES
Mice and infections
B6.129-Il12btm1lky/J (Reinhardt et al., 2006) (IL-12p40-YFP), B6.129-Ifnbtm1lky/J (Scheu et al., 2008) (IFN-β-YFP), B6.129S1-Il12rb2tm1jm/J (Il12rb2−/−), B6.129P2-Il18r1tmAki/J (Il18r1−/−) and B6.129S1-Il12atm1Jm/J (Il12p35−/−) mice were purchased from The Jackson Laboratory. B6.129S2-Ifnar1tm1Agt/Mmjax (Ifnar1−/−) mice were provided by Dr. Daniel Stetson (University of Washington) and CCR2-DTR mice (Hohl et al., 2009) were provided by Dr. Jessica Hamerman (Benaroya Research Institute). Cytokine receptor-deficient mice were crossed to OT-I TCR transgenic mice and recipient C57BL/6 mice were purchased from The Jackson Laboratory or bred at the University of Washington. For infections, mice received 1×104 naïve OT-I T cells or 5×103 each of control and cytokine receptor-deficient OT-I T cells i.v. and were then infected by oral gavage with 2×108 CFU of Yersinia pseudotuberculosis YPIII (Yptb) or YPIII expressing ovalbumin (Yptb-OVA) (Bergsbaken and Bevan, 2015). Mice were maintained in specific pathogen free facilities and all experiments were done in accordance with the Institutional Animal Care and Use Committee guidelines of the University of Washington.
For transfer experiments, C57BL/6 and Il12rb2−/− mice were infected with Yptb and 7 days after infection, CD8+ T cells were isolated to >95% purity by depleting CD4+ (RM4-5), CD19+ (1D3), and CD11b+ (M1/70) cells by incubating with biotinylated antibodies and Streptavidin-Microbeads (Miltenyi) and then passing the cells over a magnetic column. CD8+ T cells were transferred into an equal number of C57BL/6 recipent mice that were infected with Yptb 3 days prior to transfer. An equal number of YopE69–77-specific CD8+ T cells was confirmed in both donor populations by tetramer staining. Two days after T cell transfer, the spleen and ileum were homogenized in PBS and plated on cefulosidin-novobiocin-irgasan agar (Difco) to enumerate bacteria in the tissue.
For monocyte depletion experiments, C57BL/6 mice were lethally irradiated (1000 rads) and injected i.v. with a total of 106 T cell depleted CCR2-DTR and WT bone marrow cells or CCR2-DTR and Il12p35−/− bone marrow cells at a 1:1 ratio. After reconstitution, mice received 1×104 naïve OT-I T cells and were infected the next day with Yptb-OVA by oral gavage. Mice were given daily intraperitoneal injection of 150ng of DT (Sigma) beginning 5 days post infection until sacrifice.
Flow cytometry
Single cell suspensions were stained with antibodies from eBiosciences: CD8β (H35-17.2), CD8α (53–6.7), CD103 (2E7), CD69 (H1.2F3), CD45.1(A20), CD45.2 (104), CD244 (244F4), NKG2D (CX5), CXCR3 (173), KLRG1 (2F1), CD127 (A7R34), and T-bet (4B10), from Invitrogen: IFN-γ (XMG1.2), from Biolegend: Bcl-2 (BCL/10C4), or from BD Biosciences: Granzyme B (GB11) and Bcl-2 (6C8). Biotinylated YopE monomers were purchased from the Fred Hutchinson Tetramer Core Facility and tetramerized with streptavidin-PE (Thermo Fisher). Samples were acquired on a BD FACSCanto II (BD Biosciences) and analyzed using FlowJo software (Tree Star). Cell sorting was performed using a FACSAriaII (BD Bioscience).
Isolation of intestinal cells
To isolate intestinal LP cells, Peyer’s patches were removed, the small intestine was cut open longitudinally, and intestinal contents and mucus were removed. The intestine was cut into 1cm pieces and incubated in HBSS containing 1mM dithiothreitol and 10% FBS at 37°C with stirring for 20 minutes, then transferred to HBSS containing 1.3mM EDTA and stirred at 37°C for 20 minutes to remove the epithelium. Intestinal pieces were then incubated in HBSS containing 5% FBS and 150U/ml collagenase type 2 (Worthington) at 37°C with stirring for 45 minutes to isolate LP cells. LP cells were further purified by gradient centrifugation with 44% and 67% Percoll.
Immunohistochemistry
Intestinal tissues were fixed in 4% paraformaldehyde, rehydrated in 20% sucrose, and frozen in OCT media (Sakura). Tissues were cut into 7–8μm sections and treated with ice cold acetone. Sections were treated with biotin-avidin blocking (Vector labs) and stained with the following biotinylated or directly conjugated antibodies:
CD8β (YTS156.7.7, Biolegend), CD45.2 (104, eBioscience), Epcam (G8.8, Biolegend), CD11b (M1/70, eBioscience), and CD11c (HL3, eBioscience). For identification of YFP-producing cells, sections were stained with anti-GFP (Abcam, ab6556), anti-rabbit-FITC (Abcam, ab6108), and anti-FITC-AlexaFlour488 (Invitrogen, polyclonal) antibodies. No reactivity was observed in infected YFP-negative mice. Stained slides were mounted with Prolong Gold antifade reagent (Thermo Fisher Scientific), imaged using a Nikon 90i, and analyzed using Adobe Photoshop software.
Ex vivo cytokine stimulation
Mice received naïve OT-I T cells and were infected with Yptb-OVA as described, and 7 days post infection, MLNs and spleen were isolated and stimulated with TGF-β (0.5ng/ml, R&D Systems), IL-12 (20ng/ml, Peprotech), IFN-β (200U/ml, R&D Systems), and IL-18 (50ng/ml, Peprotech) for 20 hours. Expression of CD103 and CD69 were analyzed by flow cytometry.
Quantitative RT-PCR
Mice received 1×104 naïve OT-I T cells and were then infected with 2×108 CFU of Yptb-OVA. At the indicated timepoints, CD103+CD69+ and CD103−CD69+ OT-I T cells or YopE69–77 tetramer+ T cells were sorted to >95% purity. RNA was isolated using an RNeasy RNA isolation kit (QIAgen). qRT-PCR was performed using SYBR one step RT-PCR kit (QIAgen) and primers in Table S1.
Statistical Analysis
Statistical significance was determined with GraphPad Prism software using an unpaired two-tailed t test or paired two-tailed t test when analyzing two populations of cells from the same animal. Bacterial CFU and distribution of T cells in the intestinal tissue were analyzed using the Mann-Whitney test.
Acknowledgments
We are grateful to Daniel Stetson and Jessica Hamerman for providing mice and reagents and to Cody Cunningham and Marion Pepper for critical reading of this manuscript. This work was supported by grants from the NIH (R01AI064318 to P.J.F.), Howard Hughes Medical Institute (to M.J.B.), and the Cancer Research Institute Irvington postdoctoral fellowship (to T.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
T.B. designed and performed experiments, T.B., M.J.B., and P.J.F. analyzed and interpreted data and wrote the manuscript.References
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9:209–222. [Europe PMC free article] [Abstract] [Google Scholar]
- Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, Jacobs H, Haanen JB, Schumacher TN. T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science. 2014;346:101–105. [Abstract] [Google Scholar]
- Bergsbaken T, Bevan MJ. Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8(+) T cells responding to infection. Nat Immunol. 2015;16:406–414. [Europe PMC free article] [Abstract] [Google Scholar]
- Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. 2012;188:4866–4875. [Europe PMC free article] [Abstract] [Google Scholar]
- Chu T, Tyznik AJ, Roepke S, Berkley AM, Woodward-Davis A, Pattacini L, Bevan MJ, Zehn D, Prlic M. Bystander-activated memory CD8 T cells control early pathogen load in an innate-like, NKG2D-dependent manner. Cell Rep. 2013;3:701–708. [Europe PMC free article] [Abstract] [Google Scholar]
- Collins N, Jiang X, Zaid A, Macleod BL, Li J, Park CO, Haque A, Bedoui S, Heath WR, Mueller SN, et al. Skin CD4(+) memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat Commun. 2016;7:11514. [Europe PMC free article] [Abstract] [Google Scholar]
- Edelblum KL, Shen L, Weber CR, Marchiando AM, Clay BS, Wang Y, Prinz I, Malissen B, Sperling AI, Turner JR. Dynamic migration of gd intraepithelial lymphocytes requires occluding. Proc Natl Acad Sci USA. 2012;109:7097–7102. [Europe PMC free article] [Abstract] [Google Scholar]
- Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, Carbone FR, Mueller SN. Different patterns of peripheral migration by memory CD4(+) and CD8(+) T cells. Nature. 2011;477:216–219. [Abstract] [Google Scholar]
- Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, O’Malley JT, Perumal NB, Kaplan MH. Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming. J Immunol. 2009;183:3839–3847. [Europe PMC free article] [Abstract] [Google Scholar]
- Hickman HD, Reynoso GV, Ngudiankama BF, Cush SS, Gibbs J, Bennink JR, Yewdell JW. CXCR3 chemokine receptor enables local CD8+ T cell migration for the destruction of virus-infected cells. Immunity. 2015;42:524–537. [Europe PMC free article] [Abstract] [Google Scholar]
- Hohl TM, Rivera A, Lipuma L, Gallegos A, Shi C, Mack M, Pamer EG. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe. 2009;6:470–481. [Europe PMC free article] [Abstract] [Google Scholar]
- Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, Keitany GJ, Garza EN, Fraser KA, Moon JJ, et al. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity. 2015;44:155–166. [Europe PMC free article] [Abstract] [Google Scholar]
- Iijima N, Iwasaki A. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 2014;346:93–98. [Europe PMC free article] [Abstract] [Google Scholar]
- Khan TN, Mooster JL, Kilgore AM, Osborn JF, Nolz JC. Local antigen in nonlymphoid tissue promotes resident memory CD8(+) T cell formation during viral infection. J Exp Med. 2016;213:951–966. [Europe PMC free article] [Abstract] [Google Scholar]
- Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, Kaech SM. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 2014;41:633–645. [Europe PMC free article] [Abstract] [Google Scholar]
- Le Floc’h A, Jalil A, Vergnon I, Le Maux Chansac B, Lazar V, Bismuth G, Chouaib S, Mami-Chouaib F. Alpha E beta 7 integrin interaction with E-cadherin promotes antitumor CTL activity by triggering lytic granule polarization and exocytosis. J Exp Med. 2007;204:559–570. [Europe PMC free article] [Abstract] [Google Scholar]
- Mackay LK, Minnich M, Kragten NAM, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. [Abstract] [Google Scholar]
- Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. The developmental pathway for CD103(+)CD8(+) tissue-resident memory T cells of skin. Nat Immunol. 2013;14:1294–1301. [Abstract] [Google Scholar]
- Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, Braun A, Masson F, Kallies A, Belz GT, et al. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity. 2015;43:1101–1111. [Abstract] [Google Scholar]
- Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, Heath WR, Carbone FR, Gebhardt T. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci USA. 2012;109:7037–7042. [Europe PMC free article] [Abstract] [Google Scholar]
- Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. [Abstract] [Google Scholar]
- Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, Raulet DH, Lanier LL, Groh V, Spies T, et al. Coordinated induction by IL-15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. [Abstract] [Google Scholar]
- Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol. 2016;16:79–89. [Abstract] [Google Scholar]
- Natsuaki Y, Egawa G, Nakamizo S, Ono S, Hanakawa S, Okada T, Kusuba N, Otsuka A, Kitoh A, Honda T, et al. Perivascular leukocyte clusters are essential for efficient activation of effector T cells in the skin. Nat Immunol. 2014;15:1064–1069. [Abstract] [Google Scholar]
- Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063–2066. [Abstract] [Google Scholar]
- Reinhardt RL, Hong S, Kang SJ, Wang ZE, Locksley RM. Visualization of IL-12/23p40 in vivo reveals immunostimulatory dendritic cell migrants that promote Th1 differentiation. J Immunol. 2006;177:1618–1627. [Abstract] [Google Scholar]
- Scharer CD, Barwick BG, Youngblood BA, Ahmed R, Boss JM. Global DNA methylation remodeling accompanies CD8 T cell effector function. J Immunol. 2013;191:3419–3429. [Europe PMC free article] [Abstract] [Google Scholar]
- Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. [Europe PMC free article] [Abstract] [Google Scholar]
- Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346:98–101. [Europe PMC free article] [Abstract] [Google Scholar]
- Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol. 2013;14:509–513. [Europe PMC free article] [Abstract] [Google Scholar]
- Scheu S, Dresing P, Locksley RM. Visualization of IFNbeta production by plasmacytoid versus conventional dendritic cells under specific stimulation conditions in vivo. Proc Natl Acad Sci USA. 2008;105:20416–20421. [Europe PMC free article] [Abstract] [Google Scholar]
- Schietinger A, Greenberg PD. Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol. 2014;35:51–60. [Europe PMC free article] [Abstract] [Google Scholar]
- Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, Lefrançois L. Oral infection drives a distinct population of intestinal resident memory CD8(+) T cells with enhanced protective function. Immunity. 2014;40:747–757. [Europe PMC free article] [Abstract] [Google Scholar]
- Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyarto BZ, Southern PJ, Masopust D. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 2015;161:737–749. [Europe PMC free article] [Abstract] [Google Scholar]
- Szaba FM, Kummer LW, Duso DK, Koroleva EP, Tumanov AV, Cooper AM, Bliska JB, Smiley ST, Lin JS. TNFα and IFNγ but not perforin are critical for CD8 T cell-mediated protection against pulmonary Yersinia pestis infection. PLoS Pathog. 2014;10:e1004142. [Europe PMC free article] [Abstract] [Google Scholar]
- Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci USA. 2010;107:17872–17879. [Europe PMC free article] [Abstract] [Google Scholar]
- Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, Bevan MJ. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J Immunol. 2012;189:3462–3471. [Europe PMC free article] [Abstract] [Google Scholar]
- Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. [Europe PMC free article] [Abstract] [Google Scholar]
- Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. [Abstract] [Google Scholar]
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.celrep.2017.03.031
Article citations
Protective function and differentiation cues of brain-resident CD8+ T cells during surveillance of latent Toxoplasma gondii infection.
Proc Natl Acad Sci U S A, 121(24):e2403054121, 05 Jun 2024
Cited by: 2 articles | PMID: 38838017
Tissue-resident memory T cells: decoding intra-organ diversity with a gut perspective.
Inflamm Regen, 44(1):19, 17 Apr 2024
Cited by: 0 articles | PMID: 38632596 | PMCID: PMC11022361
Review Free full text in Europe PMC
CD8+ Tissue-Resident Memory T Cells: Versatile Guardians of the Tissue.
J Immunol, 212(3):361-368, 01 Feb 2024
Cited by: 2 articles | PMID: 38227907
Decoding the transcriptional heterogeneity, differentiation lineage, clinical significance in tissue-resident memory CD8 T cell of the small intestine by single-cell analysis.
J Transl Med, 22(1):203, 25 Feb 2024
Cited by: 0 articles | PMID: 38403590 | PMCID: PMC10895748
Tissue-Resident Memory T Cell: Ontogenetic Cellular Mechanism and Clinical Translation.
Clin Exp Immunol, 214(3):249-259, 01 Dec 2023
Cited by: 1 article | PMID: 37586053
Review
Go to all (90) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Human intrahepatic CD69 + CD8+ T cells have a tissue resident memory T cell phenotype with reduced cytolytic capacity.
Sci Rep, 7(1):6172, 21 Jul 2017
Cited by: 64 articles | PMID: 28733665 | PMCID: PMC5522381
Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8⁺ T cells responding to infection.
Nat Immunol, 16(4):406-414, 23 Feb 2015
Cited by: 174 articles | PMID: 25706747 | PMCID: PMC4368475
Tissue Distribution of Memory T and B Cells in Rhesus Monkeys following Influenza A Infection.
J Immunol, 195(9):4378-4386, 25 Sep 2015
Cited by: 30 articles | PMID: 26408671 | PMCID: PMC4642841
TGF-β: Many Paths to CD103+ CD8 T Cell Residency.
Cells, 10(5):989, 23 Apr 2021
Cited by: 22 articles | PMID: 33922441 | PMCID: PMC8145941
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Cancer Research Institute Irvington
Howard Hughes Medical Institute
NIAID NIH HHS (1)
Grant ID: R01 AI064318
NIH (1)
Grant ID: R01AI064318




