Abstract
Free full text

EGFR-mediated Macrophage Activation Promotes Colitis-associated Tumorigenesis
Abstract
Epidermal growth factor receptor (EGFR) signaling is a known mediator of colorectal carcinogenesis. Studies have focused on the role of EGFR signaling in epithelial cells, although the exact nature of the role of EGFR in colorectal carcinogenesis remains a topic of debate. Here, we present evidence that EGFR signaling in myeloid cells, specifically macrophages, is critical for colon tumorigenesis in the AOM-DSS model of colitis-associated carcinogenesis (CAC). In a human tissue microarray, colonic macrophages demonstrated robust EGFR activation in the pre-cancerous stages of colitis and dysplasia. Utilizing the AOM-DSS model, mice with a myeloid-specific deletion of Egfr had significantly decreased tumor multiplicity and burden, protection from high-grade dysplasia, and significantly reduced colitis. Intriguingly, mice with gastrointestinal epithelial cell-specific Egfr deletion demonstrated no differences in tumorigenesis in the AOM-DSS model. The alterations in tumorigenesis in myeloid-specific Egfr knockout mice were accompanied by decreased macrophage, neutrophil, and T cell infiltration. Pro-tumorigenic M2 macrophage activation was diminished in myeloid-specific Egfr-deficient mice, as marked by decreased Arg1 and Il10 mRNA expression and decreased IL-4, IL-10 and IL-13 protein levels. Surprisingly, diminished M1 macrophage activation was also detectable, as marked by significantly reduced Nos2 and Il1b mRNA levels and decreased IFN-γ, TNF-α, and IL-1β protein levels. The alterations in M1 and M2 macrophage activation were confirmed in bone marrow-derived macrophages from mice with the myeloid-specific Egfr knockout. The combined effect of restrained M1 and M2 macrophage activation resulted in decreased production of pro-angiogenic factors, CXCL1 and VEGF, and reduced CD31+ blood vessels, which likely contributed to protection from tumorigenesis. These data reveal that EGFR signaling in macrophages, but not in colonic epithelial cells, has a significant role in CAC. EGFR signaling in macrophages may prove to be an effective biomarker of CAC or target for chemoprevention in patients with inflammatory bowel disease.
INTRODUCTION
Colorectal cancer is the third most common cancer globally1 and accounts for approximately 10% of new cancer diagnoses annually1. The risk for colon carcinogenesis is linked to chronic inflammatory states, such as inflammatory bowel disease (IBD)2-4. Colitis-associated carcinogenesis (CAC) occurs in 20% of patients diagnosed with IBD, and mortality rates are over 50% in these patients2-4. While mechanisms by which chronic inflammation promotes colonic carcinogenesis are being investigated2-4, unanswered questions remain.
The tumor microenvironment contains various immune cell types, including macrophages2-4. Macrophages represent a heterogeneous subset of innate immune cells with roles in tissue homeostasis, pro-inflammatory and anti-microbial responses, and wound repair5-7, and are of particular interest given their highly plastic phenotypes that can both promote and inhibit tumorigenesis8,9. Macrophages can alter their function based on the activation program utilized – either M1 or M2 patterns5-7,10. M1 macrophages are pro-inflammatory, anti-microbial, and thought to be anti-tumorigenic, although this remains the subject of debate5-7. M2 macrophages are associated with wound healing and have pro-tumorigenic properties5,8,10. Macrophage activation is dependent upon the colon tumor microenvironment9,11,12 and pathways that regulate this are incompletely understood3,8. Studies have implicated nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling13-15 and other pathways16,17.
We recently demonstrated that epidermal growth factor receptor (EGFR) signaling regulates macrophage activation across various stimuli18. EGFR phosphorylation occurs in macrophages and has major effects on expression of both M1 and M2 macrophage activation markers18. Importantly, we determined that EGFR signaling occurs in human gastric macrophages from gastritis to gastric adenocarcinoma18, leading us to speculate that EGFR signaling in macrophages may also have a role in CAC. EGFR signaling has been most commonly studied within the context of epithelial cell function and has been linked to colorectal cancer initiation and progression19-21. The impact of EGFR protein levels and signaling capacity is an area of ongoing investigation19-21.
Here, we demonstrate that EGFR signaling in macrophages has a profound effect on development of CAC. Early, inflammation-mediated stages of colon carcinogenesis in humans were marked by EGFR phosphorylation in macrophages. Myeloid-specific Egfr knockout mice exhibited decreased tumor multiplicity and tumor burden, while epithelial-specific Egfr knockout mice had no differences in phenotype. Loss of Egfr in myeloid cells resulted in decreased M2 and M1 macrophage activation, and decreased angiogenesis. Thus, EGFR signaling in macrophages may represent a potential target for therapeutic intervention in CAC.
RESULTS
EGFR signaling occurs in human colonic macrophages during pre-cancerous stages of CAC
The majority of studies related to EGFR signaling during CRC have focused on epithelial cells19-23. Instead, we sought to determine if human colonic macrophages had detectable levels of phospho-EGFR (pEGFR), a marker of active EGFR signaling, during IBD and associated CAC. We utilized an IBD-associated cancer tissue microarray (TMA) from Vanderbilt University Medical Center, which contained cases of inactive and active ulcerative colitis, dysplasia, and colitis-associated carcinoma25. pEGFR levels in CD68+ macrophages were detected via immunofluorescence staining (Figure 1a) and quantified with CellProfiler image analysis software (http://www.cellprofiler.com). We observed a low percentage of CD68+pEGFR+ macrophages in inactive colitis that was significantly increased in active colitis and dysplasia (Figure 1b, c). The percentage of CD68+pEGFR+ macrophages was lower in CAC than in active colitis and dysplasia (Figure 1b, c). The histopathologic diagnosis of inactive colitis indicates an absence of neutrophils, but is typically characterized by expansion of the lamina propria immune cell compartment and relative dropout of the epithelium. As such, no differences in the overall percentage of macrophages were detected between disease groups (Supplementary Figure 1). These data indicate that pEGFR is present in macrophages during pre-cancerous events associated with colonic inflammation, implying that EGFR signaling in macrophages has an important role in macrophage function during initiation of carcinogenesis. It should be noted that, as expected, pEGFR staining was also abundant in colonic epithelial cells (CECs; Figure 1a).
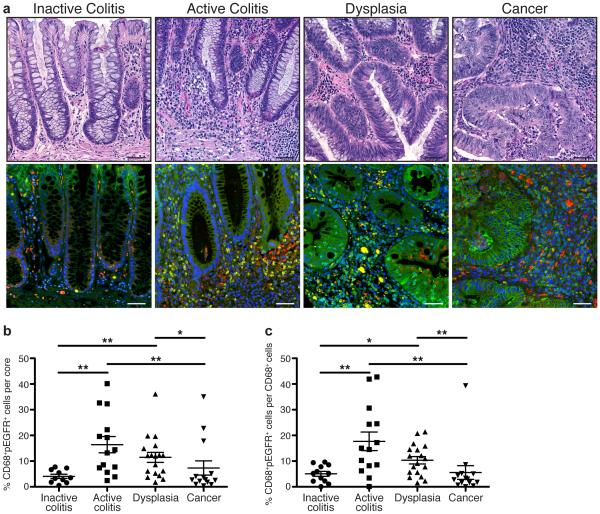
(a) Representative hematoxylin and eosin (H&E)-stained images and representative immunofluorescence images of colonic tissues from the Vanderbilt University Medical Center IBD-associated colorectal cancer TMA. Red = CD68. Green = pEGFR Y1068. Yellow = Merge. Blue = DAPI. Scale bars = 50 μm. (b) Quantification of the percentage of CD68+pEGFR+ cells among the total number of cells in each individual core in the TMA, as determined by CellProfiler. (c) Quantification of the percentage of CD68+pEGFR+ cells among the total number of CD68+ cells in each individual core in the TMA. For (a-c), n = 10 inactive colitis (normal or quiescent histology) samples, 14 active colitis (mild, moderate or severe histology) samples, 18 dysplasia samples, and 14 colorectal cancer samples. For (b-c), *P < 0.05, **P < 0.01 by one-way ANOVA with Kruskal-Wallis test, followed by Mann-Whitney U test.
EGFR signaling in macrophages contributes to AOM-DSS-induced colon tumorigenesis
We next sought to directly determine the role of EGFR signaling in macrophages during colon carcinogenesis. We utilized mice containing myeloid-specific, Egfr deletion (EgfrΔmye) driven by LysM-Cre, which we have extensively characterized in models of gastric and colonic inflammation18, and the appropriate control mice (Egfrfl/fl). Egfrfl/fl and EgfrΔmye mice were subjected to the azoxymethane (AOM)-dextran sodium sulfate (DSS) model of CAC27,28. The AOM-DSS protocol utilized is outlined in Supplementary Figure 2. No tumors were observed in the AOM-only or DSS-only groups, nor were differences in histologic colitis detectable between genotypes in the DSS-only group (data not shown). The lack of difference in the histologic colitis between genotypes in the DSS-only group is likely due to the prolonged recovery period, resulting in low colitis scores. Only data from the control and AOM-DSS groups are presented herein.
AOM-DSS-treated EgfrΔmye mice had significantly decreased tumor multiplicity and tumor burden, measured as the sum of the area of each tumor, versus AOM-DSS-treated Egfrfl/fl mice (Figure 2a, b). Further, EgfrΔmye mice were significantly protected from development of high-grade dysplasia, developing a maximum of low-grade dysplasia (Figure 2c). Representative hematoxylin and eosin (H&E)-stained images demonstrate the decrease in tumor size and protection from high-grade dysplasia in EgfrΔmye mice (Figure 2d). The myeloid-specific, Egfr knockout was maintained throughout the entire AOM-DSS protocol, as immunofluorescence images reveal the presence of many CD68+total EGFR (tEGFR)+ colonic macrophages in tissues from AOM-DSS-treated Egfrfl/fl mice, while only CD68+tEGFR− colonic macrophages are detectable in EgfrΔmye tissues (Figure 2e). Taken together, these data indicate that EGFR in myeloid cells is a potent promoter of tumorigenesis in mice.
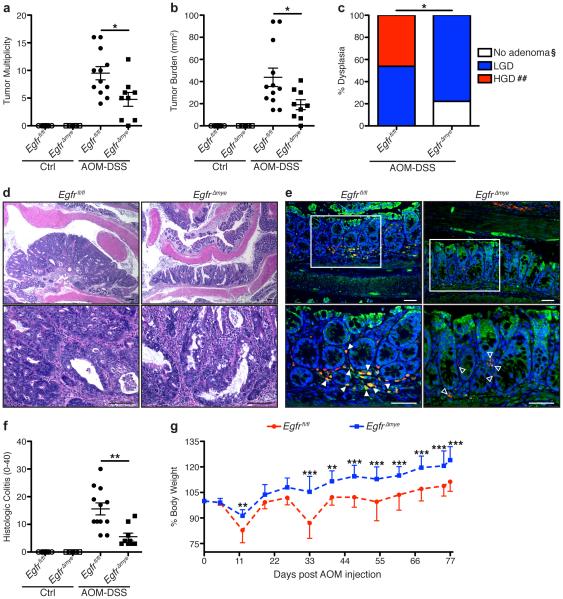
(a) Tumor multiplicity was assessed by gross visual inspection, utilizing a dissecting microscope. (b) Tumor burden was determined by the addition of the calculated area of each identified tumor, as assessed with an electronic caliper for both length and width. (c) Percentage of cases with either no adenoma, low-grade dysplasia (LGD), and high-grade dysplasia (HGD) determined by a gastrointestinal pathologist (M.K.W.) in a blinded manner. By Chi Square test, *P < 0.05. §P < 0.05 versus Egfrfl/fl; ##P < 0.01 versus Egfrfl/fl. n = 9-12 AOM-DSS-treated animals per genotype. (d) Representative H&E-stained images from AOM-DSS-treated mice. Scale bars = 100 μm. (e) Representative immunofluorescence images of tEGFR from AOM-DSS-treated mice. Red = CD68. Green = tEGFR. Yellow = Merge. Blue = DAPI. Scale bars = 50 μm. Solid arrows indicate CD68+tEGFR+ macrophages. Open arrows indicate CD68+tEGFR− macrophages. White box indicates zoomed area. n = ≥ 3 mice per genotype assessed. (f) Histologic colitis was determined by M.K.W. (g) Percentage of initial body weight was assessed at the indicated time points. *P < 0.05, **P < 0.01, ***P < 0.001 versus Egfrfl/fl AOM-DSS by two-way ANOVA with Bonferroni post-test (ANOVA significance = P < 0.001). In (a), (b), and (f), *P < 0.05, **P < 0.01 by one-way ANOVA with Kruskal-Wallis test, followed by Mann-Whitney U test. In (a), (b), (f), and (g), n = 7-9 control and 9-12 AOM-DSS-treated mice per genotype.
Additionally, EgfrΔmye mice demonstrated significantly decreased histologic colitis with AOM-DSS versus Egfrfl/fl mice (Figure 2f). In conjunction with decreased histologic colitis, EgfrΔmye mice exhibited significant protection from weight loss associated with each cycle of DSS, as compared to Egfrfl/fl mice (Figure 2g). These data indicate that EgfrΔmye mice are protected from the pro-inflammatory effects of the AOM-DSS model, which contributes to decreased CAC.
To assess the role of epithelial-specific EGFR in colon tumorigenesis, Foxa3cre/+ mice were crossed with Egfrfl/fl mice. Foxa3 is expressed in all gastrointestinal epithelial cells from mouth to anus24,29. Isolated, naïve CECs from Egfrfl/fl mice crossed with Foxa3cre/+ mice demonstrated absence of tEGFR (Supplementary Figure 3a). Despite a report indicating that Foxa3 is expressed in hematopoietic progenitor cells30, we found no detectable deletion of EGFR in naïve splenocytes (Supplementary Figure 3b), or bone marrow-derived macrophages (BMmacs; Supplementary Figure 3c). Thus, we termed these mice EgfrΔGIepi, indicative of the epithelial-specific Egfr deletion in the gastrointestinal tract. It should be noted that direct comparisons of EgfrΔmye mice and EgfrΔGIepi mice are not possible because EgfrΔGIepi mice are on a mixed background, while EgfrΔmye mice are on a congenic C57BL/6 background.
When subjected to the AOM-DSS protocol, Egfrfl/fl (littermate controls of the EgfrΔGIepi mice, and not the same as Egfrfl/fl mice utilized in studies with EgfrΔmye mice) and EgfrΔGIepi mice exhibited no differences in tumor multiplicity and tumor burden (Supplementary Figure 4a, b). Moreover, Egfrfl/fl and EgfrΔGIepi mice had similar susceptibility to low- and high-grade dysplasia (Supplementary Figure 4c). Representative H&E-stained images reveal tumors of similar size and severity of dysplasia (Supplementary Figure 4d).
Lack of differences between Egfrfl/fl and EgfrΔGIepi mice could be due to inefficient CRE-mediated excision of Egfr. However, loss of EGFR signaling was maintained in EgfrΔGIepi epithelial cells during AOM-DSS treatment (Supplementary Figure 4e). pEGFR immunoperoxidase staining is robust in epithelial cells and immune cells in Egfrfl/fl mice, but is restricted to immune cells in EgfrΔGIepi mice (Supplementary Figure 4e). In addition, no differences were observed in histologic colitis between Egfrfl/fl and EgfrΔGIepi mice (Supplementary Figure 4f). Egfrfl/fl and EgfrΔGIepi mice also displayed similar weight loss during each cycle of DSS (Supplementary Figure 4g). Taken together, EGFR signaling in immune cells, but not in epithelial cells, is critical for the promotion of tumorigenesis in mice.
EGFR signaling in macrophages enhances the innate immune response in colon tumors
The AOM-DSS model utilizes inflammation to drive tumorigenesis27,28,31. Based on decreased histologic colitis, combined with decreased tumor multiplicity and burden in EgfrΔmye mice, we sought to determine the nature of the innate immune response within the tumor microenvironment during AOM-DSS-induced CAC. We assessed 32 cytokines/chemokines via Luminex Multiplex Array. Analytes that were not different between genotypes or undetectable are in Supplementary Table 1.
The C-C ligand (CCL) chemokines CCL3 (MIP-1α) and CCL4 (MIP-1β) protein levels were significantly decreased in EgfrΔmye tumors versus Egfrfl/fl tumors (Figure 3a). CCL3 and CCL4 are chemoattractants for innate immune cells, and are produced by macrophages32. Additionally, levels of the C-X-C ligand (CXCL) chemokines, CXCL9 (MIG) and CXCL10 (IP-10), were diminished in EgfrΔmye tumors (Figure 3a). CXCL9 and CXCL10 are also primarily produced by macrophages33, but induce T cells infiltration34. CCL3, CCL4, CXCL9, and CXCL10 were induced to a similar degree in Egfrfl/fl and EgfrΔGIepi tumors, and no differences were detected between genotypes (Supplementary Figure 5a). Taken together, the decreases in CCL3, CCL4, CXCL9, and CXCL10 are indicative of decreased macrophage responses that result in decreased innate and adaptive immune cell infiltration in tumor areas. These significant decreases in macrophage-driven immune responses indicate the essential role of myeloid EGFR in driving CAC.
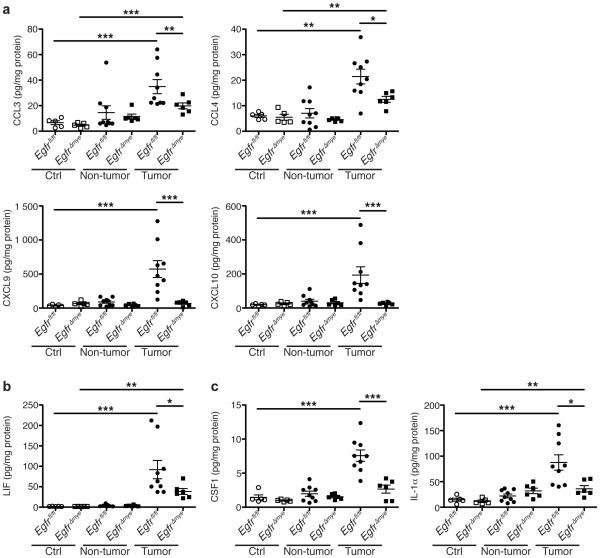
In a-c, protein levels were assessed by Luminex Multiplex Array from colonic tissues a 77 days post-AOM injection. (a) Levels of the C-C motif and C-X-C motif chemokines CCL3 (MIP-1α), CCL4 (MIP-1β), CXCL9 (MIG), and CXCL10 (IP-10). (b) Levels of the pleiotropic cytokine, LIF. (c) Levels of cytokines produced by activated macrophages, CSF1 (M-CSF) and IL-1α. In all panels, *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Kruskal-Wallis test, followed by Mann-Whitney U test. In all panels, n = 5 control tissues and 6-9 tumors with paired non-tumor area per genotype.
In addition, EgfrΔmye tumors demonstrated significant differences in cytokine levels (Figure 3b, c). Leukemia inhibitory factor (LIF), a cytokine with pleiotropic effects on immune function, was decreased in EgfrΔmye tumors (Figure 3b). LIF protein levels were induced to a similar degree in Egfrfl/fl and EgfrΔGIepi tumors (Supplementary Figure 5b). LIF overexpression is associated with poor prognosis in colorectal cancer35,36, and decreased LIF levels in EgfrΔmye tumors are consistent with the decreased tumor multiplicity and burden. Moreover, colony stimulating factor 1 (CSF1; M-CSF) and interleukin (IL)-1α were significantly decreased in EgfrΔmye tumors (Figure 3c), but not altered between Egfrfl/fl and EgfrΔGIepi tumors (Supplementary Figure 5c). CSF1 and IL-1α are produced by activated macrophages15,37-39, and CSF1 represents a key factor in macrophage activation and downstream function. Taken together, the decreases in these three cytokines indicate an overall downregulation of immune/inflammatory responses within tumors lacking EGFR in myeloid cells, and imply that macrophage EGFR is a central driver of tumorigenesis in an inflammation-dependent model.
To further investigate the effects of reduced chemokine production, we assessed populations of immune cells by immunohistochemistry with the following markers: macrophages (CD68), neutrophils (myeloperoxidase, MPO), T cells (CD3), and B cells (CD45R). Abundance of these cells was scored in non-tumor and tumor tissue, and combined for an overall score. Consistent with decreased CCL3 and CCL4 levels (Figure 3a), there were significantly diminished macrophages and neutrophils in AOM-DSS-treated EgfrΔmye mice versus Egfrfl/fl mice (Figure 4a, b). T cell abundance was also significantly decreased in AOM-DSS-treated EgfrΔmye mice (Figure 4c), consistent with diminished CXCL9 and CXCL10 in these mice (Figure 3a). B cells were not different between genotypes (Figure 4d). Taken together, these data show diminished immune cell infiltration in the EgfrΔmye mice, which is dependent upon EGFR signaling in myeloid cells.
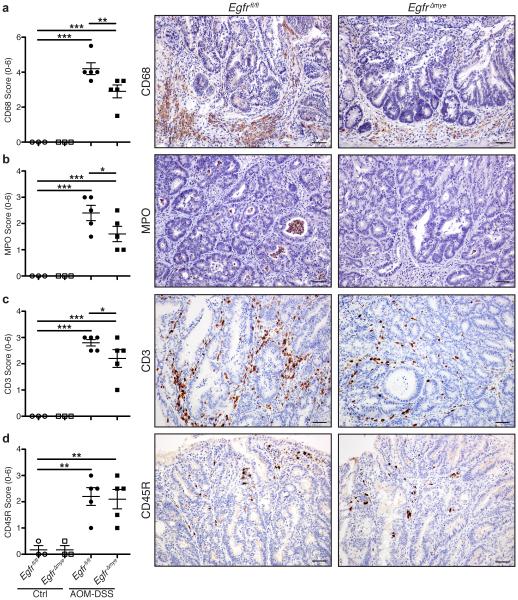
(a) Quantification of CD68+ macrophage abundance with representative immunoperoxidase images of CD68 staining in AOM-DSS-treated mice. (b) Quantification of myeloperoxidase+ (MPO) neutrophil abundance with representative immunoperoxidase images of MPO staining in AOM-DSS-treated mice. (c) Quantification of CD3+ T cell abundance with representative immunoperoxidase images of CD3 staining in AOM-DSS-treated mice. (d) Quantification of CD45R+ B cell abundance with representative immunoperoxidase images of CD45R staining in AOM-DSS-treated mice. Scoring of immune cell abundance was performed by M.B.P. as described Materials and Methods. In all panels, scale bars = 50 μm. In all panels, n = 3 control and 5 AOM-DSS-treated mice per genotype. In all panels, *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Newman-Keuls post-test after the data were square-root transformed.
EGFR signaling in macrophages enhances M2 activation in colon tumors
Because macrophages were likely the major source of several of the chemokines and cytokines which were significantly reduced in EgfrΔmye tumors, we hypothesized that loss of EGFR led to altered macrophage activation in the tumor microenvironment. M2 macrophages are tumor-associated macrophages and have pro-tumorigenic properties5-7,10. Thus, the decrease in tumor multiplicity and burden could be due to a diminished M2 macrophage response in EgfrΔmye tumors.
Indeed, protein levels of IL-4, IL-10, and IL-13, drivers of M2 activation5-7,10, were significantly upregulated in Egfrfl/fl tumors and decreased in EgfrΔmye tumors (Figure 5a). Moreover, mRNA levels of the M2 markers, arginase 1 (Arg1) and Il10, were also increased in Egfrfl/fl tumors and decreased in EgfrΔmye tumors (Figure 5b). Significant decreases in both the levels of cytokines that induce M2 activation and the expression of M2 macrophage markers indicate diminished M2 macrophage activation in EgfrΔmye mice, consistent with reduced colon tumorigenesis.
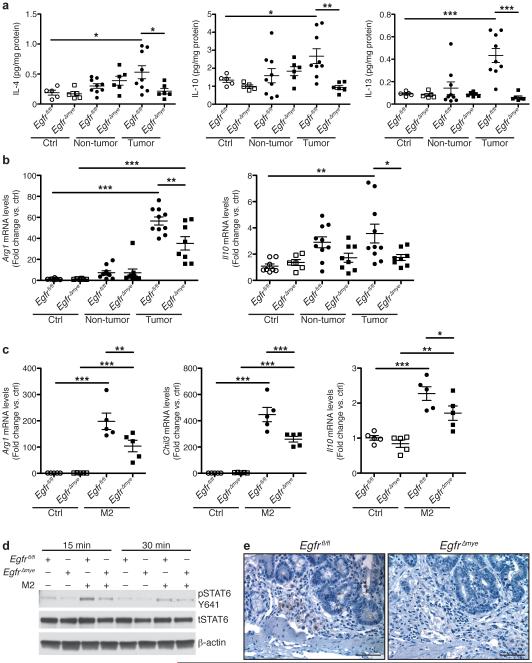
(a) Protein levels of the M2 stimuli, IL-4, IL-10, and IL-13, were assessed by Luminex Multiplex Array from colonic tissues 77 days post-AOM injection. n = 5 control tissues and 6-9 tumors with paired non-tumor area per genotype. (b) mRNA levels of M2 markers, Arg1 and Il10, were assessed by qRT-PCR from colonic tissues 77 days post-AOM injection. n = 6-8 control tissues and 8-10 tumors with paired non-tumor area per genotype. In (a) and (b), *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Kruskal-Wallis test, followed by Mann-Whitney U test. (c) mRNA levels of M2 markers, Arg1, Chil3, and Il10, were assessed by qRT-PCR in BMmacs 24 h post-treatment with classical M2 stimuli, IL-4 (10 ng/mL) and IL-10 (10 ng/mL). n = 5 biological replicates per genotype. In (c), *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Newman-Keuls post-test. (d) Representative western blot of pSTAT6 levels in BMmacs stimulated with the M2 stimulus, IL-4 (10 ng/mL), for the indicated times. n = 3 biological replicates. (e) Representative images of pSTAT6 immunoperoxidase staining in AOM-DSS-treated tissues. Scale bar = 50 μm. n = 3 mice per genotype.
Egfrfl/fl and EgfrΔGIepi tumors did not demonstrate differences in IL-4 or IL-10 levels (Supplementary Figure 6a), nor were differences detectable in Arg1 and Il10 mRNA levels (Supplementary Figure 6b). IL-13 was detected at very low levels in Egfrfl/fl and EgfrΔGIepi tumors, but no significant differences were detected (Supplementary Table 2). These data correlate with the finding that Egfrfl/fl and EgfrΔGIepi mice do not have significant differences in development of CAC.
To confirm that Egfr deletion in myeloid cells resulted in decreased M2 activation, we isolated BMmacs from Egfrfl/fl and EgfrΔmye mice and stimulated them ex vivo with IL-4 and IL-10. Markers of M2 macrophage activation were assessed by qRT-PCR. IL-4/IL-10 stimulation led to a significant induction of Arg1, chitinase-like 3 (Chil3), and Il10 in both Egfrfl/fl and EgfrΔmye BMmacs (Figure 5c). Importantly, Arg1, Chil3, and Il10 mRNA levels were significantly decreased in EgfrΔmye BMmacs (Figure 5c), consistent with the findings in EgfrΔmye tumors. Together, these data further suggest that EGFR signaling regulates M2 macrophage activation.
To address the mechanism by which macrophage EGFR signaling alters M2 activation, we isolated BMmacs from Egfrfl/fl and EgfrΔmye mice, stimulated them ex vivo with IL-4, and assessed phospho-signal transducer and activator of transcription 6 (STAT6), a known mediator of M2 activation40. EgfrΔmye BMmacs exhibited decreased pSTAT6 levels when compared to Egfrfl/fl BMmacs (Figure 5d). Similarly, AOM-DSS-treated EgfrΔmye tissues demonstrated reduced pSTAT6 levels in immune cells (Figure 5e). These data reveal a potential link between EGFR and STAT6 in regulating M2 activation in macrophages.
Macrophage-specific EGFR signaling also augments M1 activation in colon tumors
Alterations in M2 macrophage activation in EgfrΔmye mice were not unexpected, given the close association between M2 macrophages and the tumor microenvironment8,9. However, we also observed significant alterations in both the levels of cytokines that induce M1 activation and in M1 markers in EgfrΔmye tumors.
Interferon (IFN)-γ and tumor necrosis factor (TNF)-α potently induce M1 macrophage activation7. Protein levels of both IFN-γ and TNF-α were significantly upregulated in AOM-DSS-induced tumors in Egfrfl/fl mice and significantly reduced in EgfrΔmye tumors (Figure 6a), indicative of decreased capacity for M1 macrophage activation. mRNA levels of M1 markers nitric oxide synthase 2 (Nos2) and Il1b were upregulated in Egfrfl/fl tumors, and significantly decreased in EgfrΔmye tumors (Figure 6b). This decrease in M1 macrophage activation was confirmed at the protein level, as IL-1β protein was also significantly decreased in EgfrΔmye tumors (Figure 6c). Protein levels of IFN-γ and TNF-α were detectable in Egfrfl/fl and EgfrΔGIepi tumors, but were not different between genotypes (Supplementary Figure 7a). mRNA levels of Nos2 and Il1b (Supplementary Figure 7b), as well as IL-1β protein levels, were upregulated in Egfrfl/fl and EgfrΔGIepi tumors, but again were not different between genotypes.
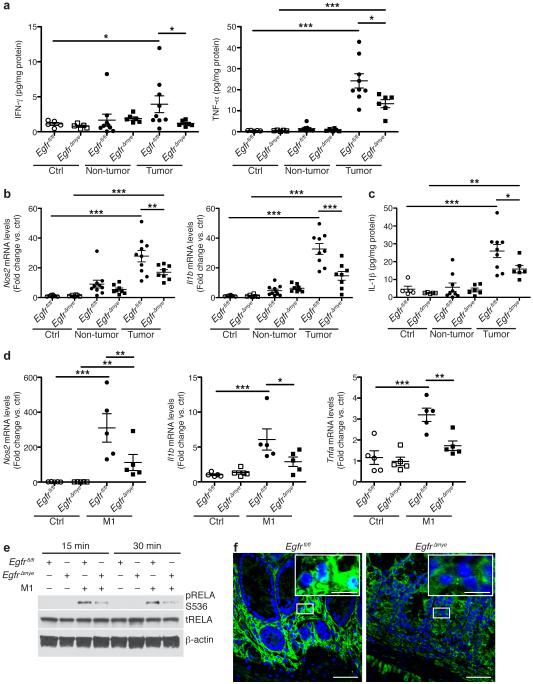
(a) Protein levels of M1 stimuli, IFN-γ and TNF-α, were assessed by Luminex Multiplex Array from colonic tissues 77 days post-AOM injection. n = 5 control tissues and 6-9 tumors with paired non-tumor area per genotype. (b) mRNA levels of M1 markers, Nos2 and Il1b, were assessed by qRT-PCR from colonic tissues 77 days post-AOM injection. n = 6-8 control tissues and 8-10 tumors with paired non-tumor area per genotype. (c) Protein levels of the M1 marker, IL-1β, were assessed by Luminex Multiplex Array from colonic tissues 77 days post-AOM injection. n = 5 control tissues and 6-9 tumors with paired non-tumor area per genotype. In (a-c), *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Kruskal-Wallis test, followed by Mann-Whitney U test. (d) mRNA levels of M1 markers, Nos2, Il1b, and Tnfa, were assessed by qRT-PCR in bone marrow derived macrophages (BMmacs) 24 h post-treatment with classical M1 stimuli, IFN-γ (200 U/mL) and TNF-α (10 ng/mL). n = 5 biological replicates per genotype. In (d), *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Newman-Keuls post-test. (e) Representative western blot of pRELA (pp65) levels in BMmacs stimulated with the M1 stimuli, IFN-γ (200 U/mL) and lipopolysaccharide (10 ng/mL), for the indicated times. n = 3 biological replicates. (e) Representative confocal immunofluorescence images of cytoplasmic and nuclear RELA (p65) in AOM-DSS-treated tissues. Green = cytoplasmic RELA. White/Aqua = nuclear RELA. Blue = DAPI. Scale bar = 40 μm, Scale bar in inset = 10 μm. n = 3 mice per genotype.
To assess the role of EGFR signaling in M1 activation, we isolated BMmacs from Egfrfl/fl and EgfrΔmye mice and stimulated ex vivo with IFN-γ and TNF-α for 24 h. M1 stimulation resulted in robust expression of M1 markers Nos2, Il1b, and Tnfa (Figure 6d). Importantly, mRNA expression of Nos2, Il1b, and Tnfa was significantly reduced in EgfrΔmye BMmacs versus Egfrfl/fl BMmacs (Figure 6d), indicating that EGFR signaling is equally critical for M1 and M2 activation. Intriguingly, M1 activation may have critical role in colon tumorigenesis in this model that is regulated by EGFR signaling.
We have previously demonstrated that EGFR signaling is upstream of NF-κB in macrophages during Helicobacter pylori infection18. Moreover, we have reported that there is enhanced nuclear translocation of RELA proto-oncogene, NF-κB subunit (RELA; also known as p65) in two different models of colitis41. Thus, we hypothesized that EGFR and NF-κB signaling may regulate M1 activation. We isolated BMmacs from Egfrfl/fl and EgfrΔmye mice, stimulated with IFN-γ and lipopolysaccharide, and assessed phospho-RELA (pRELA) levels. EgfrΔmye BMmacs had diminished pRELA levels versus Egfrfl/fl BMmacs (Figure 6e). Moreover, AOM-DSS-treated EgfrΔmye tissues had markedly decreased RELA nuclear translocation in lamina propria cells (Figure 6f). Thus, EGFR signaling activates the NF-κB pathway to regulate M1 activation.
EGFR signaling in macrophages enhances angiogenesis in colon tumors
Angiogenesis is a hallmark of carcinogenesis42, and is an essential means by which colon tumor growth is supported43,44. Several cytokines, including vascular endothelial growth factor (VEGF) A and CXCL1 (KC, GRO-α), the murine equivalent of CXCL8 (IL-8)45, contribute to angiogenesis46-49. M1 macrophages are an important source of CXCL126 and both M1 and M2 macrophages are important sources of VEGFA50.
Based on i) decreased tumor multiplicity/burden, and ii) decreased M2 and M1 macrophage activation in EgfrΔmye mice, we hypothesized that angiogenesis may be impaired, contributing to diminished tumorigenesis. EgfrΔmye tumors had decreased mRNA levels of Cxcl1 and Vegfa (Figure 7a) and altered protein levels of CXCL1 and VEGF (Figure 7b) versus Egfrfl/fl tumors. Egfrfl/fl and EgfrΔGIepi tumors did not demonstrate any differences in mRNA or protein levels of these markers (Supplementary Figure 8). Additionally, M1-activated EgfrΔmye BMmacs expressed significantly less Cxcl1 and Vegfa (Supplementary Figure 9a) and M2-activated EgfrΔmye BMmacs expressed less Vegfa (Supplementary Figure 9b) versus Egfrfl/fl BMmacs. These data confirm that macrophages are a potential source of CXCL1 and VEGF, and that EGFR signaling is critical for expression of these pro-angiogenic mediators.
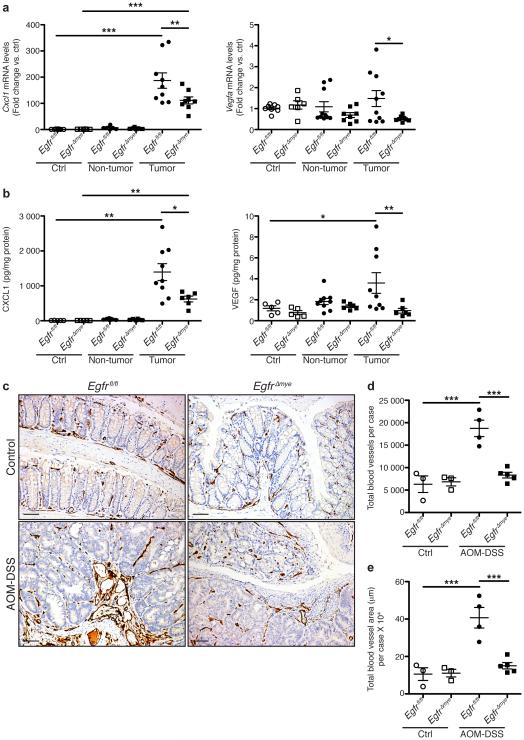
(a) mRNA levels of the pro-angiogenic chemokine, Cxcl1, and the pro-angiogenic cytokine, Vegfa, were assessed by qRT-PCR from colonic tissues 77 days post-AOM injection. n = 6-8 control tissues and 8-10 tumors with paired non-tumor area per genotype. (b) Protein levels of pro-angiogenic chemokine, CXCL1, and pro-angiogenic cytokine, VEGF, were assessed by Luminex Multiplex Array from colonic tissues 77 days post-AOM injection. n = 5 control tissues and 6-9 tumors with paired non-tumor area per genotype. In (a) and (b), *P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA with Kruskal-Wallis test, followed by Mann-Whitney U test. (c) Representative images of CD31+ blood vessel immunohistochemistry in colonic tissues. Scale bars = 50 μm. (d) Quantification of the total number of CD31+ blood vessels in (c). n = 3 control colonic tissues and 4-5 AOM-DSS-treated colonic tissues per genotype. (e) Quantification of the total CD31+ blood vessel area within tissues in (c). n = 3 control colonic tissues and 4-5 AOM-DSS-treated colonic tissues per genotype. In (d) and (e), ***P < 0.001 by one-way ANOVA with Newman-Keuls post-test.
To confirm that decreased pro-angiogenic cytokines was accompanied by decreased angiogenesis, we performed immunoperoxidase staining for CD31, a marker of vascular endothelial cells51. Representative images demonstrate significantly enhanced angiogenesis in AOM-DSS-treated Egfrfl/fl versus EgfrΔmye mice (Figure 7c). Notably, angiogenesis in AOM-DSS-treated EgfrΔmye mice was not different than angiogenesis in control Egfrfl/fl and EgfrΔmye mice (Figure 7c). Both the number of CD31+ blood vessels per case (Figure 7d) and the total area of CD31+ blood vessels (Figure 7e) were significantly decreased in AOM-DSS-treated EgfrΔmye mice versus Egfrfl/fl mice. Thus, decreases in pro-angiogenic cytokine production were associated with decreased angiogenesis in EgfrΔmye mice. Decreased angiogenesis likely contributed to diminished tumorigenesis in this CAC model.
DISCUSSION
EGFR signaling is a commonly studied pathway in carcinogenesis19,20,52-54, although most studies related to EGFR signaling have been performed in epithelial cells19,20,52-54. This study outlines an important role for EGFR signaling in macrophages during inflammatory colon tumorigenesis. Herein, we demonstrate that EGFR signaling occurs in human macrophages during pre-cancerous stages of ulcerative colitis and dysplasia, both of which are marked by active inflammation3,4. Based on diminished CD68+pEGFR+ cells in CAC, we posit that EGFR signaling in human macrophages may be essential for initiation of inflammation-associated tumorigenesis. Myeloid-specific knockout of Egfr significantly protected mice from tumorigenesis in the AOM-DSS model of CAC. Intriguingly, gastrointestinal epithelial cell-specific knockout of Egfr did not have a phenotype that was significantly different from wild-type mice. Protection from tumorigenesis in EgfrΔmye mice was accompanied by restricted M2 and M1 macrophage activation, which likely contributed to decreased angiogenesis due to less production of pro-angiogenic cytokines and chemokines. These data point to a currently underappreciated role for EGFR signaling in myeloid cells, particularly macrophages, in promoting CAC.
A previous study utilizing Wa5 mice, which carry a dominant-negative allele that impairs EGFR signaling, demonstrated that EGFR signaling inhibited CAC55, the opposite of our current result. It should be noted that differences in colon tumorigenesis were only significant when Wa5 mice were crossed with Il10−/− mice55. Il10−/− mice may have limitations as a model for CAC, because effects of the Il10 deletion can be modulated by bacterial infections56,57. The AOM-DSS model is solely dependent upon inflammation as a driver of carcinogenesis56,57. The previous work that demonstrated a protective role for EGFR in colon tumorigenesis also utilized the AOM-DSS model of colon tumorigenesis, but noted no significant differences in tumor multiplicity55. Moreover, the Wa5 allele affects all cell types, while our models used herein were myeloid- and gastrointestinal epithelial-cell-specific. Taken together, our study provides a strong body of evidence that EGFR signaling in macrophages is a critical component of CAC.
EGFR signaling appears to mediate M1 and M2 macrophage activation. Importantly, we demonstrated via multiple methods in a previously published study that EGFR signaling in macrophages had no effect on apoptosis18. Thus, the phenotypes observed are due to alterations in macrophage function, not cell viability. While the finding that decreased M2 activation, which is highly pro-tumorigenic8,10,52, correlated with decreased tumor multiplicity and burden was not unexpected, the dramatic alteration of M1 activation was somewhat surprising. However, there are potential reasons why M1 activation may have an important role in tumorigenesis. Firstly, as we demonstrated, M1 macrophages are an important source of pro-angiogenic cytokines (VEGF) and chemokines (CXCL1). Secondly, M1 macrophages are a potent source of NOS2 (as shown herein) and nitric oxide (NO)6,7,18. It has been proposed that NO, a reactive nitrogen species, promotes colon tumorigenesis by causing DNA mutations and DNA instability3,58. Our work demonstrates that EGFR signaling has a role in promoting both M1 and M2 activation, potentially working synergistically to promote colon tumorigenesis.
In sum, our study outlines a novel role for EGFR signaling in macrophages during CAC. Robust EGFR signaling in human macrophages in stages leading to colitis-associated cancer indicates that macrophage pEGFR may serve as a potential biomarker for CRC risk in colitis patients. Moreover, our previous work showed that TNF-α induced EGFR signaling in macrophages18. Thus, one potential benefit of anti-TNF treatment, frequently used in IBD patients, may be suppression of macrophage EGFR signaling. Further studies are imperative to further assess the critical role that EGFR activation plays in immune cells in patients at risk for progression to colon cancer.
MATERIALS AND METHODS
Reagents
Reagents used for cell culture were from Invitrogen (Carlsbad, CA, USA). Reagents for RNA extraction were from Qiagen (Valencia, CA, USA); those for cDNA synthesis and qRT-PCR were from Bio-Rad (Hercules, CA, USA). Murine M-CSF, IFN-γ, IL-4, and IL-10 were from PeproTech (Rocky Hill, NJ, USA). Murine TNF-α was from Thermo Fisher Scientific (Waltham, MA, USA). Azoxymethane and LPS were from Sigma-Aldrich (St. Louis, MO, USA). DSS was from TdB Consultancy (Uppsala, Sweden).
Antibodies
See Supplementary Table 4 for information regarding antibodies.
Cells and Culture Conditions
BMmacs were isolated and differentiated from murine femurs as described18,59. Red blood cells were lysed in ammonium-chloride-potassium buffer for 3 min, and 1.25 million cells were differentiated with 20 μg/mL recombinant murine M-CSF for 7 days, with media changes at days 3 and 5. Following differentiation, cells were placed in complete Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 25 mM HEPES, and 10 mM sodium pyruvate for studies with IFN-γ, TNF-α, IL-4, IL-10, or LPS. Stimulation times and doses are indicated in figure legends.
Animal Studies
Egfrfl/fl and EgfrΔmye mice were utilized as described18. C57BL/6 Egfrfl/fl mice were crossed with C57BL/6 LysMcre/cre mice to generate myeloid-specific, Egfr deletion (EgfrΔmye). C57BL/6 Egfrfl/fl mice were crossed to CD-1/DBA Foxa3cre/+ mice obtained from Timothy Wang (Columbia University Medical Center)60, to generate EgrfΔGIepi mice. As EgrfΔGIepi mice contain one Cre allele, littermates were utilized for all experiments. Animals were used under protocol M/10/155, approved by the Institutional Animal Care and Use Committee at Vanderbilt University.
Male mice, aged 6-12 weeks, were utilized for all studies. Samples sizes were based on previous AOM-DSS studies from our laboratory. Mice were not removed from the cages into which they were weaned. No other criteria were utilized for selection or randomization. Mice were subjected to the azoxymethane (AOM)-dextran sodium sulfate (DSS) colon tumorigenesis model27,61. Mice received one intraperitoneal AOM injection (12.5 mg/kg) on Day 0, and three doses of 4% DSS in their drinking water on Days 5, 26, and 47. The first two cycles of DSS lasted for 5 days, and the third for 4 days. Mice were weighed every 7 days from the start of the first DSS cycle and on Day 0 to determine the AOM dosage. Mice were sacrificed on Day 77.
Rarely, EgfrΔmye mice developed an enlarged spleen that was not attributable to AOM-DSS treatment. These mice were excluded from further analysis.
Tumor multiplicity was determined by visual inspection via dissecting microscope. Tumor burden was determined by the summation of tumor area, assessed by electronic caliper. Histologic colitis and dysplasia were determined by a gastrointestinal pathologist, M.K.W., in a blinded manner.
Human Tissues
The IBD-associated CAC human TMA was utilized as described25.
Real-Time PCR
RNA was isolated from cells and tissues as described18,59. cDNA was prepared and PCR was performed as described18,59. See Supplementary Table 3 for primers utilized.
Luminex Multiplex Array
Luminex Multiplex Array was performed as described18,59. Tissues were harvested at time of sacrifice, homogenized via a handheld homogenizer in 300 μL CellLytic MT Cell Lysis Reagent (Sigma) and centrifuged at 14,000g at 4°C twice. All kit instructions were followed in the performance of the Luminex Multiplex Array assay.
Immunofluorescence Staining for tEGFR and CD68, and RELA
Staining for tEGFR and CD68 in both the TMA and in murine tissues was performed as described18, with the following exceptions in murine tissue. An initial blocking step was performed with Background Sniper (Biocare Medical), followed by incubation with anti-tEGFR. A second blocking step was performed with the Fab fragment of murine IgG for 15 min, followed 3% goat serum for 15 min, with subsequent incubation with anti-CD68. All other steps were as described. RELA staining and confocal microscopy was performed as described41.
Immunoperoxidase Staining for pEGFR Y1068
Staining for pEGFR in human colonic biopsies was performed as described52.
Immunoperoxidase Staining
CD68, MPO, and pSTAT6
After overnight heating at 37°C, sections were deparaffinized in xylene and rehydrated in graded alcohols. Sections were incubated with primary antibody overnight at room temperature. After washing, incubation with anti-rabbit HRP-polymer was performed for 30 min. Sections were rinsed and incubated with streptavidin-HRP (Biocare Medical, Concord, CA, USA) for 30 min.
CD3, CD45R, and CD31
Heat-induced antigen retrieval was performed on the Leica Bond Max using Epitope Retrieval 2 solution for 10 min (CD3, CD31) and 20 min (CD45R). Slides were incubated with primary antibody for 1 h and incubated with secondary antibody for 15 min. CD31 Images were captured using a high throughput Leica SCN400 Slide Scanner automated digital image system. Tissue samples were mapped using Ariol® Review software. The numbers and areas of CD31+ blood vessels were determined in the Ariol® software.
Scoring of CD68, MPO, CD3, and CD45R Immunoperoxidase Staining
Slides were scored in a blinded manner by a gastrointestinal pathologist (M.B.P.). Samples were evaluated for staining in non-tumor and tumor areas using the following scale: 0=scarce positive cells, 1=low abundance of positive cells, 2=moderate abundance of positive cells, and 3=high abundance of positive cells. The two scores were combined for a highest possible score of 6.
Purification of colonic epithelial cells
Purification of colonic epithelial cells was performed as described18.
Statistical Analysis
Data represent mean ± S.E.M. Where data were normally distributed, two-tailed Student’s t test was used to determine significance in experiments with only two groups, and one-way ANOVA with the Newman-Keuls test was used to determine significant differences between multiple test groups. In Figure 4, the data were square root transformed to ensure normal distribution prior to statistical analysis. In other cases where data were not normally distributed, a one-way ANOVA with Kruskal-Wallis test, followed by a Mann-Whitney U test, was performed. Please see Supplementary Table 5 for all relevant P values as determined by Kruskal-Wallis test. Statistical analysis of all weight loss curves was performed by two-way ANOVA with Bonferroni post-test. All statistics were performed in Prism 5.0 (GraphPad Software, San Diego, CA, USA). A P value of < 0.05 was considered significant.
ACKNOWLEDGEMENTS
CD3, CD45R, and CD31 staining was performed by the Vanderbilt University Medical Center Translational Pathology Shared Resource supported by NIH grant P30CA068485 and the Vanderbilt Mouse Metabolic Phenotyping Center, supported by NIH grant U24DK059637. Slide imaging and quantification was performed in the Digital Histology Shared Resource (www.mc.vanderbilt.edu/dhsr). This work was funded by NIH grants R01DK053620, R01AT004821, R01CA190612, P01CA116087, and P01CA028842 (K.T.W.), Veterans Affairs Merit Review grant I01BX001453 (K.T.W.), the Thomas F. Frist Sr. Endowment (K.T.W.), and the Vanderbilt Center for Mucosal Inflammation and Cancer (K.T.W.). The human TMA was supported by NIH grant P50CA095103 and the DDRC Tissue Morphology Subcore (NIH grant P30DK058404). D.M.H. was supported by T32GM008554 and F31DK10715. L.A.C. was supported by Veterans Affairs Career Development Award 1IK2BX002126.
Footnotes
CONFLICT OF INTEREST
The authors declare that no conflict of interest exists.
SUPPLEMENTARY INFORMATION
Supplementary Information accompanies the paper on the Oncogene website (http://nature.com/onc).
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/onc.2017.23
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/onc201723.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/onc.2017.23
Article citations
Identification and Functional Analysis of Ras-Related Associated with Diabetes Gene (<i>rrad</i>) in <i>Edwardsiella piscicida</i>-Resistant Individuals of Japanese Flounder (<i>Paralichthys olivaceus</i>).
Int J Mol Sci, 25(19):10532, 01 Oct 2024
Cited by: 0 articles | PMID: 39408905 | PMCID: PMC11476895
The reverse transsulfuration pathway affects the colonic microbiota and contributes to colitis in mice.
Amino Acids, 56(1):63, 19 Oct 2024
Cited by: 0 articles | PMID: 39427081 | PMCID: PMC11490428
Advances in molecular mechanisms of inflammatory bowel disease‑associated colorectal cancer (Review).
Oncol Lett, 27(6):257, 09 Apr 2024
Cited by: 1 article | PMID: 38646499 | PMCID: PMC11027113
Review Free full text in Europe PMC
Progress of research on the relationship between efferocytosis and tumor.
Front Oncol, 14:1361327, 09 Apr 2024
Cited by: 0 articles | PMID: 38655133 | PMCID: PMC11035832
Review Free full text in Europe PMC
IL-17RA-Mediated Epithelial Cell Activity Prevents Severe Inflammatory Response to Helicobacter pylori Infection.
Immunohorizons, 8(4):339-353, 01 Apr 2024
Cited by: 0 articles | PMID: 38639570 | PMCID: PMC11066722
Go to all (54) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Ornithine Decarboxylase in Macrophages Exacerbates Colitis and Promotes Colitis-Associated Colon Carcinogenesis by Impairing M1 Immune Responses.
Cancer Res, 78(15):4303-4315, 31 May 2018
Cited by: 41 articles | PMID: 29853605 | PMCID: PMC6072585
Activation of the epidermal growth factor receptor in macrophages regulates cytokine production and experimental colitis.
J Immunol, 192(3):1013-1023, 03 Jan 2014
Cited by: 55 articles | PMID: 24391216 | PMCID: PMC4006992
EGFR in Tumor-Associated Myeloid Cells Promotes Development of Colorectal Cancer in Mice and Associates With Outcomes of Patients.
Gastroenterology, 153(1):178-190.e10, 09 Apr 2017
Cited by: 60 articles | PMID: 28400195 | PMCID: PMC5766132
Epithelial Nuclear Factor-x03BA;B Activation in Inflammatory Bowel Diseases and Colitis-Associated Carcinogenesis.
Digestion, 93(1):40-46, 14 Jan 2016
Cited by: 6 articles | PMID: 26789263
Review
Funding
Funders who supported this work.
BLRD VA (2)
Grant ID: I01 BX001453
Grant ID: IK2 BX002126
NCCIH NIH HHS (1)
Grant ID: R01 AT004821
NCI NIH HHS (5)
Grant ID: P30 CA068485
Grant ID: P50 CA095103
Grant ID: R01 CA190612
Grant ID: P01 CA028842
Grant ID: P01 CA116087
NIDDK NIH HHS (4)
Grant ID: P30 DK058404
Grant ID: R01 DK053620
Grant ID: F31 DK107159
Grant ID: U24 DK059637
NIGMS NIH HHS (1)
Grant ID: T32 GM008554





