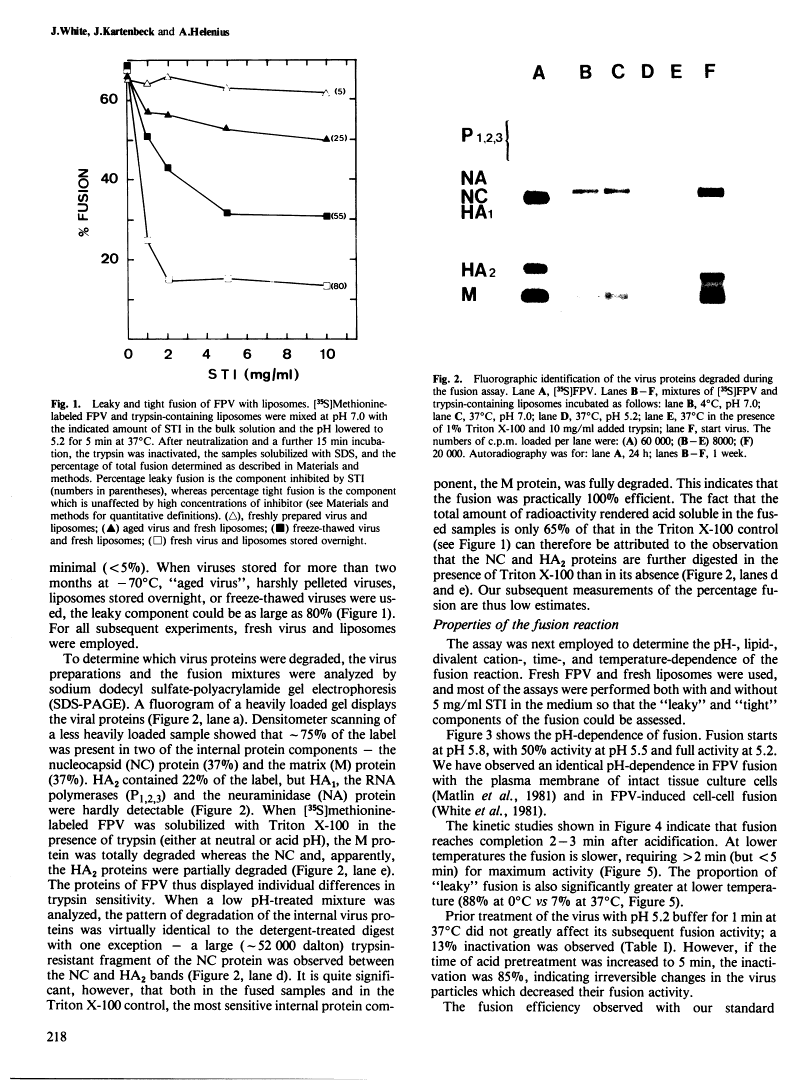
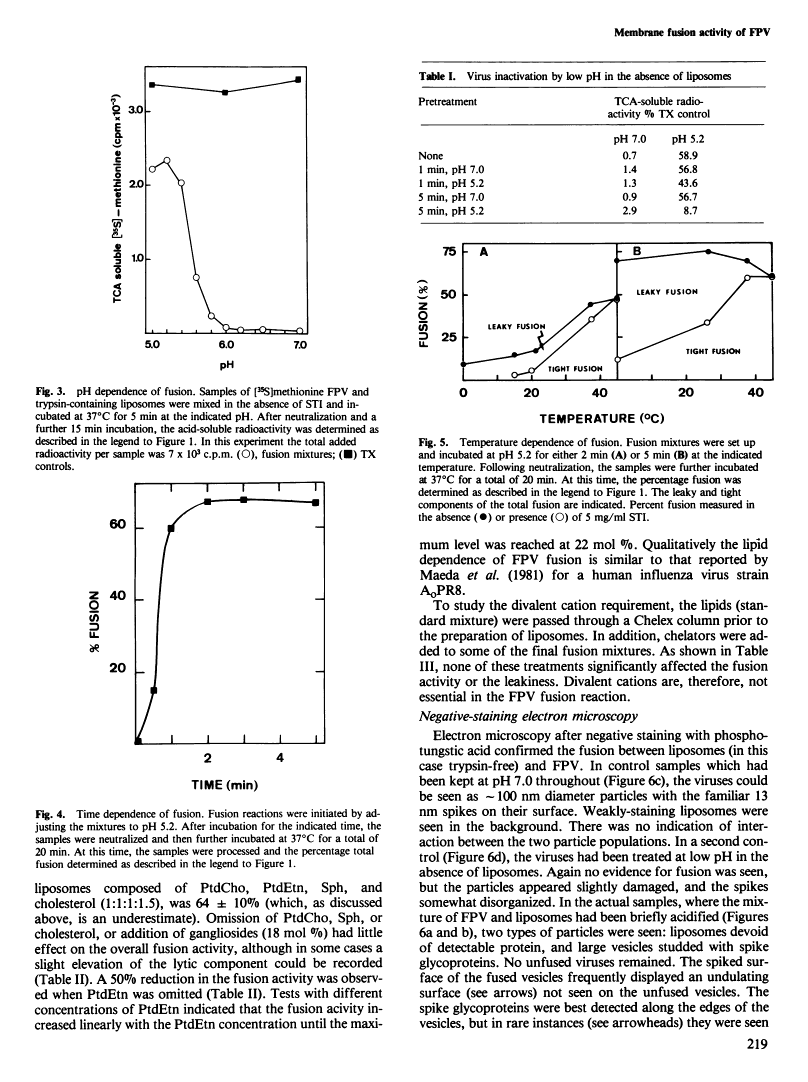
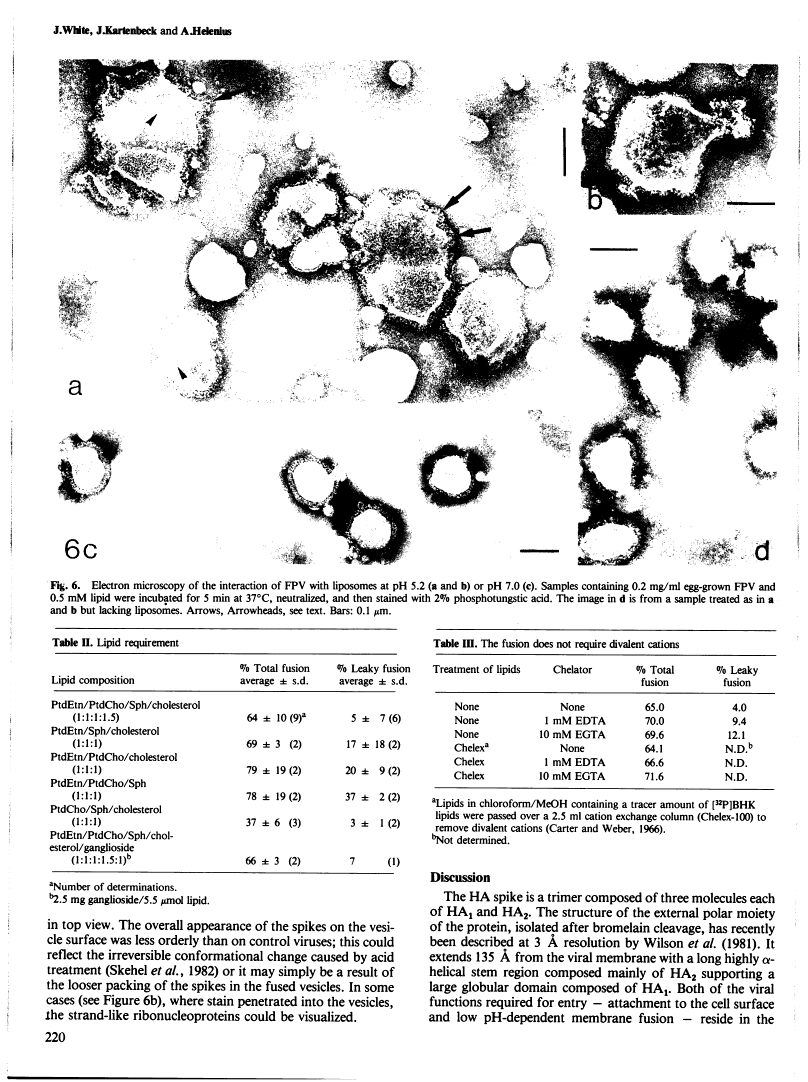
Abstract
Free full text

Membrane fusion activity of influenza virus.
Abstract
A simple assay is described to monitor fusion between fowl plague virus (FPV, an avian influenza A virus) and liposomes which allows the simultaneous quantitation of both lytic and non-lytic fusion events. As in fusion between viruses and the plasma membrane and in FPV-induced cell-cell fusion, the reaction only occurs at pH 5.5 or below, and it is fast, highly efficient, and essentially non-lytic when fresh virus and liposomes are used. The fusion occurs over a broad temperature range, and has no requirement for divalent cations. The fusion factor of influenza virus is a hemagglutinin (HA) spike which protrudes from the virus membrane and which is also responsible for virus binding to the host cell. The finding that fusion occurs as efficiently with liposomes containing or lacking virus receptor structures, further emphasizes the remarkable division of labor in the HA molecule: the receptor-binding sites are located in the globular HA1 domains and the fusion activation peptide is found at the N-terminal of HA2 in the stem region of the protein. The mechanism of fusion is discussed in terms of the three-dimensional structure of the HA and the conformational change which the protein undergoes at the fusion pH optimum.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.9M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Huang RT, Rott R, Klenk HD. Influenza viruses cause hemolysis and fusion of cells. Virology. 1981 Apr 15;110(1):243–247. [Abstract] [Google Scholar]
- Klenk HD, Rott R, Orlich M, Blödorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975 Dec;68(2):426–439. [Abstract] [Google Scholar]
- Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975 Dec;68(2):440–454. [Abstract] [Google Scholar]
- Lenard J, Miller DK. pH-dependent hemolysis by influenza, Semliki, Forest virus, and Sendai virus. Virology. 1981 Apr 30;110(2):479–482. [Abstract] [Google Scholar]
- LOWRY OH, ROSEBROUGH NJ, FARR AL, RANDALL RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [Abstract] [Google Scholar]
- Maeda T, Kawasaki K, Ohnishi S. Interaction of influenza virus hemagglutinin with target membrane lipids is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4133–4137. [Europe PMC free article] [Abstract] [Google Scholar]
- Maeda T, Ohnishi S. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 1980 Dec 29;122(2):283–287. [Abstract] [Google Scholar]
- Marsh M, Helenius A. Adsorptive endocytosis of Semliki Forest virus. J Mol Biol. 1980 Sep 25;142(3):439–454. [Abstract] [Google Scholar]
- Matlin KS, Reggio H, Helenius A, Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol. 1981 Dec;91(3 Pt 1):601–613. [Europe PMC free article] [Abstract] [Google Scholar]
- Jou WM, Verhoeyen M, Devos R, Saman E, Fang R, Huylebroeck D, Fiers W, Threlfall G, Barber C, Carey N, et al. Complete structure of the hemagglutinin gene from the human influenza A/Victoria/3/75 (H3N2) strain as determined from cloned DNA. Cell. 1980 Mar;19(3):683–696. [Abstract] [Google Scholar]
- Rand RP. Interacting phospholipid bilayers: measured forces and induced structural changes. Annu Rev Biophys Bioeng. 1981;10:277–314. [Abstract] [Google Scholar]
- Richardson CD, Scheid A, Choppin PW. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology. 1980 Aug;105(1):205–222. [Abstract] [Google Scholar]
- Skehel JJ, Waterfield MD. Studies on the primary structure of the influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 1975 Jan;72(1):93–97. [Europe PMC free article] [Abstract] [Google Scholar]
- Vänänen P, Käriäinen L. Haemolysis by two alphaviruses: Semliki Forest and Sindbis virus. J Gen Virol. 1979 Jun;43(3):593–601. [Abstract] [Google Scholar]
- Vänänen P, Käriäinen L. Fusion and haemolysis of erythrocytes caused by three togaviruses: Semliki Forest, Sindbis and rubella. J Gen Virol. 1980 Feb;46(2):467–475. [Abstract] [Google Scholar]
- White J, Helenius A. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3273–3277. [Europe PMC free article] [Abstract] [Google Scholar]
- White J, Kartenbeck J, Helenius A. Fusion of Semliki forest virus with the plasma membrane can be induced by low pH. J Cell Biol. 1980 Oct;87(1):264–272. [Europe PMC free article] [Abstract] [Google Scholar]
- White J, Matlin K, Helenius A. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J Cell Biol. 1981 Jun;89(3):674–679. [Europe PMC free article] [Abstract] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. [Abstract] [Google Scholar]
- Yamamoto K, Suzuki K, Simizu B. Hemolytic activity of the envelope glycoproteins of western equine encephalitis virus in reconstitution experiments. Virology. 1981 Mar;109(2):452–454. [Abstract] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. [Europe PMC free article] [Abstract] [Google Scholar]
- Carter HE, Weber EJ. Preparation and properties of various salt forms of plant phosphatidyl inositols. Lipids. 1966 Jan;1(1):16–20. [Abstract] [Google Scholar]
- Fang R, Min Jou W, Huylebroeck D, Devos R, Fiers W. Complete structure of A/duck/Ukraine/63 influenza hemagglutinin gene: animal virus as progenitor of human H3 Hong Kong 1968 influenza hemagglutinin. Cell. 1981 Aug;25(2):315–323. [Abstract] [Google Scholar]
- Gething MJ, Bye J, Skehel J, Waterfield M. Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature. 1980 Sep 25;287(5780):301–306. [Abstract] [Google Scholar]
- Gething MJ, White JM, Waterfield MD. Purification of the fusion protein of Sendai virus: analysis of the NH2-terminal sequence generated during precursor activation. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2737–2740. [Europe PMC free article] [Abstract] [Google Scholar]
- Helenius A, Fries E, Kartenbeck J. Reconstitution of Semliki forest virus membrane. J Cell Biol. 1977 Dec;75(3):866–880. [Europe PMC free article] [Abstract] [Google Scholar]
- Helenius A, Kartenbeck J, Simons K, Fries E. On the entry of Semliki forest virus into BHK-21 cells. J Cell Biol. 1980 Feb;84(2):404–420. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1002/j.1460-2075.1982.tb01150.x
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc553023?pdf=render
Citations & impact
Impact metrics
Article citations
Plant-Derived Epi-Nutraceuticals as Potential Broad-Spectrum Anti-Viral Agents.
Nutrients, 15(22):4719, 08 Nov 2023
Cited by: 1 article | PMID: 38004113 | PMCID: PMC10675658
Review Free full text in Europe PMC
Initiation and evolution of pores formed by influenza fusion peptides probed by lysolipid inclusion.
Biophys J, 122(6):1018-1032, 27 Dec 2022
Cited by: 4 articles | PMID: 36575795 | PMCID: PMC10111278
A Single Amino Acid Residue R144 of SNX16 Affects Its Ability to Inhibit the Replication of Influenza A Virus.
Viruses, 14(4):825, 15 Apr 2022
Cited by: 1 article | PMID: 35458555 | PMCID: PMC9032038
Negatively charged residues in the membrane ordering activity of SARS-CoV-1 and -2 fusion peptides.
Biophys J, 121(2):207-227, 18 Dec 2021
Cited by: 5 articles | PMID: 34929193 | PMCID: PMC8683214
How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity.
Viruses, 13(11):2309, 19 Nov 2021
Cited by: 10 articles | PMID: 34835115 | PMCID: PMC8619935
Review Free full text in Europe PMC
Go to all (152) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change.
J Biol Chem, 260(5):2973-2981, 01 Mar 1985
Cited by: 210 articles | PMID: 3972812
Fusion activity of influenza virus PR8/34 correlates with a temperature-induced conformational change within the hemagglutinin ectodomain detected by photochemical labeling.
Biochemistry, 30(9):2432-2438, 01 Mar 1991
Cited by: 19 articles | PMID: 2001371
Evidence for H(+)-induced insertion of influenza hemagglutinin HA2 N-terminal segment into viral membrane.
J Biol Chem, 269(28):18353-18358, 01 Jul 1994
Cited by: 63 articles | PMID: 8034580
Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin.
Annu Rev Biochem, 69:531-569, 01 Jan 2000
Cited by: 1678 articles | PMID: 10966468
Review