Abstract
Free full text

An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma
Abstract
The incidence of pediatric adrenal cortical carcinoma (ACC) in southern Brazil is 10–15 times higher than that of pediatric ACC worldwide. Because childhood ACC is associated with Li-Fraumeni syndrome, we examined the cancer history and p53 status of 36 Brazilian patients and their families. Remarkably, 35 of 36 patients had an identical germ-line point mutation of p53 encoding an R337H amino acid substitution. Differences within intragenic polymorphic markers demonstrated that at least some mutant alleles arose independently, thus eliminating a founder effect. In tumor cells, the wild-type allele was deleted, and mutant p53 protein accumulated within the nuclei. Although these features are consistent with Li-Fraumeni syndrome-associated adrenal tumors, there was no history of increased cancer incidence among family members. Therefore, this inherited R337H p53 mutation represents a low-penetrance p53 allele that contributes in a tissue-specific manner to the development of pediatric ACC.
Pediatric adrenal cortical carcinoma (ACC) tumors are rare. The estimated annual incidence in the U.S. is 0.3 per million children under 15 years of age (1). In southern Brazil, however, the incidence is ≈10–15 times greater (2). Most cases occur in the contiguous states of Sao Paulo and Paraná, but the cause of this higher rate has not been identified. The region has no known endemic transmissible or occupational diseases. The population is heterogeneous and is mainly of European extraction. New cases of ACC occur in all ethnic groups. Familial genetic predisposition to cancer has not been identified among the relatives of affected Brazilian children (3).
ACC occurs with increased frequency in children with Beckwith-Wiedemann syndrome (4) or inherited germ-line p53 gene mutations associated with the Li-Fraumeni familial cancer syndrome (5, 6). Childhood ACC is often the first indicator of familial predisposition to cancer (7). Typically, the p53 mutations in these families are located in the highly conserved DNA binding domain of p53 (exons 5–8), clustering at codons 175, 248, 273, and 282. However, some children who have ACC in the absence of familial cancer syndrome carry germ-line mutations in other regions of the p53 gene (8, 9). The high incidence of childhood ACC in southern Brazil prompted us to examine all coding regions of the p53 genes of 36 patients from 34 families. Here we describe a germ-line mutation found in exon 10, codon 337 of p53 in 35 of the 36 children.
Methods
Subjects and DNA Samples.
Between 1966 and 1999, 92 children were treated for ACC at the Clinics Hospital in Curitiba, southern Brazil. Tumors were classified histopathologically as adenoma or carcinoma according to established criteria (10). After p53 sequencing revealed that many of these patients had an identical, germ-line point mutation in exon 10 of p53 encoding an R337H amino acid substitution, 55 families of these patients were located and invited to participate. Study eligibility required a willingness to provide blood samples for p53 sequencing and a signed statement of informed consent. One hundred eighty-six family members, including 21 parents and 29 siblings of 36 patients, were tested for the presence of the mutation (Table (Table1).1). The tumor histology of the 36 index cases was confirmed by review at St. Jude Children's Research Hospital.
Table 1
Distribution of the germ-line R337H p53 mutation in children with ACC, their relatives, and unrelated subjects
subjects
| Test subjects | No. | Germ-line p53 (R337H) mutation (%) |
|---|---|---|
| Study patients (34 families) | 36 | 35 (97.2) (97.2) |
| Both parents (7 couples) | 14 | 7 (50) (50) |
| One parent | 7 | 4 (57.1) (57.1) |
| Siblings | 29 | 13 (44.8) (44.8) |
| Both grandparents (affected parental line) | 4 | 2 (50) (50) |
| One grandparent (affected parental line) | 7 | 3 (42.8) (42.8) |
| Other second-degree relatives (affected parental line) | 69 | 21 (30.4) (30.4) |
| Second-degree relatives (nonaffected parental line) | 56 | 0 |
| Control subjects unrelated to ACC families | 22 | 0 |
Pediatric endocrinologists obtained all family medical histories by personal interview with at least one parent. During a scheduled follow-up visit, a standardized questionnaire was completed for each child with ACC. Parents provided information about any cancer among first-, second-, and third-degree relatives; when possible, age at the time of diagnosis, tumor site, and type of cancer were recorded. Medical records were reviewed when available. Histologic review of tumor tissue from family members was not attempted. The number of cancers was ascertained separately for each parental line.
The control group comprised 22 children receiving follow-up care at the same institution for diabetes mellitus, constitutional growth hormone deficiency, or congenital hypothyroidism. Children in the study and control groups had been born and reared in the same geographic region. None of the control subjects had tumors or a family history suggesting increased cancer incidence. With informed consent, blood was drawn from control subjects for p53 sequencing at the time of their follow-up laboratory studies. An additional 56 family members from the noncarrier parental side of the index patients' families were tested (Table (Table11).
DNA Isolation and Analysis.
Genomic DNA was extracted from frozen tumor tissue or peripheral blood cells with the QIAamp kit (Qiagen, Valencia, CA). DNA quality was assessed by agarose gel electrophoresis (2% SeaKem, FMC). DNA was quantified by measuring absorbance with a Beckman DU-650 spectrophotometer (Beckman Instruments, Fullerton, CA). Genomic DNA was amplified by multiplex PCR as specified by the manufacturer of the GeneChip p53 probe array (Affymetrix, Santa Clara, CA), under conditions optimized by Gene Logic (Gaithersburg, MD). The 10 fragments that make up the p53 coding region were amplified with p53-specific primers and AmpliTaq Gold polymerase (Applied Biosystems). Each reaction contained 300 ng of DNA; a placental DNA control was run in each set. PCR products were evaluated on a 4% NuSieve 3:1 agarose gel (FMC) in 1× TAE buffer (0.4 M Tris acetate, 1 mM EDTA, pH 8.0). p53 exons were identified by fragmenting the amplified DNA and hybridizing it to the p53 gene probe array according to the manufacturer's instructions.
All positive p53 gene probe results were confirmed by sequencing. The p53 exons were amplified as described above. The QIAquick 96-well PCR Purification Kit (Qiagen) was used to purify double-stranded PCR products, which were evaluated by electrophoresis on a 2% agarose gel (FMC). The samples were labeled with fluorescent dyes (Big Dye Terminator Sequencing Kit, Applied Biosystems), purified with the AGTC 96-well dye purification kit (Edge Biosystems, Gaithersburg, MD), and electrophoresed on an ABI 377 DNA sequencer (Applied Biosystems) equipped with sequence navigator version 1.01 software.
Samples also were analyzed by a PCR-based assay. Amplified DNA was digested with restriction endonuclease HhaI, which cleaves wild-type p53 but not the R337H mutant. Briefly, a 447-bp DNA fragment encompassing exon 10 of p53 was amplified with the primers 5′-CTG AGG CAC AAG AAT CAC-3′ (forward) and 5′-TCC TAT GGC TTT CCA ACC-3′ (reverse). Samples devoid of DNA were used as negative controls. Samples were denatured at 95°C for 45 seconds. Thirty amplification cycles (annealing at 62°C for 45 sec and primer extension at 72°C for 45 sec) followed. Amplified products (20 μl) were digested with HhaI. Digestion products were separated on a 1.8% agarose gel prepared in Tris-borate-EDTA buffer containing ethidium bromide. Digested wild-type alleles revealed two bands (154 and 293 bp), whereas mutant alleles revealed only one (447 bp).
Immunohistochemical Studies.
The biotin-avidin-horseradish peroxidase immunohistochemical staining protocol was used to identify p53 in tumor cells. Deparaffined sections were incubated with a primary mouse anti-p53 mAb (clone DO7; Dako) at a 1:50 dilution for 30 min at room temperature. The sections were incubated similarly with a secondary biotinylated antibody (Dako). The remaining steps were performed as recommended by the antibody manufacturer. For each staining procedure, a section of squamous-cell lung carcinoma with known high expression of p53 was used as a positive control, and a tumor section stained without primary antibody was used as a negative control.
Functional Characterization of the Mutant.
We used a PCR-based strategy (11) to create a CGC → CAC point mutation at codon 337 of a human wild-type p53 cDNA; this cDNA encoded a p53 protein with the arginine-to-histidine substitution observed in ACC. The fidelity of the reaction and the presence of the intended mutation were confirmed by sequencing the entire coding region. The mutant R337H cDNA was cloned into the BamHI site of pCMV-NEO-Bam under the transcriptional control of a potent cytomegalovirus (CMV) promoter (pCMV-53–337H) (12). As controls, wild-type p53 and a mutant p53 cDNA encoding an R248W substitution also were cloned into this plasmid (pCMV-53 and pCMV-53–248W, respectively).
Immortal murine (10)1 BALB/c fibroblasts (13) were used for promoter transactivation studies and human SaOs-2 osteosarcoma cells (14) for colony reduction assays; neither cell line expresses endogenous p53. Cells were cultured at 37°C and 5% CO2 in DMEM supplemented with 10% FBS [(10)1 cells] or 20% FBS (SaOs-2 cells).
The wild-type p53-responsive promoter-luciferase reporter construct (50–2 Luc) was used for the transactivation assays (15). This reporter contains a luciferase reporter gene under the transcriptional control of the adenovirus major late TATA box and terminal deoxynucleotidyltransferase initiator element. In addition, two copies of the p53 response element from the murine muscle creatine kinase gene were cloned upstream of the TATA box and both were arranged in the forward orientation. The (10)1 cells were transiently transfected in duplicate with 250 ng of the 50–2 Luc reporter construct alone or with 50–2 Luc and the indicated amount of pCMV-NEO-Bam, pCMV-53, or pCMV-53–337H, by the calcium phosphate method (14). Cells were harvested, protein extracts were prepared and quantified, and equal amounts of protein were used in a standard luciferase assay (Promega). Western blot analysis was used to quantify p53 protein (16).
In the colony reduction assay, SaOs-2 cells were transfected in duplicate with 100 ng or 1 μg of pCMV-NEO-Bam, pCMV-53, pCMV-53–248W, or pCMV-53–337H by the calcium phosphate method. Twenty-four hours later, cells were plated in medium containing 1 mg/ml G418 (Life Technologies, Gaithersburg, MD) and cultured for 10–14 days to select for productively transfected cells. Colonies were fixed in methanol, stained with a modified Giemsa dye (Sigma), and counted.
We also assayed the function of the R337H mutant in an apoptosis assay using human H1299 lung carcinoma cells, which lack endogenous p53. The cells were microinjected with a green fluorescent protein expression vector to indicate productively injected cells and with expression plasmids (50 ng DNA/sample) encoding CMV only, wild-type p53, or R337H mutant p53. On average, 100–150 cells were injected with each type of plasmid. Cells were scored by phase-contrast and fluorescent microscopy for morphological features of apoptosis and were photographed to document phenotypic changes.
Analysis of p53 Polymorphism.
Four polymorphisms within or flanking the p53 gene were used to determine genotypes of the participants. The flanking markers were D17S1832, which is 13 centimorgans (cM) or ≈70 centirays (cR) from the telomere of 17p, and D17S786, which is ≈18 cM or 90 cR from the 17p telomere. The p53 gene is located 82 cR from the 17p telomere. The primers, PCR conditions, and exact positions of these markers have been described (Stanford Human Genome Center, http://shgc-www.stanford.edu.; Whitehead Institute for Biomedical Research/Massachusetts Institute of Technology Center for Genome Research, http://www.genome.wi.mit.edu). The two intragenic markers were a dinucleotide repeat and a pentanucleotide repeat for which the primers and conditions have been reported (17). Findings were interpreted as described (18).
Results
Patients and Families.
The median age of the 29 girls and seven boys at the time of diagnosis of ACC was 3 years (range, 4 months to 13.5 years). Thirty-four patients had signs of virilization (precocious development of secondary masculine features); 13 of these had signs of Cushing's syndrome (obesity, hypertension, muscular weakness, and/or excessive hair growth). One patient had signs of Cushing's syndrome only, and one showed no evidence of adrenal hyperfunction. Histologic review confirmed the tumors as carcinomas (n = 27) or adenomas (n = 9). Six patients, all with carcinomas, had advanced-stage disease (distant metastasis or unresectable tumors), and 30 had resectable localized tumors.
Only one occurrence of cancer (lung cancer) was reported among the 126 first-degree relatives (parents and siblings) of the 36 index cases. For 25 of the families, we also obtained cancer histories for three or more generations of both parental lines. There was no evidence of increased predisposition to cancer in any of the parental lines of 24 of these 25 families. On the maternal side of the remaining family, for which three generations were characterized, at least one member of each generation had cancer, including the great grandfather (intestinal cancer), two great aunts and one aunt (breast cancer), and one cousin (leukemia). This family's spectrum of cancers was consistent with a variant of Li-Fraumeni syndrome (19). Sarcoma of soft tissue or bone, which is typical of Li-Fraumeni syndrome (6), was not reported.
Remarkably, there were multiple cases of ACC in four families. In each of two families, two half-siblings were affected; in another, two first cousins. In these three families, all children with ACC were index cases. In the remaining family (Fig. (Fig.1),1), five children had ACC; two were index cases, two were deceased children identified through family history and medical records, and one was a girl identified only through family history who died before the age of 3 years of an abdominal tumor with virilizing syndrome.
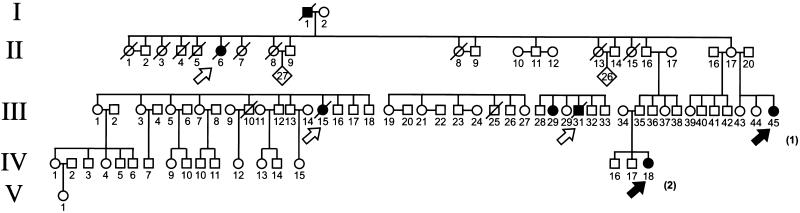
Pedigree of family with five cases of ACC. Five generations are represented. Two of the children with ACC (closed arrows) were index cases; their ages at diagnosis are noted in parentheses. The three others are indicated by open arrows. There were two additional cases of cancer in the family: an unspecified intraabdominal carcinoma (I-1) and an astrocytoma (III-29). Deceased individuals are indicated by an oblique bar.
Identification of a Germ-Line p53 Mutation Associated with Adrenal Cortical Tumor.
The p53 status of the patients with ACC was determined by genechip p53 gene probe array and sequence analysis of genomic DNA isolated from peripheral blood lymphocytes. Thirty-five of the 36 patients had an identical, germ-line p53 point mutation in exon 10 at codon 337 (CGC → CAC) encoding an arginine-to-histidine substitution (R337H) (Fig. (Fig.2).2). The mutation is referred to as the R337H mutation for the purpose of this report. The remaining patient, an 11-year-old girl, had only wild-type p53 germ-line sequences. No other p53 mutations were detected in this group. The control subjects were found to carry only wild-type p53 germ-line sequences. The R337H mutation was found in at least one parent in each of the seven sets of parents tested and in the affected parental line of each family but was not found in any of 56 individuals analyzed from the noncarrier side of the patients' families (Table (Table1).1). These results provide compelling evidence that this mutation is a potent risk factor associated with ACC rather than a benign polymorphism commonly found in southern Brazil.
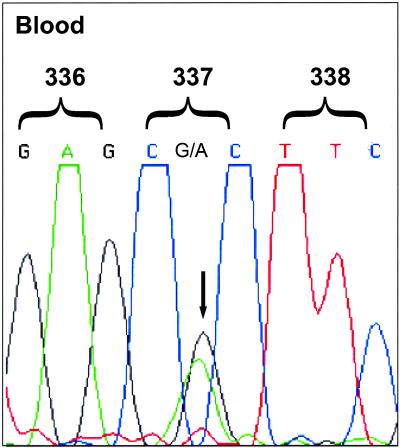
Identification of germ-line R337H p53 mutation in patients with ACC. p53 was sequenced in genomic DNA prepared from peripheral blood cells. A representative sequence from a patient with ACC is shown. Sequences correspond to the p53 sense strands between codons 336 and 338 of exon 10. The G at position 1010 (codon 337) in the wild-type strand is replaced by A in the mutated strand (arrow).
The appearance of this germ-line mutation in one parent of each of seven complete sets of parents tested suggests that the mutation is inherited. The oldest carrier tested was a 70-year-old man.
Independent Origin of the Mutation.
Although the pedigree of most of these patients do not overlap, the remarkable frequency with which the R337H germ-line mutation occurred in these patients is consistent with their having had a common ancestor. To directly investigate this possibility, we performed a detailed polymorphism analysis using intergenic and intragenic p53 markers. Four polymorphic markers were characterized in 17 unrelated patients (Fig. (Fig.3)3) and 10 relatives. Although the genetic phase could not be definitively established in all subjects, these analyses clearly identified differing alleles of the p53 gene and its surrounding markers in several patients. For example, the haplotypes of patients 3 and 8 in Fig. Fig.33 differed at the p53 locus of both chromosomes. These results eliminate the possibility of a single founder.
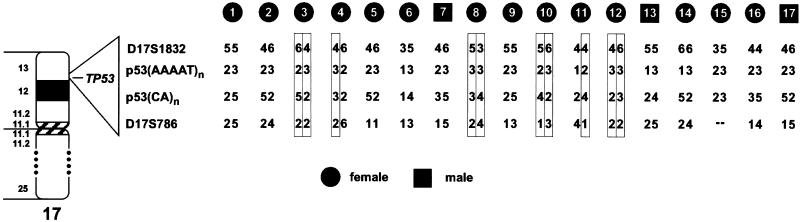
Analysis of p53 polymorphisms in patients with ACC. Genotypes were determined by using four markers, including two intragenic p53 polymorphisms. Their positions relative to the p53 gene and their order on chromosome 17 are shown next to an ideogram of chromosome 17. Allelic order of the boxed markers was determined by studies of at least one parent or sibling. Because the markers clearly differed even among patients for whom the allelic order could not be confirmed, a founder effect is highly unlikely to account for the presence of the mutation. D17S1832 and D17S786 = dinucleotide repeat markers; p53(AAAAT)n = intragenic p53 pentanucleotide polymorphism located in intron 1; p53(CA)n = intragenic p53 dinucleotide polymorphism located in codon 213.
Loss of Heterozygosity in Tumor Cells.
Loss of function by tumor suppressor genes, often caused by mutation of one allele and deletion of the other, is a hallmark of tumorigenesis. To determine the status of the wild-type p53 allele in the adrenal tumors, we isolated total genomic DNA for analysis by DNA sequencing and a PCR-based assay. Only the mutated allele was observed by DNA sequencing in five of six patients' tumor cells (see Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org). PCR amplification of exon 10 followed by digestion with HhaI (which cleaves only the wild-type p53 sequence) confirmed the sequencing analysis (data not shown). Therefore, these tumors commonly underwent loss of heterozygosity while retaining the R337H mutant allele.
Nuclear Accumulation of p53 in Tumor Cells.
Mutant p53 tends to accumulate in detectable quantities within the nuclei of affected tumor cells. We investigated the expression and localization of the R337H mutant in 11 tumor samples by immunohistochemistry. Eight of the 11 tumors (seven carcinomas and one adenoma, including samples shown to have undergone loss of heterozygosity) showed nuclear accumulation of p53. No p53 accumulation was seen in the single tumor with wild-type p53 sequences (see Fig. 6, which is published as supplemental data on the PNAS web site). Interestingly, the pattern of nuclear accumulation of the mutant protein was comparable to that observed in classic Li-Fraumeni-associated tumors.
Functional Analysis of the Mutant p53.
Mutation of p53 usually disrupts DNA binding activity, precluding the induction of cell-cycle arrest. To determine the functional effect of the R337H mutation, we conducted a transient transfection promoter-reporter assay (15). Wild-type p53 and the R248W mutant (frequently observed in Li-Fraumeni families) were positive and negative controls, respectively. The R337H mutant activated the reporter as efficiently as did wild-type p53, whereas the R248W mutant, previously shown to be defective in DNA binding and transactivation of reporter constructs (20), was inactive (Fig. (Fig.44A). The extent of reporter activation was correlated with the level of p53 protein expression, as shown by Western blot analysis (Fig. (Fig.44B). Wild-type p53 and the R337H mutant, but not the R248W mutant, were able to suppress colony growth of SaOs-2 cells, which lack endogenous p53 (see Fig. 7, which is published as supplemental data on the PNAS web site). In human H1299 lung carcinoma cells, which lack endogenous p53, ectopically expressed R337H mutant p53 was competent to induce apoptosis (see Fig. 8, which is published as supplemental data on the PNAS web site). These results indicate that the R337H mutant retains p53 activity when expressed at supraphysiological levels.
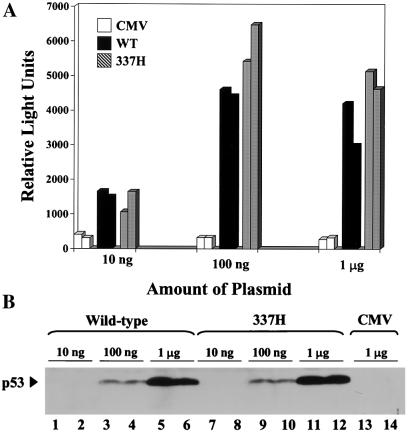
The R337H mutant retains transactivation function when overexpressed. (A) Wild-type p53 and the R337H mutant showed comparable relative transactivation of a wild-type p53-responsive promoter. (B) Western blot analysis showed that the wild-type and R337H p53 proteins were equally expressed in this assay. WT = wild type; 337H = R337H p53 mutant; Neo = neomycin; 248 = R248W p53 mutant associated with Li-Fraumeni syndrome.
Discussion
Southern Brazilian children with ACC consistently carry an inherited germ-line p53 mutation in exon 10 at codon 337 that causes an arginine-to-histidine substitution. The striking frequency of the mutation (35 of 36 patients) suggested that it might be a common benign polymorphism among southern Brazilians; however, none of the 22 children in the control group nor any of the more than 50 individuals from the noncarrier side of the patients' families harbored this mutation. Chompret et al. (21) recently reported an identical inherited mutation in a patient with ACC. Although the mutation might have represented a polymorphism, it had not been observed in more than 200 random alleles tested in that laboratory. Another germ-line p53 mutation involving the first position of codon 337 that encoded an arginine-to-cysteine missense mutation has been described in two families with a predisposition to cancer (22–24).
The cause of the R337H p53 mutation is presently unknown. Our demonstration of different markers surrounding and within the mutated p53 gene in various unrelated subjects argues strongly against its derivation from a common ancestor (i.e., there is no “founder effect”). To date, we have tested members of several generations of the affected parental line in four families for the R337H mutation. Although these studies are incomplete, the age of the oldest carrier identified was 70 years. Thus, the mutation arose at least 70 years ago.
Interestingly, this mutation is reminiscent of the somatic p53 “signature” mutation at codon 249 observed in ≈50% of hepatocellular carcinomas in southern Africa and in Qidong, China, where exposure to dietary aflatoxin B1 and hepatitis B virus is common (25, 26). In endemic hepatocellular carcinoma, however, the p53 mutation is somatically acquired by hepatocytes, whereas in children with ACC, it is inherited. In southern Brazil, which is predominantly agricultural, the unregulated long-term use of pesticides and other industrial chemicals may be implicated in the origin of this abnormality. Alternatively, spontaneous or oxidant-induced deamination of 5-methylcytosine at a CpG site of codon 337 could result in a G:C→A:T transition. It remains to be determined whether environmental mutagens, directly or through epigenetic mechanisms, cause the germ cell mutation.
The loss of the wild-type allele and the abnormal nuclear accumulation of the mutant protein in these tumors (a common finding in p53-associated cancers) support the premise that this mutation plays an essential role in the development of ACC. Although we cannot dismiss the possibility that the R337H mutation is a surrogate marker for another gene flanking the region, the diversity of the intragenic p53 polymorphic markers found among the unrelated study subjects argues strongly against that conclusion.
Although childhood ACC, particularly at young ages, has been used to identify cancer-prone families that carry p53 mutations, our study group was unlike these families in several ways. First, the frequency and types of cancers that occurred in children and adolescents in our patients' families were not typical of those seen in Li-Fraumeni or Li-Fraumeni-like families (6, 27). In Li-Fraumeni families, the most common tumors in children below age 14 years include sarcoma of soft tissue or bone, brain tumor, leukemia, and ACC (8). In our series, no relatives of the study subjects had leukemia or sarcoma of soft tissue or bone at a young age, although one had had a brain tumor. Second, the incidence of cancer in our patients' families does not appear to be abnormally high, although the absence of a Brazilian cancer registry prevents statistical comparisons. In striking contrast, Li-Fraumeni families in which there is a proband with childhood ACC have a pervasive history of cancer, and carriers of the germ-line p53 mutation present with typical Li-Fraumeni tumors at exceptionally young ages (28). Our findings suggest that the overall malignant potential of the R337H mutation is low, yet markedly organ-specific.
The R337H mutation lies within the p53 dimerization domain (amino acids 336–353), which is considered essential for p53 function (29–31). Specifically, arginine-337 of one subunit forms a salt bridge with aspartate-352 in the corresponding monomer to stabilize the complex (29–32). From the pKa of the imidazole ring of histidine (≈6.0–6.5, independent of dipole moment), it is predicted that the R337H mutant will not efficiently form the salt bridge at physiologic pH and therefore will be functionally impaired. Moreover, the results of functional analysis of separated alleles in yeast (FASAY) assays have suggested that mutations in the oligomerization domain are associated with partial or complete loss of transactivation activity (24, 32). In our assays, R337H had wild-type p53 activity when expressed at supraphysiological levels in fibroblasts and osteosarcoma cells. Growing evidence indicates that although these in vitro transfection assays are useful for detecting profound defects in p53 (e.g., in the Li-Fraumeni R248W mutant), they are not sufficiently sensitive to detect the effects of subtle alterations, such as mutations that target specific posttranslational modification sites (33, 34). Thus, the currently available functional studies of p53 may not identify all clinically meaningful mutations. The presence of the R337H germ-line mutation in 35 of 36 patients, the deletion of the wild-type allele in most of the analyzed tumors, and the nuclear accumulation of the mutant p53 protein in ACC tumor cells strongly imply that the protein is functionally impaired under physiological conditions.
Impaired p53 activity, rather than complete abrogation of function, in the R337H mutant could predispose individuals to certain types of tumors (e.g., ACC) without producing a highly penetrant and diverse cancer phenotype such as that associated with Li-Fraumeni syndrome. This premise is supported by the recent work of Varley et al. (8), who observed p53 mutations in nine of 14 cases of childhood ACC in the absence of a family history of cancer. It is therefore reasonable to speculate that decreased p53 apoptotic function and/or cell cycle regulation could significantly contribute to tumorigenesis of the adrenal cortex, a tissue that undergoes extensive cellular remodeling during development (35). Conclusive evidence of defective function of the R337H mutant will require establishment of a gene knock-in animal model.
Birch and colleagues (36) have noted an association between the type of p53 mutation and the cancer phenotype. Our findings add to this observation the possibility that some germ-line p53 mutations may not be associated with familial clustering of cancer and that the malignant potential of the mutant protein may be highly tissue-specific.
Acknowledgments
We are indebted to Drs. M. B. Kastan and J. L. Cleveland for critical review; S. H. Naron and Dr. J. C. Jones for editorial assistance; Dr. J. J. Jenkins for immunohistochemical studies; Drs. M. E. Conley and J. Rohrer for confirmatory testing of tumor-cell DNA; C. Baer for technical assistance; and C. Alvares, J. Boland, P. Murphy, and E. Pettit of the DNA profiling laboratory at Gene Logic Inc., for their contribution to this study. This work was supported in part by Grants RO1-CA63230, P30-CA-21765 and PO1-CA-20180 from the National Cancer Institute and by the American Lebanese Syrian Associated Charities (ALSAC).
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.161479898
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/doi/pdf/10.1073/pnas.161479898
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.161479898
Article citations
Pediatric Adrenocortical Carcinoma: The Nuts and Bolts of Diagnosis and Treatment and Avenues for Future Discovery.
Cancer Manag Res, 16:1141-1153, 07 Sep 2024
Cited by: 0 articles | PMID: 39263332 | PMCID: PMC11389717
Review Free full text in Europe PMC
AI-guided identification of risk variants for adrenocortical tumours in TP53 p.R337H carrier children: a genetic association study.
Lancet Reg Health Am, 38:100863, 23 Aug 2024
Cited by: 0 articles | PMID: 39258234 | PMCID: PMC11386259
Importance of genetic cancer risk assessment as a strategy to stratify risk and provide precision prevention in high-risk patients and families.
Rev Assoc Med Bras (1992), 70(suppl 1):e2024S117, 07 Jun 2024
Cited by: 0 articles | PMID: 38865537
Current prospects of hereditary adrenal tumors: towards better clinical management.
Hered Cancer Clin Pract, 22(1):4, 26 Mar 2024
Cited by: 0 articles | PMID: 38532453 | PMCID: PMC10964668
Review Free full text in Europe PMC
The genetic risk of acute lymphoblastic leukemia and its implications for children of Latin American origin.
Front Oncol, 13:1299355, 09 Jan 2024
Cited by: 0 articles | PMID: 38264740 | PMCID: PMC10805326
Review Free full text in Europe PMC
Go to all (276) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Li-Fraumeni and Li-Fraumeni-like syndrome among children diagnosed with pediatric cancer in Southern Brazil.
Cancer, 119(24):4341-4349, 07 Oct 2013
Cited by: 29 articles | PMID: 24122735
Novel germline mutation of the p53 tumor suppressor gene in a child with incidentally discovered adrenal cortical carcinoma.
Am J Pediatr Hematol Oncol, 16(4):341-347, 01 Nov 1994
Cited by: 10 articles | PMID: 7978053
Penetrance of adrenocortical tumours associated with the germline TP53 R337H mutation.
J Med Genet, 43(1):91-96, 20 Jul 2005
Cited by: 84 articles | PMID: 16033918 | PMCID: PMC2564508
The Inherited p53 Mutation in the Brazilian Population.
Cold Spring Harb Perspect Med, 6(12):a026195, 01 Dec 2016
Cited by: 49 articles | PMID: 27663983 | PMCID: PMC5131754
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (5)
Grant ID: P01-CA-20180
Grant ID: R01-CA63230
Grant ID: R01 CA063230
Grant ID: P30 CA021765
Grant ID: P30-CA-21765





