| PMC full text: | Nat Biotechnol. Author manuscript; available in PMC 2017 Nov 1. Published in final edited form as:
|
Figure 6
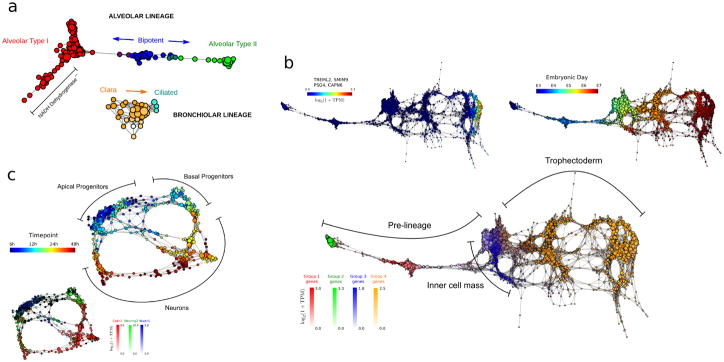
a Topological representation of 80 embryonic (E18.5) mouse lung epithelial cells35 labeled according to cell type. scTDA resolves the alveolar and bronchiolar lineages that were originally identified using PCA, and identifies a putative set of cells with low NADH dehydrogenase expression that were not identified in the original analysis.
b. Topological representation of 1,529 individual cells from 88 human pre-implantation embryos36. Top-left, bottom: topological representation (labeled by expression levels of genes) associated with cellular populations identified during differentiation. scTDA resolved, without supervision, the segregation of the trophectoderm and inner cell mass from pre-lineage cells (bottom), as well as a polar trophectoderm (top-left) that were originally identified using a supervised analysis based on PCA and k-medoid clustering. Top-right: topological representation labeled by embryonic day.
c. Topological representation of 272 newborn neurons from the mouse neocortex37, labeled by sampling time after mitosis (top) and expression levels of Cntr2, Neurog2, and Wwtr1 (bottom). scTDA recapiltulated the converging developmental relations between apical and basal progenitors, and neurons that were originally identified using hierarchical clustering and additionally found lineage convergence.