Abstract
Free full text

Kaposi's sarcoma-associated herpesvirus infection promotes differentiation and polarization of monocytes into tumor-associated macrophages
ABSTRACT
Tumor associated macrophages (TAMs) promote angiogenesis, tumor invasion and metastasis, and suppression of anti-tumor immunity. These myeloid cells originate from monocytes, which differentiate into TAMs upon exposure to the local tumor microenvironment. We previously reported that Kaposi's sarcoma-associated herpes virus (KSHV) infection of endothelial cells induces the cytokine angiopoietin-2 (Ang-2) to promote migration of monocytes into tumors. Here we report that KSHV infection of endothelial cells induces additional cytokines including interleukin-6 (IL-6), interleukin-10 (IL-10), and interleukin-13 (IL-13) that drive monocytes to differentiate and polarize into TAMs. The KSHV-induced TAMs not only express TAM-specific markers such as CD-163 and legumain (LGMN) but also display a gene expression profile with characteristic features of viral infection. More importantly, KSHV-induced TAMs enhance tumor growth in nude mice. These results are consistent with the strong presence of TAMs in Kaposi's sarcoma (KS) tumors. Therefore, KSHV infection of endothelial cells generates a local microenvironment that not only promotes the recruitment of monocytes but also induces their differentiation and polarization into TAMs. These findings reveal a new mechanism of KSHV contribution to KS tumor development.
Introduction
Kaposi's sarcoma (KS) is a vascular malignancy of endothelial cell origin that is associated with infection by Kaposi's sarcoma-associated herpesvirus (KSHV).1,2 Although KSHV infection has been experimentally demonstrated to directly induce malignant transformation of the infected cells,3 early stage KS is not a true tumor but a hyperplasia driven by KSHV-induced proliferation, angiogenesis and inflammation.4,5 A hallmark of KS is the presence of significant amounts of infiltrating inflammatory cells including monocytes and macrophages..6,7
Several studies reported that KSHV establishes a latent infection in immortalized monocytic cell lines and deregulates expression of several inflammatory cytokines.8-11 We notice that KSHV effectively infects primary monocytes. However, most of the infected cells undergo KSHV lytic replication and die days after infection. Nevertheless, KSHV infection of other types of cells such as endothelial and epithelial cells may indirectly influence monocytes. Indeed, we recently reported that KSHV infection of endothelial cells induces production and secretion of the angiogenic and inflammatory cytokine angiopoietin-2 (Ang-2), which promotes migration and recruitment of monocytes into KS tumors.12 In addition, a recent study demonstrated that endothelial cells ectopically expressing KSHV vFLIP protein produce cytokines that remodel myeloid cells toward a “pro-tumor” phenotype.13 Nevertheless, little is known on how KSHV acute infection of endothelial cells impacts monocytes, and the role of monocytes/macrophages on KS tumor development has never been studied.
In many cancer models, the contribution of tumor-associated macrophages (TAMs), to tumor development and progression is well established.14,15 Peripheral blood monocytes, depending on local presence of particular cytokines, differentiate and polarize into either M1 macrophages to clear up pathogens and infected cells, or M2 macrophages for tissue repair and wound healing.16-19 Lipopolysaccharide (LPS), interferon-gamma (IFN-γ), and granulocyte-macrophage colony stimulating factor (GM-CSF) induce differentiation and polarization of monocytes into M1 macrophages during infection. In contrast, interleukin-4 (IL-4), interleukin-6 (IL-6), interleukin-10 (IL-10), interleukin-13 (IL-13), and macrophage colony stimulating factor (M-CSF) induce differentiation and polarization of monocytes into M2 macrophages. A subset of M2 macrophages display characteristics of TAMs,20-23 which produce multiple cytokines and matrix metallopeptidases (MMPs) to promote proliferation, angiogenesis, tumor invasion and metastasis, suppression of anti-tumor immunity, and development of resistance to chemotherapy.24-26 TAMs express common monocyte markers such as CD-14 and CD-68 and unique markers such as CD-11b,27,28 CD-163,29 CD-206,30 and legumain (LGMN), an asparaginyl endopeptidase that degrades fibronectin to activate pro-oncogenic MMPs such as MMP-2, MMP-9, and cathepsins B, L, and V.31-34
We have found strong presence of TAMs in KS tumors. Mechanistically, we demonstrate that KSHV acute infection of endothelial cells strongly induces expression of IL-6, IL-10, and IL-13 that are known to promote differentiation and polarization of monocytes into M2 macrophages/TAMs.16,35,36 Indeed, incubation of monocytes with culture supernatants from KSHV-infected endothelial cells results in macrophages that express TAMs-specific markers such as LGMN and CD-163. Data from RNA-seq analysis indicate that the KSHV-induced TAMs display a gene expression profile with characteristic features of viral infection. More importantly, co-inoculation of KS tumor cells with KSHV-induced TAMs significantly enhances tumor growth in nude mice.
To our knowledge, this is the first study to demonstrate how KSHV acute infection of endothelial cells generates a local micro-environment that promotes differentiation and polarization of monocytes into TAMs. Our results reveal a new mechanism by which KSHV infection contributes to KS tumor development.
Results
TAMs are strongly present in KS tumors
To detect TAMs in KS tumors, we conducted fluorescence antibody staining on paraffin section from 9 KS tumors with antibodies to TAMs-specific marker LGMN and monocyte/macrophage marker CD-68. All 9 KS tumors display significant numbers of cells expressing both markers as shown in Fig-1A, ,B,B, and andC.C. The average percentage (%) of TAMs from the 9 tumors is 9.2 ± 2.8 %. As described previously,37 LGMN is mainly present in the cytoplasm in CD-68 positive TAMs (Fig. 1B). As a typical feature of KS, most tumor cells express KSHV latent nuclear antigen (LANA) (Fig. 1D). These results indicate that TAMs are strongly present in KS tumors.
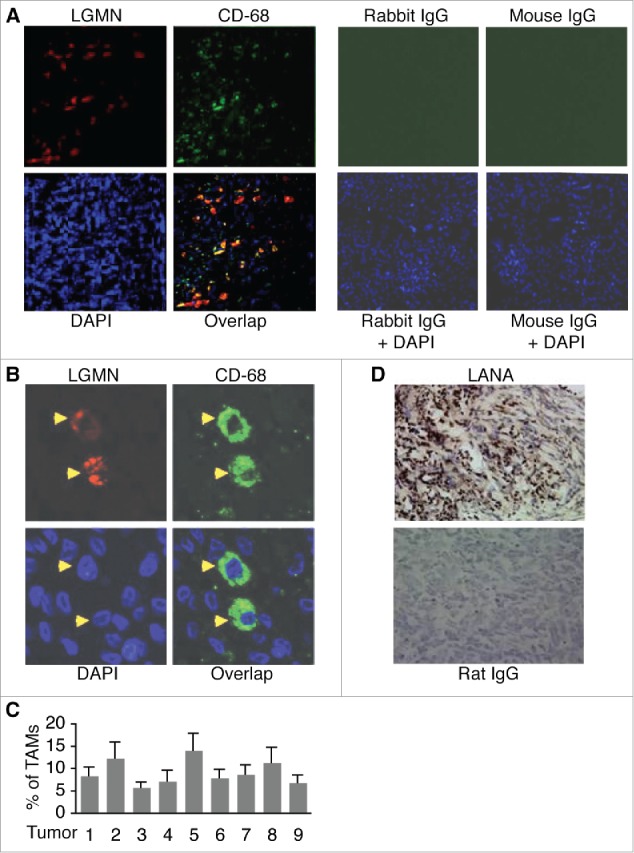
Detection of TAMs in KS tumor sections by immuno-fluorescence antibody staining (IFA). A, representative image of KS tumor sections co-stained with a rabbit anti-human LGMN polyclonal antibody and a mouse anti-human CD-68 monoclonal antibody, which were revealed with an Alexa Fluor®-633 conjugated goat anti-rabbit secondary antibody and a FITC-conjugated goat anti-mouse secondary antibody respectively. Rabbit IgG and mouse IgG were used as negative controls, and DAPI was used for nuclear staining of all cells. LGMN and CD-68 positive cells were imaged under a confocal microscope with a 10x objective. B, LGMN and CD-68 positive cells described in (A)that were imaged under a confocal microscope with a 40x oil objective. C, percentage (%) of TAMs in KS tumors. The numbers of LGMN+/CD-68+ cells and total cells per field under 40x oil objective from 4 fields per tumor were counted and used for the calculation of % of TAMs. D, representative image of KS tumor sections stained with a rat monoclonal antibody to KSHV latent protein LANA, which was revealed with a biotinylated secondary antibody and streptavidin-horseradish peroxidase, and DAB (3,3′-diaminobenzidine) detection system from Biolegend (San Diego, California, USA). Rat IgG was used as a negative control. Images were captured under a microscope (Carl Zeiss, Inc., Thornwood, NY).
KSHV infection of endothelial cells induces TAMs-promoting cytokines
KS tumors result from KSHV infection of endothelial cells. Thus, we next attempted to examine if KSHV infection of endothelial cells contributes to the strong presence of TAMs in KS tumors. We previously showed that KSHV infection of endothelial cells induces Ang-2 to promote migration and recruitment of monocytes into KS tumors.12 We suspected that KSHV infection of endothelial cells induce additional cytokines that drive the recruited monocytes to differentiate and polarize into TAMs. To test this hypothesis, we infected human umbilical vein endothelial cells (HUVECs) with mock or purified recombinant KSHV, BAC16,38 for various time points, followed by measurement of expression of TAM-promoting cytokines such as IL-4, IL-6, IL-10, IL-13, and M-CSF.
As shown in Fig-2A, ,B,B, and andC,C, KSHV infection of HUVECs strongly induces expression of IL-6, IL-10, and IL-13 at the mRNA level, which peaks at 48 hours post-infection (hpi). However, the mRNA of neither IL-4 nor M-CSF was detected, suggesting that HUVECs do not express these 2 cytokines. Consistent with previous studies,39-41 KSHV infection induces expression of Ang-2 and VEGF at the mRNA level (Fig. 2D and andE).E). Notably, IL-6, IL-10, and IL-13 proteins are highly present in the supernatants of KSHV-infected HUVECs, and the protein levels of these cytokines also peak at 48 hpi (Fig. 2F). In contrast, strongest expression of Ang-2 is detected at 72 hpi while enhanced expression of VEGF starts as early as 3 hpi. Therefore, KSHV infection of HUVECs induces at least 3 different TAMs-promoting cytokines, which may act alone or in combination on the recruited monocytes to induce their differentiation and polarization into TAMs.
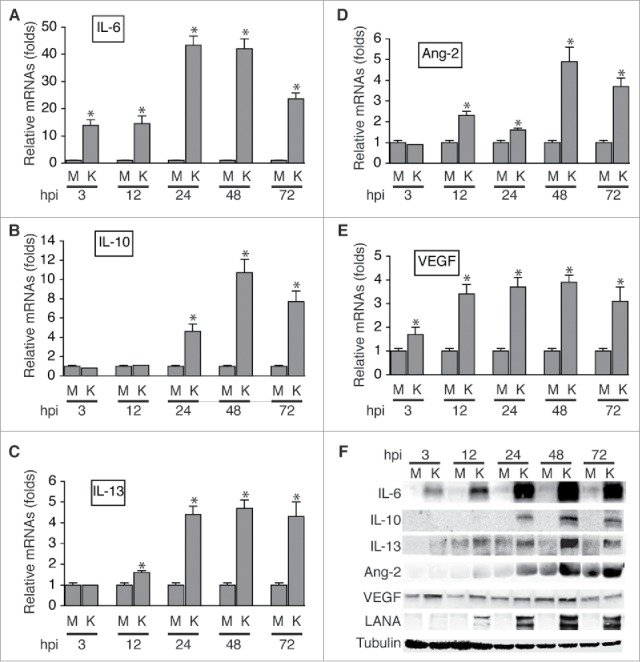
KSHV acute infection of HUVECs induces expression of cytokines that are known to promote differentiation and polarization of monocytes into M2 macrophages/TAMs. A, B, C, D, and E, relative levels of the mRNAs encoding IL-6, IL-10, IL-13, Ang-2, and VEGF in HUVECs infected with mock (M) or KSHV (K) at different hours post-infection (hpi), measured by qRT-PCR. Differences in mRNA levels between M and K with a P value < 0.05 are marked with a star. F, Western blot detection of IL-6, IL-10, IL-13, Ang-2, and VEGF proteins in the supernatants of mock (M) or KSHV (K)-infected HUVECs. The protein level of β-tubulin in cells corresponding to each supernatant was measured to demonstrate that equal numbers of cells were used for the comparison between mock (M) and KSHV (K) at time point (hpi).
Incubation of monocytes with supernatant from KSHV-infected HUVECs generates macrophages expressing TAMs-specific markers
To examine if KSHV-induced IL-6, IL-10, and IL-13 drive differentiation and polarization of monocytes into TAMs, we purified CD-14+ monocytes (> 90% purity) from blood of healthy donors, seeded the cells in culture plates, and cultured the cells in RPMI 1640 medium plus 5% fetal bovine serum (FBS) mixed with equal volume of supernatant from mock or KSHV-infected HUVECs for 2 d. Un-treated monocytes were used as a reference. Upon collecting the cells, we analyzed expression of TAMs-specific markers at both mRNA and protein levels by qRT-PCR, fluorescence antibody staining and flow cytometry analysis, and Western blot detection.
As shown in Fig. 3A, the mRNA levels of TAMs-specific markers LGMN, CD-163, CD-206, and CD-11b are significantly higher in cells resulting from incubation with supernatant of KSHV-infected HUVECs (ϕ-KSHV) than in cells resulting from incubation with supernatant of mock-infected HUVECs (ϕ-mock). Data from Western blot detection demonstrate higher protein levels of LGMN and CD-163 in ϕ-KSHV than in ϕ-mock (Fig. 3B). To further confirm the Western blot detection results, we stained the un-treated monocytes, ϕ-mock, and ϕ-KSHV with fluorescently labeled antibodies to CD-16, CD-163, and LGMN, and isotype control IgGs, followed by flow cytometry analysis of the expression levels of CD-163 and LGMN in these cells (Fig. S1). Clearly, ϕ-KSHV express significantly higher levels of LGMN and CD-163 than ϕ-mock (Fig. 3C and andD).D). Together, these results demonstrate that supernatant from KSHV-infected HUVECs indeed promotes differentiation and polarization of monocytes into TAMs.
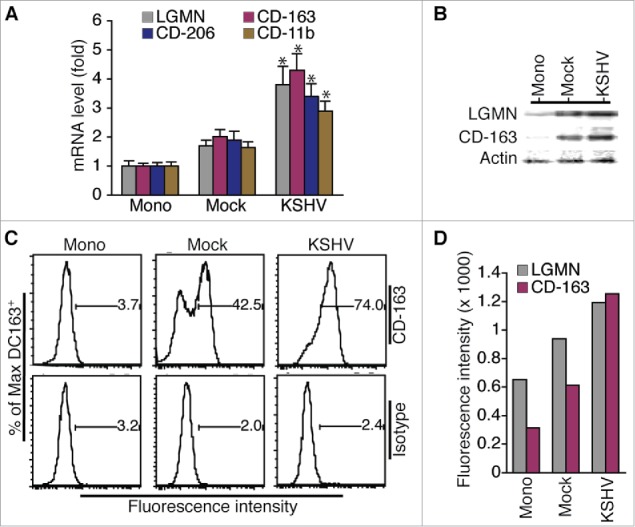
Incubation of monocytes with supernatant from KSHV-infected HUVECs results in cells expressing TAMs-specific markers. A, relative levels of mRNAs encoding LGMN, CD-163, CD-206, and CD-11b in un-treated monocytes (Mono) and macrophages (ϕ) resulting from incubation with supernatant from mock or KSHV-infected HUVECs. Differences in mRNA levels between ϕ-mock (Mock) and ϕ-KSHV (KSHV) with a P value < 0.05 are marked with a star. B, Western blot detection of LGMN, CD-163, and α-actin in cells described in A. C, representative histograms of cells described in (A) that were stained with fluorescently labeled anti-CD-163 antibody or control isotype IgG and subsequently analyzed by flow cytometry as described in Materials and Methods. D, mean values from 2 independent experiments of the fluorescence intensities in cells described in (A)that were stained with fluorescently labeled anti-LGMN or CD-163 and analyzed by flow cytometry as shown in C.
To determine if KSHV-induced IL-6, IL-10, and IL-13 contribute to the conversion of monocytes into TAMs, we incubated purified monocytes with supernatant from KSHV-infected HUVECs in the presence of neutralizing antibodies specific for each of these cytokines. As shown in Fig. 4A and andB,B, each neutralizing antibody reduces the protein expression levels of LGMN and CD-163. In addition, a remarkable additive effect is seen when all 3 antibodies are present. These results strongly suggest that KSHV-induced IL-6, IL-10, and IL-13 from HUVECs are responsible for inducing differentiation and polarization of monocytes into TAMs.
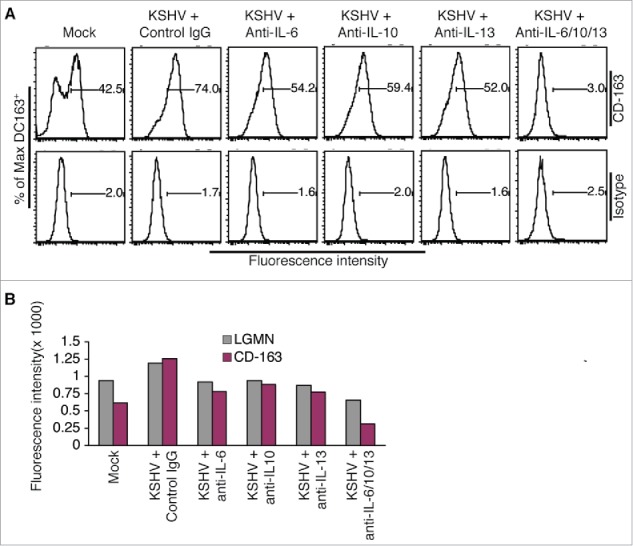
KSHV-induced IL-6, IL-10, and IL-13 are responsible for inducing differentiation and polarization of monocytes into TAMs. A, representative flow cytometry histograms of ϕ-mock (Mock) and ϕ-KSHV (KSHV) generated by incubation of monocytes with supernatant from mock or KSHV-infected HUVECs in the presence of control IgG or neutralizing antibodies specific for human IL-6, IL-10, and IL-13, alone or in combination. The cells were stained with fluorescently labeled anti-CD-163 antibody or control isotype IgG and subsequently analyzed by flow cytometry. Similar experiment was also done on these cells with fluorescently labeled anti-LGMN antibody or control IgG. B, mean values from 2 independent experiments of the fluorescence intensities in cells that were stained with fluorescently labeled anti-LGMN or CD-163 and analyzed as described in A.
Notably, compared with un-treated monocytes, incubation of monocytes with supernatant from mock-infected HUVECs also results in modest increases in the expression levels of LGMN and CD-163 (Fig. 3), which might result from stimulation by low levels of IL-6 and IL-13 in the supernatant of mock-infected HUVECs (Fig. 2B).
KSHV-induced TAMs display characteristic features of viral infection
To further characterize KSHV-induced TAMs, we isolated monocytes from 3 donors and stimulated them with supernatant from mock or KSHV-infected HUVECs as described earlier. We then purified total RNAs from un-treated monocytes and the resulting ϕ-mock and ϕ-KSHV, and conducted RNA-seq analysis. The average levels of each individual transcript in un-treated monocytes, ϕ-mock, and ϕ-KSHV from 3 donors were used for the comparison. As shown in Fig. 5A, monocytes, ϕ-mock, and ϕ-KSHV display very distinctive transcription profiles. In total, 524 transcripts are differentially expressed in ϕ-KSHV when compared with ϕ-mock, among which 325 are upregulated (≥ 2 folds with a P value ≤ 0.05) and 199 are downregulated (≤ 2 folds with a P value ≤ 0.05) (Table S1). The 10 most upregulated and 10 most downregulated transcripts are shown in Fig. 5B. We next validated the RNA-seq data by conducting qRT-PCR with the same RNA samples used for RNA-seq. The mRNA levels of interferon induced transmembrane protein 1 (IFIT1), interferon-α inducible protein 27 (IFI27), and radical S-adenosyl methionine domain containing 2 (RSAD2), which are among the most upregulated transcripts, are indeed significantly higher in ϕ-KSHV than in ϕ-mock (Fig. 6A). In contrast, the mRNA levels of G antigen 1 (GAGE1), G antigen 12J (GAGE12J), and G antigen 8 (GAGE8), which are among the most downregulated transcripts, are substantially lower in ϕ-KSHV than in ϕ-mock (Fig. 6B).
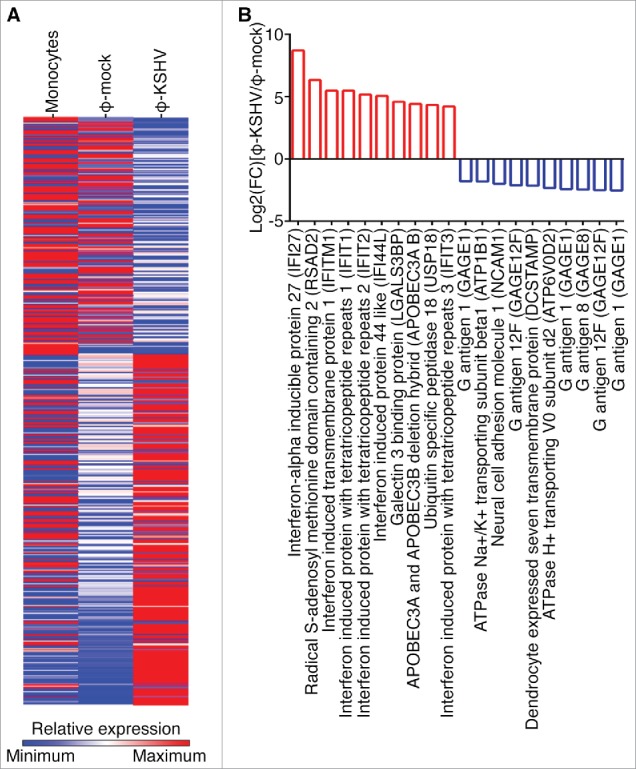
Transcription profile of KSHV-induced TAMs. A, transcription profiles of un-treated monocytes and macrophages resulting from incubation of monocytes with supernatant from mock (ϕ-mock) or KSHV-infected (ϕ-KSHV) HUVECs, generated by RNA-seq analysis, using monocytes from 3 healthy donors. B, lists of 10 most upregulated and 10 most downregulated transcripts in ϕ-KSHV, when compared with ϕ-mock.
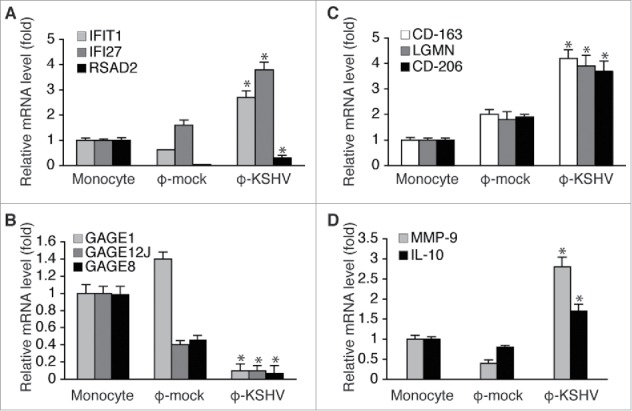
Validation of RNA-seq data by qRT-PCR. A, mRNA levels of IFIT1, IFI27, and RSAD2, which are 3 of the 10 most upregulated transcripts in ϕ-KSHV, in the same RNA samples from un-treated monocytes, ϕ-mock, and ϕ-KSHV that were used for RNA-seq analysis. B, mRNA levels of GAGE1, GAGE12J, and GAGE8, 3 of the 10 most downregulated transcripts ϕ-KSHV, in the RNA samples described in A. C, mRNA levels of TAMs-specific markers LGMN, CD-163, and CD-206 in the RNA samples described in A. D, mRNA levels of MMP-9 and IL-10 in the RNA samples described in A. Differences between ϕ-mock and ϕ-KSHV with a P value < 0.05 are marked with a star.
Consistent with qRT-PCR data shown in Fig. 3A, CD-163 and other M2 macrophage/TAMs markers such as CD-209, CCL-18, and CCR-5 are among the upregulated transcripts. However, based on the RNA-seq data, the mRNA levels of LGMN, CD-206, and CD-11b only display modest increases (~ 1.2 to 1.5 folds) in ϕ-KSHV. This discrepancy prompted us to verify the mRNA levels of these transcripts in the same RNAs samples used for RNA-seq by qRT-PCR. As shown in Fig. 6C, by qRT-PCR analysis, the mRNA levels of LGMN, CD-206, and CD-163 are over 2.0 folds higher in ϕ-KSHV than in ϕ-mock. Since qRT-PCR is a more accurate quantification method than RNA-seq, we conclude that these TAMs-specific markers are upregulated in ϕ-KSHV. In addition, the mRNA levels of MMP-9 and IL-10, which are highly expressed by TAMs,24-26 are also higher in ϕ-KSHV (Fig. 6D). Collectively, data from RNA-seq and qRT-PCR consistently demonstrate that ϕ-KSHV display a gene expression profile characteristic of M2 macrophage/TAMs.
The transcriptomic changes in ϕ-KSHV appear to involve diverse biologic signaling pathways (Fig. S2). To further annotate the set of genes differentially expressed in ϕ-KSHV, we analyzed both up and downregulated genes using ingenuity pathway analyses. As shown in Fig. S3, the most significant canonical pathway upregulated in ϕ-KSHV are interferon signaling pathways. Consistent with this result, the most significant pathway suppressed in ϕ-KSHV is eukaryotic translation initiation actor 2 (eIF2) required for initiation of protein synthesis, suppression of which is a hallmark of interferon signaling.42 This gene expression feature of ϕ-KSHV is likely a hallmark of KSHV-induced TAMs, which likely results from stimulation by high levels of interferon in the supernatant of KSHV-infected HUVECs. Indeed, data from qRT-PCR and ELISA (Fig. S4) indicate that KSHV infection substantially increases expression of all members of type-1 interferon in HUVECs. Since viral particles in the supernatant of KSHV-infected HUVECs had been pre-cleared by high-speed centrifugation and subsequent filtration through 0.22 μm filters before incubation with monocytes, and no KSHV transcript in ϕ-KSH, we rule out the possibility that interferon is induced in monocytes as a result of KSHV infection. Thus, the supernatant of KSHV-infected HUVECs is likely the main source responsible for activation of the interferon signaling pathways in ϕ-KSHV. Intriguingly, despite the presence of interferon in the supernatant of KSHV-infected HUVECs, data from RNA-seq analysis show no obvious differences in the mRNA levels of M1 macrophage specific markers CD-80 and CD-86, suggesting that KSHV infection has little effect on M1 macrophage production.
KSHV-induced TAMs enhance tumor growth in nude mice
We next tested if KSHV-induced TAMs promote tumor growth by using telomerase-immortalized human umbilical vein endothelial cells with latent KSHV infection (TIVE-KSHV), which grow “KS-like” tumors in nude mice.43 Briefly, we mixed and subcutaneously injected equal numbers (1 × 107/injection site) of TIVE-KSHV cells with RPMI-1640 medium (Control) or ϕ-KSHV (2 × 105/per injection site; TIVE-KSHV to ϕ-KSHV ratio is 50 to 1) into nude mice, followed by measurement of tumor volume on a weekly basis. To make certain that the added human macrophages alone do not grow tumors, 2 mice were injected with the same numbers of ϕ-KSHV only.
As shown in Fig. 7A and andB,B, tumors grow significantly faster and larger in mice injected with TIVE-KSHV plus ϕ-KSHV than in mice injected with TIVE-KSHV plus medium. No tumor occurred in mice injected with ϕ-KSHV only. These results strongly suggest that the co-injected ϕ-KSHV enhance tumor growth and thus display the “pro-tumor” feature of TAMs.
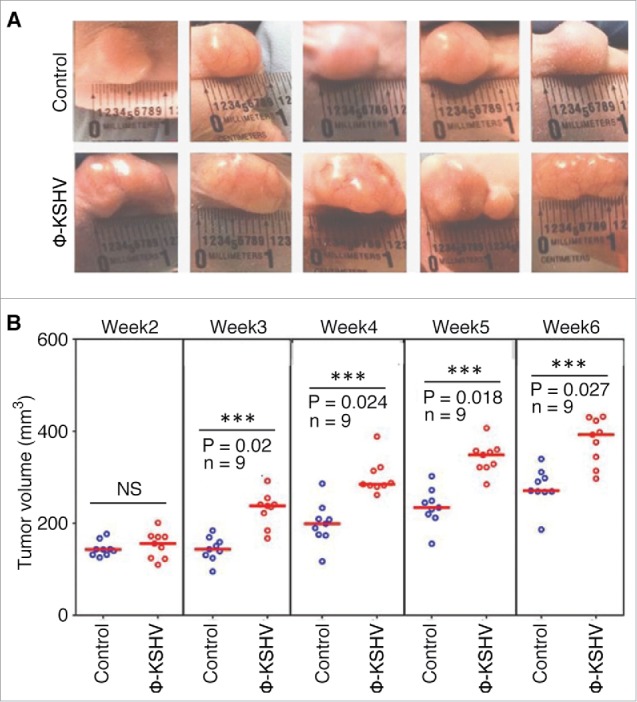
KSHV-induced TAMs enhance tumor growth in nude mice. A, representative images of tumors resulting from subcutaneous inoculation of equal numbers of (1 × 107/injection site) of TIVE-KSHV cells mixed with medium (control) or KSHV-induced TAMs (ϕ-KSHV) with a TIVE-KSHV to ϕ-KSHV ratio of 50 to 1 into nude mice at 6 weeks post-inoculation. B, average tumor volumes of the 2 groups of tumors described in (A) at different weeks post-inoculation. Statistically significant differences (P value < 0.05) are marked with 3 stars. NS, not significant; n, number of tumors per group.
To examine if KSHV-induced TAMs promote tumor angiogenesis as described in other cancer models,24-26 we conducted immuno-chemical staining on tumor sections with an antibody specific for mouse CD-31, which is a marker for blood vessel endothelial cells. As shown in Fig. 8A, ,B,B, and andC,C, tumors containing ϕ-KSHV, which express the human monocyte/macrophage marker CD-68, display increased numbers of blood vessels. These results suggest that KSHV-induced TAMs indeed promote tumor angiogenesis.
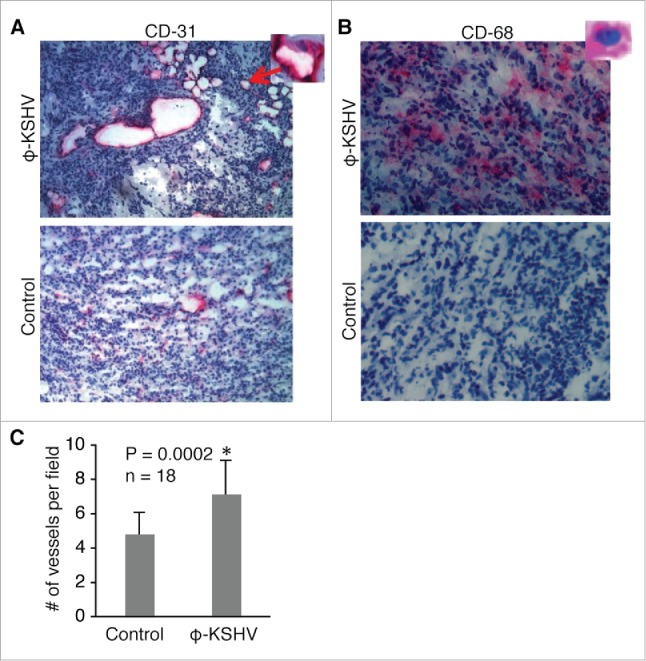
KSHV-induced TAMs enhance tumor angiogenesis. (A) and (B), immunochemical staining of blood vessels with an antibody specific for mouse endothelial cell marker CD-31 and ϕ-KSHV with a mouse anti-human CD-68 antibody in tumors resulting from subcutaneous inoculation of TIVE-KSHV cells mixed with medium (control) or KSHV-induced TAMs (ϕ-KSHV), which were collected 43 d upon inoculation. DAPI was used for nuclear staining of all cells. Enlarged representative blood vessels and ϕ-KSHV are indicated with an arrow. C, average numbers of vessels per field (10x magnification) from 3 fields per tumor and 6 tumors per group (n = 18). Significant differences (P value < 0.05) are marked with a star.
Discussion
KS is a highly angiogenic and inflammatory neoplasia.44 Hallmarks of KS include “spindle-shaped” KS tumor cells with mostly latent KSHV infection, extensive “slit-like” blood vessel networks, and large numbers of infiltrating inflammatory cells.1,2 The roles of KSHV infection in proliferation and malignancy transformation of the infected cells and tumor angiogenesis have been well established.45,46 However, little is known on how KSHV infection contributes to tumor inflammation and whether the infiltrating inflammatory cells play a role in KS tumor development and progression.
We previously reported that KSHV infection of endothelial cells induces the angiogenic and inflammatory cytokine Ang-2 to promote migration and recruitment of monocytes into tumors.12 In the present study, we found that KSHV infection of endothelial cells strongly induces expression and release of IL-6, IL-10, and IL-13 that individually and collectively promote differentiation and polarization of monocytes into TAMs. The KSHV-induced TAMs express well-known TAM-specific markers such as CD-163 and LGMN, promote angiogenesis, and significantly enhance tumor growth in nude mice. Therefore, our data suggest a pivotal role of KSHV acute infection of endothelial cells in not only the recruitment of monocytes but also their conversion into TAMs.
Mechanistically, KSHV-induced IL-6, IL-10, and IL-13 from endothelial cells likely play a major role in the process of monocyte differentiation as neutralizing antibodies to these cytokines effectively reduce the numbers of cells expressing TAMs-specific markers CD-163 and LGMN. Nevertheless, the neutralizing antibodies did not completely block formation of TAMs. One possibility for the incomplete blocking is that the efficiencies of the cytokine neutralization antibodies at the used dose are not 100%. A more likely reason is the presence of additional factors that contribute to the conversion of monocytes into TAMs as well. The most likely such factor is vIL-6, a KSHV-encoded homolog of IL-6 that is strongly expressed in KSHV-infected HUVECs (data not shown). Given its high homology with IL-6,47 the role of vIL-6 in KSHV induction of monocyte differentiation and polarization into TAMs warrants further investigation.
In the present study, we mainly examined how KSHV acute infection of HUVECs induces IL-6, IL-10, and IL-13. The expression levels of these cytokines in TIVE-KSHV cells appear to be much lower than in KSHV acutely infected HUVECs (data not shown), suggesting that the effect of KSHV latently infected KS tumor cells on generation of TAMs exists but is less pronounced. However, expression of these TAMs-promoting cytokines in KS tumor cells may increase when latent KSHV reactivates to undergo lytic replication. Therefore, the impacts of latent KSHV and its reactivation on monocytes for generation of TAMs need to be further studied as well.
The roles of TAMs in tumor invasion and metastasis, angiogenesis, suppression of anti-tumor innate immunity, and development of resistance to chemotherapy have been well established in various cancer models. Although TAMs generally belong to a sub-group of M2 macrophages and express certain TAMs-specific markers, the molecular features of TAMs may differ in different types of cancers due to differences in the local tumor microenvironment. Data from both qRT-PCR and RNA-seq analyses demonstrate that ϕ-KSHV display a gene transcription profile characteristic of M2 macrophages/TAMs, with significantly higher expression of TAMs-specific markers such as CD-163, CD-206, and LGMN. However, KSHV infection of endothelial cells also induces expression of interferon in addition to IL-6, IL-10, IL-13, and vIL-6. This unique microenvironment confers ϕ-KSHV a feature of viral infection with increased expression of interferon-stimulated genes.
In summary, our study demonstrates that KSHV acute infection of endothelial cells creates a local microenvironment that not only recruits monocytes but also induces their differentiation and polarization into “pro-tumor” TAMs. These findings definitely uncover a new mechanism of KS tumorigenesis as a result of KSHV infection.
Materials and methods
KSHV virion production and purification
iSLK-BAC16 cells, which carry the recombinant KSHV, BAC16, and a tetracycline-inducible cassette of KSHV lytic switch protein RTA,38 were cultured in Dulbecco's Modified Eagle Medium (DMEM) medium plus 10% fetal bovine serum (FBS). To induce virion production, iSLK-BAC16 cells were stimulated with sodium butyrate (NaB) and doxycycline as described previously.38 Three days after stimulation, the culture supernatants were collected and centrifuged at 3,000 g for 15 minutes at 4°C to eliminate cellular debris, then loaded onto a 30% sucrose cushion and subjected to high-speed (28,000 g, 4 hours) centrifugation. The virions were re-constituted in basic endothelial cell culture medium EBM2 without growth factor supplements.
Infection of HUVECs and preparation of supernatants
HUVECs were grown in 6-well plates in EBM2 medium with growth factor supplements. The cells were infected with mock (EBM2 medium without growth factor supplements, 0.5 ml per well) or purified KSHV (0.5 ml per well) for various time points. Under these conditions, KSHV infection rate was close to 100% at 72 hpi. The supernatants were collected, centrifuged at 3,000 g for 10 minutes, and then filtered through a 0.22 μm filter. Cytokines in the supernatants were detected by Western blot analysis, using antibodies to human IL-4 (Cell Signaling Technology, Danvers, MA, USA), IL-6 (Cell Signaling Technology), IL-10 (Cell Signaling Technology), IL-13 (R&D Systems, Minneapolis, MN, USA), M-CFS (R&D Systems), Ang-2 (Santa Cruz Biotechnology, Dallas, TX, USA), and VEGF (Santa Cruz Biotechnology), respectively. To assess cytokine activity, neutralizing antibodies to IL-6 (Mabtech, Inc., Mariemont, OH, USA), IL-10 (R&D Systems), and IL-13 (BD Biosciences, San Rose, CA, USA), or control IgG, were added to the supernatants, alone or in combination, to the final concentration of 2.5 μg/ml/antibody.
Isolation, culture, and treatment of human monocytes
Blood from healthy donors were collected by strictly following an IRB protocol (06–14–23) approved by the Institutional Review Board (IRB) of Case Western Reserve University and University Hospital. Monocytes were isolated from the blood by using the EasySep™ human monocyte isolation kit from STEM CELL Technologies Canada Inc. (Vancouver, BC, Canada) and following instructions by the manufacturer. Purity of the monocytes were verified by fluorescent antibody staining with anti-human CD-14 PerCP-Cyanine5.5 and anti-human CD-68-APC antibodies (BD Biosciences), followed by flow cytmetry analysis. The purified monocytes were first seeded in culture flasks in macrophage attachment medium (PromoCell, Heidelberg, Germany) for one hour. After removing the attachment medium, the cells were cultured in a mixture containing one volume of RPMI 1640 medium plus 5% FBS and one volume of supernatant from mock or KSHV-infected HUVECs for 48 h. The resulting macrophages (ϕ-mock and ϕ-KSHV) were collected by using macrophage detachment medium (PromoCell), washed with ice-cold PBS, and used for RNA purification or subjected to fluorescent antibody staining with antibodies to TAMs-specific markers and subsequent flow cytometry analysis.
Fluorescent antibody staining and flow cytometry analysis of TAMs
Un-treated monocytes, ϕ-mock, and ϕ-KSHV were collected and washed with PBS containing 1% BSA and 1% EDTA, followed by live surface staining with a rabbit (IgG) anti-human LGMN (Bioss Antibodies, Inc., Woburn, Massachusetts, USA), and a mouse (IgG1) anti-human CD-16 labeled with PE (BD Biosciences) for one hour. Upon washing with PBS plus 1% BSA and 1% EDTA, the cells were further incubated with a goat anti-rabbit secondary antibody that is labeled with APC (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 40 minutes on ice. As background controls, a small portion of each sample was stained the same way with mouse IgG1 labeled with PE, rabbit IgG, and secondary anti-rabbit IgG labeled with APC. Similar dual staining was also done with a APC-labeled mouse anti-human CD-163 (Jackson ImmunoResearch Laboratories) and PE-labeled mouse anti-human CD-16, and the corresponding isotype control IgGs. The cells were then washed with PBS and fixed with a solution containing one volume of PBS and one volume of Intracellular Fixation buffer (ThermoFisher Scientific) for 45 minutes. Upon washing with PBS plus 1% BSA and 1% EDTA, the cells were analyzed by flow cytometry using BD Fluorescence Activated Cell Sorting (FACS) Calibur or BD Fortessa Cytometers. The data were analyzed with FlowJo 9.7.6 software.
RNA isolation and qRT-PCR
Total RNAs were isolated using the RNeasy® Plus Mini Kit from Qiagen (Valentia, California, USA), which begins with removal of contaminating genomic DNA. The mRNAs were converted into cDNAs by using a reverse transcription kit from Qiagen. Standard procedure for qRT-PCR was followed to measure mRNA levels of TAMs-specific genes as well as various cytokines, using primers described in Table S2. Primers used for qRT-PCR measurement of different members of interferon-α (IFN-A) were as described previously.48 All qRT-PCR reactions were performed in triplicate.
RNA-seq and data analysis
Monocytes from 3 donors (biologic replicates) were isolated and stimulated with supernatant from mock or KSHV-infected HUVECs as described above. Total RNAs from un-treated monocytes, the resulting Φ-mock, and Φ-KSHV from 3 donors were purified using the RNeasy® Plus Mini Kit from Qiagen and used for subsequent cDNA library preparation and RNA-seq analysis using the Illumina HiSeq 2500 instrument. RNA-Seq service was provided by Cofactor Genomics (St. Louis, Missouri, USA). Functional annotation of the gene sets were performed using GENE-E (Broad Institute, MA), Ingenuity Pathway Analysis (IPA) (Qiagen) and N2C (Network2Canvas).49
Tumor growth in nude mice
Equal numbers (1 × 107/injection site) of TIVE-KSHV cells were mixed with RPMI-1640 medium (Control) or ϕ-KSHV (2 × 105/per injection site), and then subcutaneously injected into 4–6 weeks old female nude mice. To make certain that the added human macrophages alone do not grow tumors, 2 mice were injected with the same numbers of ϕ-KSHV. Tumor volume (width × length × height) was determined on a weekly basis by using a caliper. All procedures were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and following a protocol (2013–0067) that was approved by the Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University.
Acknowledgments
We thank the Immunology Core Facility of CFAR at CWRU for patient recruitment, blood collecting, purification of monocyte, and flow cytometry analysis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by Case Western Reserve University (CWRU) / University Hospital (UH) Center for AIDS Research (CFAR): NIH Grant P30 AI036219.
References
Articles from Cell Cycle are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1080/15384101.2017.1356509
Read article for free, from open access legal sources, via Unpaywall:
https://www.tandfonline.com/doi/pdf/10.1080/15384101.2017.1356509?needAccess=true
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1080/15384101.2017.1356509
Article citations
Monocyte and Macrophage Functions in Oncogenic Viral Infections.
Viruses, 16(10):1612, 15 Oct 2024
Cited by: 0 articles | PMID: 39459945 | PMCID: PMC11512331
Review Free full text in Europe PMC
Analysis of tumor infiltrating immune cells in Kaposi sarcoma lesions discovers shifts in macrophage populations.
Glob Health Med, 6(5):310-315, 01 Oct 2024
Cited by: 0 articles | PMID: 39483448 | PMCID: PMC11514630
Virally encoded interleukin-6 facilitates KSHV replication in monocytes and induction of dysfunctional macrophages.
PLoS Pathog, 19(10):e1011703, 26 Oct 2023
Cited by: 4 articles | PMID: 37883374 | PMCID: PMC10602306
Macrophages drive KSHV B cell latency.
Cell Rep, 42(7):112767, 12 Jul 2023
Cited by: 5 articles | PMID: 37440412 | PMCID: PMC10528218
Kaposi Sarcoma, a Trifecta of Pathogenic Mechanisms.
Diagnostics (Basel), 12(5):1242, 16 May 2022
Cited by: 4 articles | PMID: 35626397 | PMCID: PMC9140574
Review Free full text in Europe PMC
Go to all (11) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Suppression of KSHV-induced angiopoietin-2 inhibits angiogenesis, infiltration of inflammatory cells, and tumor growth.
Cell Cycle, 15(15):2053-2065, 13 Jun 2016
Cited by: 15 articles | PMID: 27294705 | PMCID: PMC4968962
KSHV induces aerobic glycolysis and angiogenesis through HIF-1-dependent upregulation of pyruvate kinase 2 in Kaposi's sarcoma.
Angiogenesis, 18(4):477-488, 20 Jun 2015
Cited by: 61 articles | PMID: 26092770 | PMCID: PMC4659376
Role of heme oxygenase-1 in the pathogenesis and tumorigenicity of Kaposi's sarcoma-associated herpesvirus.
Oncotarget, 7(9):10459-10471, 01 Mar 2016
Cited by: 9 articles | PMID: 26859574 | PMCID: PMC4891132
Pathological Features of Kaposi's Sarcoma-Associated Herpesvirus Infection.
Adv Exp Med Biol, 1045:357-376, 01 Jan 2018
Cited by: 13 articles | PMID: 29896675
Review




