Abstract
Free full text

Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS
Abstract
Approximately 200 BRAF mutant alleles have been identified in human tumours. Activating BRAF mutants cause feedback inhibition of GTP-bound RAS, are RAS-independent and signal either as active monomers (class 1) or constitutively active dimers (class 2)1. Here we characterize a third class of BRAF mutants—those that have impaired kinase activity or are kinase-dead. These mutants are sensitive to ERK-mediated feedback and their activation of signalling is RAS-dependent. The mutants bind more tightly than wild-type BRAF to RAS–GTP, and their binding to and activation of wild-type CRAF is enhanced, leading to increased ERK signalling. The model suggests that dysregulation of signalling by these mutants in tumours requires coexistent mechanisms for maintaining RAS activation despite ERK-dependent feedback. Consistent with this hypothesis, melanomas with these class 3 BRAF mutations also harbour RAS mutations or NF1 deletions. By contrast, in lung and colorectal cancers with class 3 BRAF mutants, RAS is typically activated by receptor tyrosine kinase signalling. These tumours are sensitive to the inhibition of RAS activation by inhibitors of receptor tyrosine kinases. We have thus defined three distinct functional classes of BRAF mutants in human tumours. The mutants activate ERK signalling by different mechanisms that dictate their sensitivity to therapeutic inhibitors of the pathway.
Some BRAF mutants, first described by Marais and colleagues2 are kinase-dead (D594G/N) or have lower activity (G466V/E) than wild-type BRAF (Extended Data Fig. 1a). In contrast to tumours harbouring activating BRAF mutants, RAS is active in cells expressing these mutants (Extended Data Fig. 1b). Expression of these mutants increases the levels of phosphorylated MEK (p-MEK) and cyclin D1, but to a much lesser extent than do activating BRAF mutants (V600E, K601E or G469A) (Fig. 1a). Moreover, whereas activating mutants decrease RAS–GTP and CRAF phosphorylation, low-activity or kinase-dead mutants do not (Fig. 1a). Thus, ERK activation by these mutants is less pronounced than that by activating mutants and induces insufficient feedback to inhibit RAS.
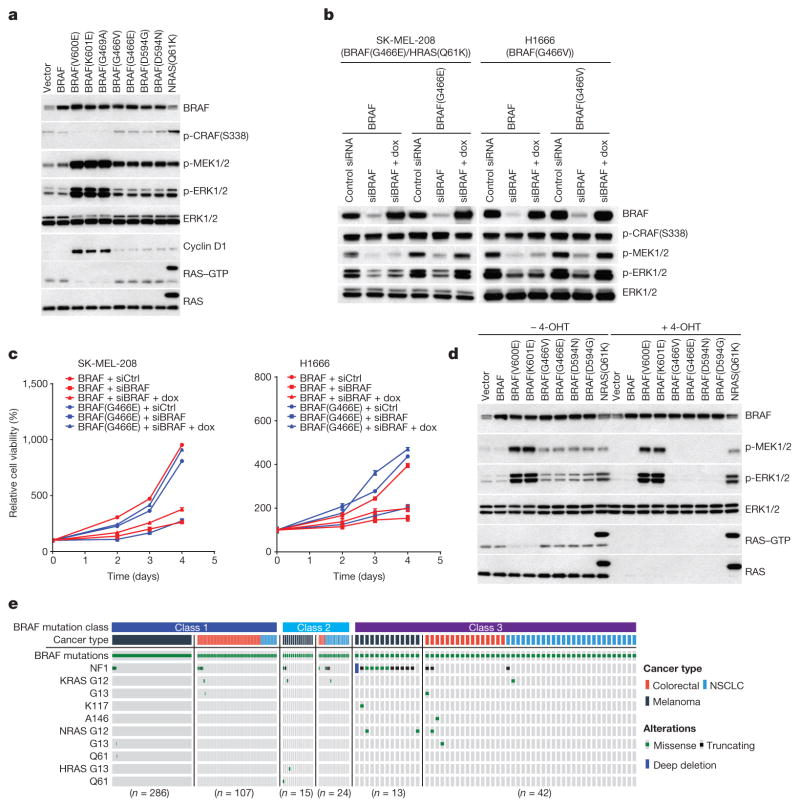
a, ERK signalling was assessed in NIH3T3 cells expressing the indicated BRAF proteins (30 ng ml−1 doxycycline, 24 h). b, c, Inducible wild-type BRAF or mutant BRAF (G466E or G466V) was introduced into H1666 or SK-MEL-208 cells. The indicated cells were transfected with control siRNA or siRNA against the human BRAF gene. b, After 1 day, 106 cells of each cell line were treated with doxycycline (dox; 30 ng ml−1, for 24 h) and ERK was assessed. c, 3,000 cells of each siRNA transfected cell line were then plated in 96-well plates in medium with doxycycline. Cell growth was determined by ATP-Glo assay. Growth curves were generated with Prism 6 (mean ± s.d., n = 8). d, Expression of indicated BRAF proteins was induced (10 ng ml−1 doxycycline, 24 h) in the conditional RAS-less cells that were pre-treated with 4-hydroxytamoxifen (4-OHT) to knock out the last RAS allele. In a, b and d, Erk signalling was examined by western blot and RAS–GTP levels were determined by the active RAS pull-down assay. The gel source data are provided in Supplementary Fig. 1. e, Oncoprint showing co-mutation of class 3 BRAF mutants with RAS/NF1 in samples from cancer patients. The data were collected from http://cbioportal.org.
SK-MEL-208 is a melanoma cell line with mutant HRAS(Q61K) and the low-activity BRAF mutant G466E. H1666 is a non-small-cell lung cancer (NSCLC) cell line with the low-activity BRAF mutant G466V. Knocking down BRAF expression inhibited ERK activation and the proliferation of both cell lines (Fig. 1b, c). As both wild-type and mutant BRAF were knocked down, we performed a rescue experiment. Introduction of the low-activity mutants into SK-MEL-208 and H1666 in which BRAF was knocked down restored ERK signalling and cell proliferation whereas introduction of wild-type BRAF did not (Fig. 1b, c). Thus, low-activity BRAF mutants amplify ERK signalling and drive the proliferation of tumour cells.
The failure of hypoactive BRAF mutants to reduce RAS–GTP suggested that they may signal in a RAS-dependent manner. We confirmed this in ‘RAS-less’ cells3 in which MEK/ERK signalling was rescued by BRAF(V600E), BRAF(K601E) or NRAS(Q61K) but not by wild-type, G466V/E or D594N/G BRAF (Fig. 1d). We have characterized 31 different mutant BRAF alleles found in human tumours, 16 of which are kinase-impaired or kinase-dead (13 are shown in Fig. 1d, Extended Data Fig. 1c, d, class 3 in Table 1). All were shown to be RAS-dependent (unlike activating BRAF mutants).
Table 1
Classification of cancer-associated BRAF mutants
| BRAF mutation | Kinase activity | RAS dependency | Dimer dependency | Sensitivity to vemurafenib | |
|---|---|---|---|---|---|
| Wild type | Neutral | Yes | Yes | Insensitive | |
| Class 1 | V600E | High | No | No | Sensitive |
| V600K | High | No | No | Sensitive | |
| V600D | High | No | No | Sensitive | |
| V600R | High | No | No | Sensitive | |
| V600M | Intermediate | No | No | Sensitive | |
| Class 2 | K601E | High | No | Yes | Insensitive |
| K601N | High | No | Yes | Insensitive | |
| K601T | High | No | Yes | Insensitive | |
| L597Q | High | No | Yes | Insensitive | |
| L597V | Intermediate | No | Yes | Insensitive | |
| G469A | High | No | Yes | Insensitive | |
| G469V | High | No | Yes | Insensitive | |
| G469R | Intermediate | No | Yes | Insensitive | |
| G464V | Intermediate | No | Yes | Insensitive | |
| G464E | Intermediate | No | Yes | Insensitive | |
| Fusions* | High | No | Yes | Insensitive | |
| Class 3 | D287H | Low | Yes | Yes | Insensitive |
| V459L | Low | Yes | Yes | Insensitive | |
| G466V | Low | Yes | Yes | Insensitive | |
| G466E | Low | Yes | Yes | Insensitive | |
| G466A | Low | Yes | Yes | Insensitive | |
| S467L | Low | Yes | Yes | Insensitive | |
| G469E | Low | Yes | Yes | Insensitive | |
| N581S | Low | Yes | Yes | Insensitive | |
| N581I | Low | Yes | Yes | Insensitive | |
| D594N | None | Yes | Yes | Insensitive | |
| D594G | None | Yes | Yes | Insensitive | |
| D594A | None | Yes | Yes | Insensitive | |
| D594H | None | Yes | Yes | Insensitive | |
| F595L | Low | Yes | Yes | Insensitive | |
| G596D | Low | Yes | Yes | Insensitive | |
| G596R | Low | Yes | Yes | Insensitive | |
In NIH3T3 cells expressing wild-type BRAF, BRAF(G466V/E) or BRAF(D594N/G), serum starvation reduced, whereas EGF administration activated, RAS–GTP levels and ERK signalling (Extended Data Fig. 1e). These manipulations had no effect in cells expressing RAS-independent activating BRAF mutants or mutant NRAS. Thus, ERK signalling driven by low-activity or kinase-dead BRAF mutants is RAS-dependent and regulated by upstream signalling. When ERK activity was suppressed in wild-type BRAF or mutant cells with the ERK inhibitor (SCH772984), RAS–GTP levels rose suggesting relief of ERK-dependent feedback inhibition of RAS. However, levels of phosphorylated MEK rose only in cells expressing RAS-dependent wild-type BRAF or low-activity or kinase-dead BRAF mutants (G466V/E or D594N/G) (Extended Data Fig. 1f). Thus, ERK signalling driven by BRAF(G466V/E) and BRAF(D594N/G) is limited by ERK-dependent feedback inhibition of RAS, and signalling of RAS-independent BRAF mutants is not.
Elevated induction of ERK output by low-activity or kinase-dead BRAF mutants therefore requires adequate RAS activity despite ERK-dependent feedback. Kinase-dead BRAF mutants have been noted to coexist and synergize with mutant RAS4. Analysis of 24,000 sequenced human tumours shows that, in melanoma, low-activity and kinase-dead BRAF always coexist with genetic lesions that dysregulate RAS (NF1 deletion/mutation or RAS mutation). By contrast, activating BRAF mutants rarely coexist with these alterations (Fig. 1e).
The most common class of BRAF mutants in NSCLC are hypo-active: low activity or kinase-dead (Extended Data Fig. 1g and refs 5–8). In NSCLCs and colorectal cancers (CRCs) with these mutants, coexistent NF1 or RAS lesions are uncommon (Fig. 1e and Extended Data Fig. 1h). Consistent with this observation, an accompanying paper by Santamaria and colleagues shows that a kinase-dead BRAF mutant does cause lung cancers in mice in the absence of RAS or NF1 mutations9. In these tumours, we hypothesized that RAS activation by receptor tyrosine kinases (RTKs) was sufficient. Three tumour cell lines with low-activity BRAF mutations were studied: NSCLC H1666 (BRAF(G466V)), NSCLC CAL-12T (BRAF(G466V)) and CRC H508 (BRAF(G596R)). All expressed high levels of phosphorylation of one or more RTKs, including the insulin and IGF1R receptors, MET, ERBB2 and EGFR. The last was detected in all three cell lines. By contrast, low levels of phosphorylated RTKs were detected in SK-MEL-208, a melanoma cell line with coexistent BRAF(G466E) and HRAS(Q61K) (Extended Data Fig. 1i). RAS activation, ERK signalling and the growth of all three cell lines with low-activity BRAF mutations were sensitive to the EGFR antibody cetuximab (Extended Data Fig. 1j, k). By contrast, SK–MEL-208 and tumour cells with activating BRAF mutants (BRAF(V600E), BRAF(G469A) and BRAF(L485–P490>Y))1,10 were insensitive (Extended Data Fig. 1j, k). Expression of mutant but not wild-type NRAS in H1666 reduced its sensitivity to cetuximab (Extended Data Fig. 1l, m), but its sensitivity to the MEK inhibitor trametinib was unaffected (Extended Data Fig. 1m). Similar results were obtained in the CRC cell line H508 harbouring BRAF(G596R) mutant (Extended Data Fig. 1n, o). These data suggest that tumours with low activity or kinase-dead BRAF mutants may be classified by the mechanism whereby RAS is activated in the specific tumour. Apparently, RAS activation in melanocytes is too low to cooperate with these BRAF mutations. In support of this idea, kinase-dead BRAF does not cause melanomas in a GEMM model4.
Previous work11 suggested that low-activity BRAF mutants catalyse MEK phosphorylation in a RAS-independent manner, which we show here is not the case. Other work by the same group4 showed that kinase-dead mutants function differently, in a RAS-dependent manner. They suggested that RAF inhibitors selectively inhibit BRAF and that this causes RAS-dependent activation of CRAF (paradoxical activation). From this idea they concluded that kinase-dead BRAF mutants activated CRAF in an analogous manner. However, we have previously shown that RAF inhibitors activate CRAF in cells that lack BRAF and activate truncated CRAF homodimers12. Thus, selective inhibition of BRAF is not required for paradoxical activation and the mechanism by which kinase-dead BRAF activates CRAF remains unclear. A more detailed account of the evolution of our ideas on how the low-activity mutants work, from their discovery to this paper, is provided in the Supplementary Discussion, part 2.
By contrast, we show that the functional properties of low-activity and kinase-dead BRAF mutants are very similar. Both activate ERK signalling in a RAS-dependent fashion and are sensitive to ERK-dependent feedback inhibition of RAS. In melanoma, they both commonly coexist with mutants that activate RAS. In epithelial tumours, they often rely on RTK-activated RAS. We therefore call low-activity or kinase-dead BRAF mutants class 3 mutants, to distinguish them from activated, RAS-independent class 1 and 2 mutants.
The common properties of these mutants suggest that they activate ERK signalling by the same mechanism. Our data indicate that class 3 mutants serve to amplify the ERK signal in tumours in which they coexist with activated RAS. Expression of NRAS(Q61K) in 293H cells induces p-MEK and p-ERK. Overexpression of wild-type BRAF enhances this induction and class 3 BRAF mutants do so to a much greater degree (Fig. 2a and Extended Data Fig. 2a). We explored the mechanism by which this occurs. Expression of class 3 BRAF mutants enhanced the binding of BRAF but not CRAF to mutant NRAS (Fig. 2a). A concomitant increase in class 3 mutant BRAF–wild-type CRAF but not in wild-type BRAF–CRAF dimers was observed (Extended Data Fig. 2b).
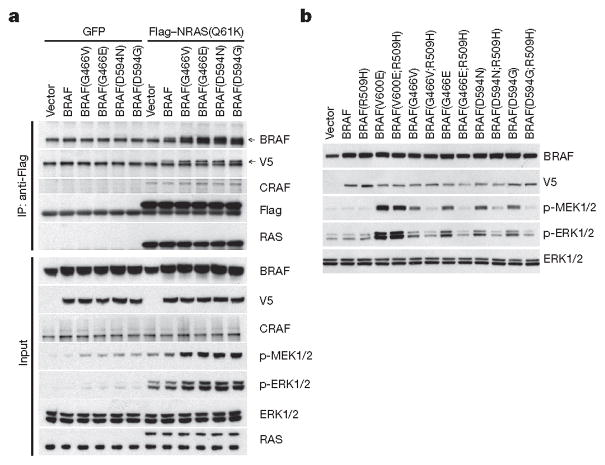
a, 293H cells stably expressing GFP or Flag-tagged NRAS(Q61K) were transfected with empty pcDNA3 vector or pcDNA3 encoding the indicated V5-tagged BRAF proteins. 24 h after transfection, cells were collected and NRAS(Q61K) was immunoprecipitated with anti-Flag antibody. b, 293H cells were transfected with pcDNA3 plasmids encoding the indicated BRAF proteins or the same proteins into which the R509H mutation was inserted. After 24 h, the cells were treated with serum-free medium for 4 h and then collected. For gel source data, see Supplementary Fig. 2.
These data suggested that increased binding of class 3 BRAF mutants to activated RAS is associated with formation of more heterodimers of mutant BRAF and wild-type CRAF. In agreement with this model and the work of others, ERK activation by these mutants is CRAF-dependent4,11. In Raf1-knockout mouse embryonic fibroblasts, class 3 BRAF mutants have almost no effect on p-MEK, nor does co-expression of wild-type BRAF and CRAF. However co-expression of class 3 BRAF mutants with wild-type CRAF in these cells induced ERK signalling (Extended Data Fig. 2c).
Disruption of BRAF dimerization with the R509H mutation almost completely abolishes induction of p-MEK and p-ERK by class 3 BRAF mutants, but not by dimer-independent BRAF(V600E) (Fig. 2b and Extended Data Fig. 2d). Moreover, R509H abolished the kinase activity of wild-type BRAF but enhanced its binding to active RAS. By contrast, the binding of BRAF(D594N) to RAS was not further enhanced by the R509H mutation (Extended Data Fig. 2e). Thus BRAF binding to activated RAS did not require the dimerization of the former and was enhanced by mutations that reduce kinase activity. Moreover, drugs that inhibit BRAF or CRAF activity also enhanced their binding to active RAS (Extended Data Fig. 2f); but did not affect the binding of BRAF or CRAF into which gatekeeper mutations have been introduced (Extended Data Fig. 2g). These results support the idea that reduction of the catalytic activity of RAF proteins by mutation or pharmacologic inhibition enhances their binding to active RAS.
Class 3 BRAF mutants amplify ERK signalling in tumours, and the MEK inhibitor trametinib inhibits the proliferation of tumour cells harbouring these mutants (Fig. 3a and Extended Data Fig. 3a, b). MEK inhibitors may have some utility for treating these tumours, but their toxicity is likely to limit their effectiveness. It is unlikely that current RAF inhibitors will suppress ERK signalling in class 3 mutant-driven tumours, since they preferentially inhibit activated BRAF monomers (V600 mutants)1,13–15. Tumours with class 3 BRAF mutants and activated RAS express both mutant BRAF–wild-type RAF heterodimers and wild-type RAF dimers (Extended Data Fig. 2b). Current RAF inhibitors will optimally inhibit 50% of the activity of the former when bound to the wild-type protomer and paradoxically activate the wild-type dimers (for further details see Supplementary Discussion, part 1). Thus, current RAF inhibitors are not expected to effectively inhibit the ERK pathway in these tumours. As predicted, the RAF inhibitor vemurafenib failed to inhibit ERK signalling in tumour cells and NIH3T3 that express class 3 BRAF mutants (Fig. 3b, c). All such mutants tested thus far are insensitive to vemurafenib (Table 1).
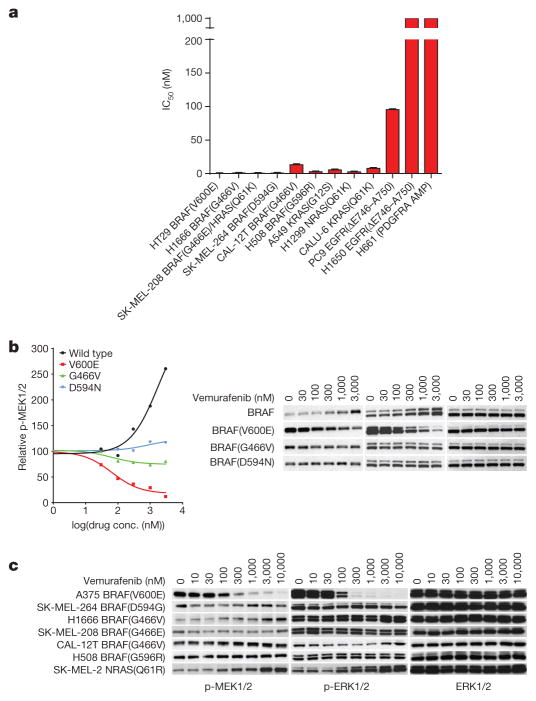
a, IC50 values for growth inhibition by trametinib of the indicated cell lines (mean ± s.d., n =8). IC50 is calculated as shown in Extended Data Fig. 3b. b, NIH3T3 cells expressing the indicated BRAF proteins were exposed to 20 ng ml−1 doxycycline for 24 h. Cells were then treated with vemurafenib (1 h) at various concentrations in the presence of doxycyclin. Cells were collected and cell lysates were examined. The relative p-MEK levels on each blot as shown in the curves were determined by densitometry analysis using ImageJ. The dose–response curves were generated by GraphPad Prism 6. c, Cells were treated with vemurafenib at the indicated concentrations for 1 h. For gel source data, see Supplementary Fig. 3.
Thus, in tumours in which RAS activation is due to coexistent RAS mutation or NF1 loss, the only way to effectively inhibit ERK signalling is with MEK or ERK inhibitors, which have a narrow therapeutic index. By contrast, in most lung, colorectal and other epithelial tumours with class 3 mutants, RAS is activated by other mechanisms, including activation of RTKs. Our model predicts that tumours with class 3 BRAF mutants in which RAS activation is RTK-dependent should be sensitive to inhibitors of that RTK, alone or in combination with MEK/ERK inhibition. In support of this hypothesis, we studied a tumour from a patient with metastatic colorectal cancer with BRAF(G466V) and wild-type RAS and NF1 (Extended Data Table 1). Treatment with the anti-EGFR antibody panitumumab and irinotecan caused tumour regression (Fig. 4a), and a 2.9 × 2.1 cm hepatic lesion was undetectable 5 months after treatment. A patient-derived xenograft (PDX) generated from this lesion shared the same genotype. In PDX-derived cells, ERK signalling was sensitive to cetuximab and resistant to vemurafenib (Extended Data Fig. 4a). The PDXs were sensitive to cetuximab treatment with complete regression of the tumour and were insensitive to vemurafenib, whereas trametinib was able to delay tumour growth (Fig. 4b, c).
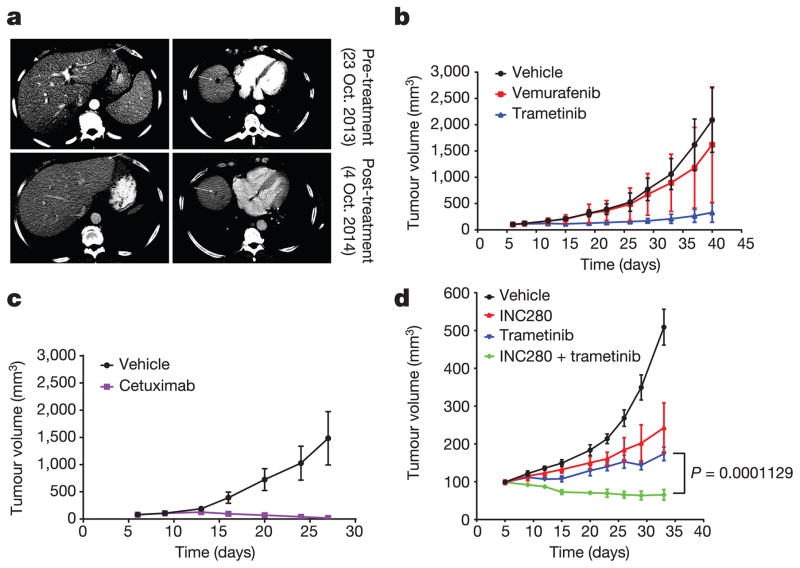
a, A patient with metastatic colorectal cancer (BRAF(G466V)) involving the liver was treated with panitumumab plus irinotecan. Their liver lesions (marked with arrows on CT scan) were compared before or after 4 months of drug treatment. b, c, PDX was established from a tumour biopsy specimen from the patient indicated in a. Drug response was monitored by measurement of the tumour sizes. Vemurafenib was given at 75 mg kg−1 twice per day, trametinib 1.5 mg kg−1 once daily. Cetuximab (50 mg kg−1) was given twice per week. Data in graphs are mean ± s.d., n =10. d, PDX was established from the progressing ovarian metastasis from a CDC patient. INC280 was given at 10 mg kg−1 twice per day, trametinib 1.5 mg kg−1 once daily. Data in graphs are mean ±s.d., n = 5. P value calculated by unpaired t-test.
A second patient with a metastatic collecting duct carcinoma of the kidney (CDC) responded to combination chemotherapy except for growth of one ovarian lesion, which was resected. A PDX was generated and a clonal class 3 BRAF(G466A) mutation was detected in both the primary tumour and the ovarian metastasis, whereas another class 3 mutant, BRAF(D594N), was only detected in the ovarian metastasis. No mutations in RAS family members, RTKs or alterations in NF1 were detected (Extended Data Table 2). Mass spectrometry revealed high levels of MET and EGFR expression in the tumour. Phosphorylation of these receptors was detected in lysates of tumour cells isolated from the PDX (Extended Data Fig. 4b). In PDX-derived cells, RAS activation and ERK signalling were sensitive to MET inhibitors (crizotinib, INC280), but not to cetuximab, suggesting that MET was the dominant driver of RAS activation in this tumour (Extended Data Fig. 4c, d). ERK signalling and cell growth were sensitive to both MET (INC280) and MEK (trametinib) inhibition, and a combination of both proved even more potent (Extended Data Fig. 4e, f). Signalling inhibition of either target was associated with slowing of tumour growth in vivo, but the combination was more effective and caused tumour regression (Fig. 4d).
Thus, based on their mechanisms of activation, oncogenic BRAF mutants may be divided into three categories that determine their sensitivity to inhibitors. Class 1 BRAF mutations (BRAF V600 mutations) are RAS-independent, signal as monomers and are sensitive to current RAF ‘monomer’ inhibitors. Class 2 BRAF mutants are RAS-independent, signal as constitutive dimers and are resistant to vemurafenib. Such mutants may be sensitive to novel RAF dimer inhibitors or MEK inhibitors1,13. In this work, we define a third class of BRAF mutants consisting of those with low or absent kinase activity. They are RAS-dependent and sensitive to ERK-dependent feedback of RAS. They activate ERK by increasing their binding to RAS and require coexistent mechanisms for RAS activation in the tumour in order to function. They are thus not independent drivers; they act as amplifiers of the RAS signal induced by RAS mutation, NF1 loss, or activation of receptors. Class 3 BRAF mutants coexist with mutations in RAS and NF1 in melanomas, and these tumours may be treated with MEK inhibitors. The great majority of class 3 mutants in epithelial tumours, however, are not associated with RAS/NF1 alterations and may be effectively treated with combinations that include inhibitors of the RTKs responsible for driving RAS activation.
METHODS
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Cell culture and reagents
Cell lines were obtained from MSKCC cell collection or American Type Culture Collection (ATCC catalogue numbers: A549, CCL-185; Calu-6, HTB-56; H1666, CRL-5885; HT-29, HTB-38; H508, CCL-253; H1395, CRL-5868; H2405, CRL-5944; H1650, CRL-5883; H661, HTB-183; and H1299, CRL-5803). The conditional Ras-knockout cell line was from M. Barbacid. Raf1-knockout MEFs from M. Baccarini. SK-MEL-2, SK-MEL-264, SK-MEL-208, H1395, H1666, CAL-12T and H508 were cultured in RPMI, 10% FBS. H2405 in DMEM/F12 + 10% FBS, other cell lines in DMEM with glutamine, antibiotics, 10% FBS. Inducible-expression cells were maintained in medium with 50 μgml−1 hygromycin and 0.2 μgml−1 puromycin. NIH3T3 cells were used to construct the stable lines with inducible expression of mutant BRAF or NRAS, for the study of BRAF-mutant-mediated feedback regulation of RAS. Conditional Ras-knockout and Raf1-knockout MEFs were employed to determine the RAS and CRAF dependency of mutant BRAF-driven ERK signalling. Co-expession and co-immunoprecipitation experiments were performed with 293H cells (ThermoFisher Scientific, no. 11631017), in which more than two exogenous proteins were transiently expressed. All cell lines tested negative for mycoplasma. Gene alterations in cell lines from the MSKCC cell collection were confirmed by IMPACT sequencing. Drugs were obtained from: Vemurafenib, Plexxikon; Trametinib, GlaxoSmithKline; SCH772984, Merck; INC280, Novartis. Cetuximab was purchased from the MSK hospital pharmacy. Doxycycline and 4-OHT from Sigma Aldrich; EGF, puromycin and hygromycin stock solution from Invitrogen. Drugs were stored at −20 °C, in 10 mM DMSO stocks.
Antibodies
Western blot, immunoprecipitation and in vitro kinase assays were performed as described1. Antibodies used include: anti-p217/p221-MEK1/2 (p-MEK1/2) (no. 9154), anti-p202/p204-ERK1/2 (p-ERK1/2) (no. 4370), anti-MEK1/2 (no. 4694), anti-ERK1/2 (no. 4696), anti-p-RSK (no. 9346) from Cell Signaling; anti-V5 (R960-25), ThermoFisher Scientific; anti-BRAF (sc-5284), Santa Cruz Biotechnology; anti-Flag (F3165) Sigma; anti-CRAF (610152), BD Transduction Laboratories; anti-p-CRAF S338 (05-538), Millipore; anti-RAS from the active RAS pull-down and detection kit (ThermoFisher Scientific, no. 16117). For immunoprecipitations of tagged proteins we used: anti-V5 agarose affinity gel (Sigma, A7345), anti-Flag M2 affinity gel (Sigma, F1804), protein G agarose gel (ThermoFisher Scientific, no. 15920010).
Plasmids
The pcDNA3-BRAF-V5/Flag, pcDNA3-CRAF-V5/Flag were constructed as previously described1. Plasmids for retroviral-based inducible expression system were provided by S. Lowe. The Braf and Nras genes were sub-cloned into TTIGFP-MLUEX vector harbouring the tet-regulated promoter. Mutations were introduced using the site-directed Mutagenesis Kit (Stratagene). The oligos for mutagenesis of Braf are listed in the Supplementary information.
Transfection
Cells were seeded in 60-mm or 100-mm plates and transfected the following day using Lipofectamine 2000 (ThermoFisher Scientific, no. 11668019) according to the instructions. A DNA (μg) to lipofectamine (μl) ratio of 1:3 was employed.
Gene knockdown
ON-TARGET plus Non-targeting Pool (D-001810-10) and BRAF siRNA SMART pool (L-003460-00) were purchased from Dharmacon. Target cells was transfected with 200 pmol siRNA pool by lipofectamin2000 24 or 48 h before further treatments or assays. Suppression of gene expression was determined by western blot.
Immunoprecipitation and in vitro kinase assay
Cells were collected and stored on ice and lysed in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP40, 1 mM EDTA) supplemented with protease and phosphatase inhibitor cocktail tablets (Roche). Immunoprecipitations were carried out at 4 °C for 2 h, followed by five washes with lysis buffer. Pull-down complexes were eluted with 1 × SDS loading buffer or V5 (V7754, Sigma-Aldrich) or Flag (F3290, Sigma-Aldrich) and assayed by western blotting.
When the in vitro kinase assay was performed after immunoprecipitation, 0.02% SDS was added to the wash buffer and one extra wash with kinase buffer12 was required. RAF kinase assays were conducted in the presence of 200 μM ATP, at 30 °C for 20 min. Recombinant inactive K97R MEK (Millipore) was used as a substrate and the reaction was terminated with 1 × SDS loading buffer and boiling. Kinase activity was estimated by immunoblotting for p-MEK.
Active RAS assay
Cells were cultured in 10-cm plates until 70–80% confluent. GTP-bound RAS was quantified using RAF1 Ras-binding domain (RBD) pull-down from Detection Kit (Thermo Scientific, no. 16117), as per the manufacturer.
Generation of inducible-expression cells
Retrovirus encoding rtTA or Braf or Nras was packaged in Phoenix-AMPHO cells obtained from ATCC. Medium containing virus was filtered with 0.45 PVDF filters followed by incubation with the target cells for 6 h. Cells were then maintained in virus free media for 2 days. Cells were selected using puromycin (2 μgml−1) or hygromycin (250 μgml−1) for 3 days. Cell populations that tested positive for infection were further sorted using GFP as a marker after overnight exposure to 1 μgml−1 doxycycline. GFP-positive cells were then cultured and expanded in medium without doxycycline but with antibiotics. For the conditional Ras-knockout cells (Hras−/−;Nras−/−;Kraslox/lox; Rertert/ert MEF cells), the inducible-expression cells were created in the absence of 4-OHT. The Kras gene was removed with 1 μM 4-OHT before the induction of wild-type or mutant BRAF expression.
RTK array
Human Phospho-RTK arrays (R&D Systems, ARY001B) were used according to manufacturer’s instructions. Cells were washed with cold PBS and lysed in 1% NP40 lysis buffer, 200 μg lysates were incubated with blocked membranes overnight. Membranes were subsequently washed and exposed to chemiluminescent reagent and exposed to X-ray film.
Cell growth assay
Cells were seeded into 96-well plates at 1,000 cells per well. Cell growth was quantified using the ATP-Glo assay (Promega, G7572) every 24 h. For each condition, 8 replicates at each concentration were measured. IC50 values were calculated using GraphPad Prizsm 6.
Mutational data and enrichment analysis
Genetic data were obtained from: The Cancer Genome Atlas (TCGA), publicly available published studies, and an ongoing prospective clinical sequencing initiative at Memorial Sloan Kettering that utilizes targeted capture-based sequencing of protein-coding exons and select introns of 314 or 410 cancer-associated genes (MSK-IMPACT)16. As mutation calling algorithms and mutation filtering and reporting practices varied from study to study, somatic mutational data review and correction were undertaken where possible as previously described6. Mutation calls were uniformly re-annotated to gene transcripts in Ensembl release 75 (Gencode release 19), and a single canonical effect per mutation was reported using Variant Effect Predictor (VEP) version 81 and vcf2maf version 1.6.2. To exclude putative germline variants misattributed as somatic mutations, we exclude any variant identified in ExAC r0.3 with a minor population allele frequency greater than 0.06%. We sought to determine the enrichment of co-occurring Ras/RTK lesions in tumours possessing BRAF mutants of low kinase activity in cancer types of presumed high or low basal levels of RTK signalling. Low kinase activity BRAF mutants were those identified in this study.
Sequencing of patient samples
Liver metastasis and normal tissue DNA from the CRC patient were analysed with MSK-IMPACT. Mean overall coverage in this sample was 834×. Primary tumour tissue from the CDC patient was sent to Foundation Medicine for deep sequencing. The ovarian metastasis tumour tissue was sequenced and analysed with MSK-IMPACT16. Tumour analysis was conducted under Institutional Review Board (IRB) protocol and waiver and Human Biospecimen Utilization Committee protocol (waiver WA0178-14, protocol 06-107, MATCH-R NCT02517892 and HBS2014026).
Targeted protein quantification by SRM–MS
Target proteins were quantitated by single reaction monitoring–mass spectrometry (SRM–MS) as previously described17. In brief, a tissue section (5 μM) was cut from each FFPE block, mounted on a Plus microcrope slide, deparaffinized, and stained with haematoxylin. Tumour areas were marked by a board-certified pathologist to achieve a cumulative tumour area of 12 mm2 (from multiple sections of a single tumour if necessary). Sequential tissue sections (10 μM) were cut from the same FFPE block, mounted onto DIRECTOR microdissection slides, and deparaffinized. Marked tumour areas were microdissected using a non-contact laser method, and captured tumour cells were solubilized to tryptic peptides using Liquid Tissue technology. Total protein concentration of each tryptic peptide mixture was measured by microBCA (ThermoFisher Scientific). Starting with on-column injection of 1 μg of tryptic peptide mixture, each sample was subjected to triplicate SRM–MS analysis using stable isotope-labelled internal standards for accurate quantitation of analytical targets. Instrumental analyses were performed on TSQ series (Vantage or Quantiva) triple quadrupole mass spectrometer (Thermo Scientific). The SRM–MS and chromatography conditions have been previously described18. Data analysis performed using Pinpoint software.
Patient-derived xenograft studies
Tumour tissue was transplanted orthotopically into NSG mice to make the PDX (IRB protocols 06-107, 14-091 and 06-107 A (13)). Once a tumour became visible in the mouse, it was transplanted and expanded in mice. The tumour tissue was implanted subcutaneously in the flank of 4–6-week-old NSG female mice and treatment of the mice began when tumour reached 100–150 mm3 in size. Mice were randomized (n = 10 mice per group in the CRC PDX experiment, n = 5 mice per group in the CDC PDX experiment) to receive drug treatment or vehicle as control. Studies were performed in compliance with institutional guidelines under an IACUC approved protocol. The animals were immediately euthanized as soon as we were notified that the tumours reached the IACUC set limitations. Investigators were not blinded when assessing the outcome of the in vivo experiments.
Statistical analysis
Results are mean values ± s.d. Investigators were not blinded when assessing the outcome of the in vivo experiments. All cellular experiments were repeated at least three times.
Data availability
The genetic data are available at http://cbioportal.org. All the source data for western blots and graphs are included in the Supplementary Information.
Extended Data
Extended Data Figure 1
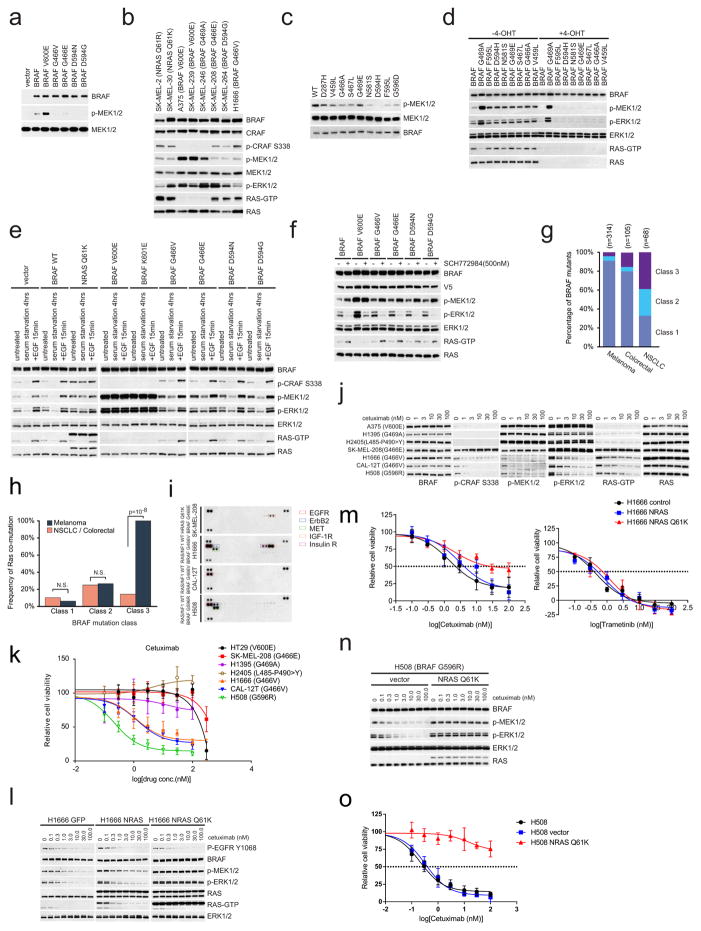
a, V5-tagged wild-type (WT) or mutant BRAF kinases were expressed in 293H cells that stably express NRAS(Q61K). These BRAF protein kinases were isolated with anti-V5 agarose. The in vitro kinase assay was performed with kinase-dead MEK1(K97R). The phosphorylation of MEK1 was determined by western blot. For gel source data, see Supplementary Fig. 4. b, Western blot analysis for components of the RAS/RAF/ERK signalling pathway in a panel of cancer cell lines harbouring the indicated mutations. Cellular RAS–GTP levels were determined by the active RAS pull-down assay. For gel source data, see Supplementary Fig. 4. c, In vitro kinase activity of the indicated BRAF proteins which were isolated from 293H cells that stably express NRAS(Q61K) was assessed as described in a. For gel source data, see Supplementary Fig. 4. d, MEK/ERK activation mediated by indicated BRAF proteins was assayed as described in Fig. 1d. For gel source data, see Supplementary Fig. 4. e, NIH3T3 cells expressing the indicated BRAF proteins were stimulated with 100 ng ml−1 EGF for 15 min, serum deprived for 6 h, or left untreated. RAS/RAF/MEK/ERK signalling activation of these cells was examined by western blot. Cellular RAS–GTP levels were determined by the active RAS pull-down assay. For gel source data, see Supplementary Fig. 5. f, Cells as indicated in Fig. 1a were cultured in doxycycline (30 ng ml−1) containing medium for 24 h and then treated with 500 nM ERK inhibitor SCH772984 for 8 h. Whole-cell lysates were then prepared and examined by western blot. RAS–GTP levels were determined using the active RAS pull-down assay. For gel source data, see Supplementary Fig. 5. g, The frequency distribution of the three classes of BRAF mutants in human BRAF-mutant melanoma tumours, or colorectal or non-small cell lung carcinomas. The data were collected from http://cbioportal.org. h, The frequency of coexistent RAS or NF1 alterations in human BRAF mutant melanomas compared to that in NSCLC and colorectal cancers.. The calculation was based on the sample sets as shown in Fig. 1e. The P values were calculated by using a paired t-test. N.S., not significant. i, The phosphorylation of multiple RTKs in the indicated cell lines was assayed using the Human Phospho-RTK Array Kit. Phosphorylated RTKs are highlighted with boxes in different colours. j, Cells were treated with increasing concentrations of cetuximab for 4 h. Levels of ERK signalling intermediates were determined by western blot. Cellular RAS–GTP levels were determined by the active RAS pull-down assay. For gel source data, see Supplementary Fig. 6. k, Cells were cultured and exposed to cetuximab at concentrations of 0, 0.3, 1, 3, 10, 30, 100 and 300 nM for 3 days. The effects of drug on cell growth were quantified using the ATP-Glo assay. Graphs were generated using Prism 6 (mean ±s.d. are represented by the dots and error bars, n =8). l, GFP, wild-type NRAS or NRAS(Q61K) were stably expressed in H1666 cells. Then the indicated cells were treated with cetuximab at increasing doses for 2 h. Cells were collected and cell lysates were examined by western blot. RAS–GTP levels were determined using the active RAS pull-down assay. For gel source data, see Supplementary Fig. 7. m, The effects of cetuximab or trametinib on the growth of the cells described in l was determined by ATP-Glo assay after 3 days treatment. Graphs were generated using Prism 6 (mean ± s.d., n = 8). n, NRAS(Q61K) or vector was stably expressed in H508 cells. The cells were treated with cetuximab at increasing doses for 2 h. Cells were collected and cell lysates were examined by western blot. For gel source data, see Supplementary Fig. 7. o, Growth inhibition of the cells described in n after three days exposure to varying doses of cetuximab on day 3 was determined by ATP-Glo assay. Graphs were generated using Prism 6 (mean ± s.d., n =8)
Extended Data Figure 2
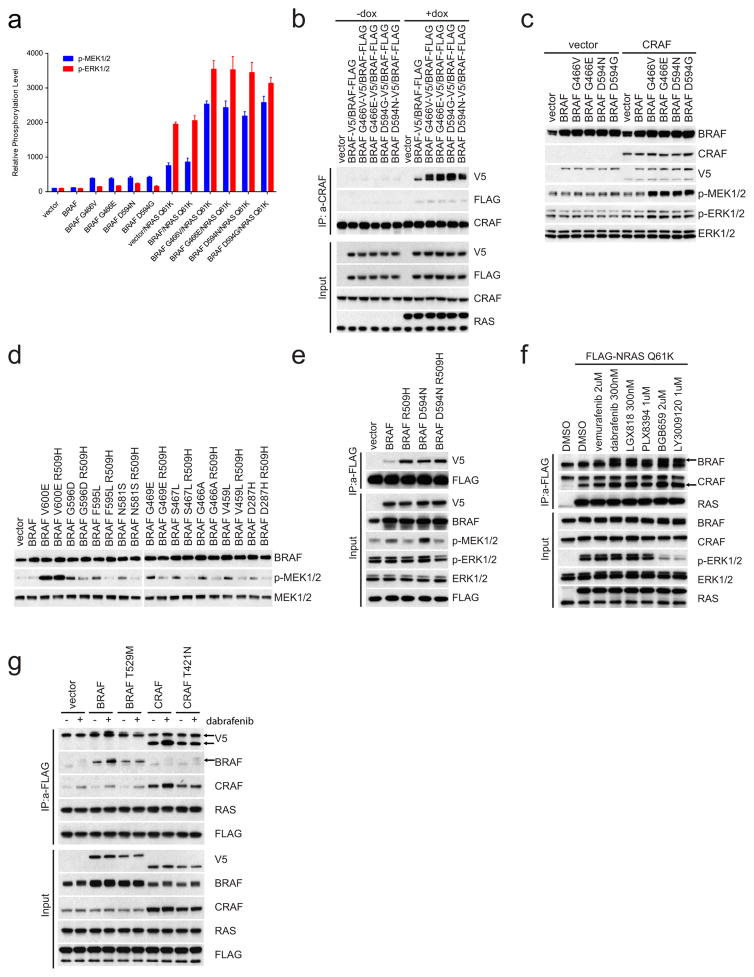
a, The levels of p-MEK1/2 and p-ERK1/2 in each transfectant in Fig. 2a were quantified by densitometry analysis using ImageJ software. The columns represent the relative levels of p-MEK/12 and p-ERK1/2 as normalized to the levels in cells transfected with vector plasmid and the expression levels of V5-tagged BRAF proteins (mean ± s.d. calculated on the basis of four independent experiments). b, Flag-tagged wild-type BRAF was co-expressed with the indicated V5-tagged BRAF proteins in 293H cells that were induced to express NRAS(Q61K). CRAF-bound BRAF proteins were determined by immunoprecipitation followed by western blot analysis. The results show RAS-dependent enhanced binding of CRAF to hypoactive BRAF mutants compared to their binding to wild-type BRAF. For gel source data, see Supplementary Fig. 8. c, V5-tagged wild-type or mutant BRAF kinases were expressed in Raf1-knockout cells with or without V5-tagged CRAF expression. For gel source data, see Supplementary Fig. 8. d, 293H cells were transfected with plasmids that encode the indicated BRAF proteins. After 24 h, the cells were collected. Cell lysates were then analysed by western blot with the indicated antibodies. For gel source data, see Supplementary Fig. 8. e, 293H cells stably expressing Flag-tagged NRAS(Q61K) were transfected with pcDNA3 plasmids expressing indicated proteins. The interaction between active RAS and the indicated BRAF proteins was determined by immunoprecipitation with anti-Flag beads. For gel source data, see Supplementary Fig. 9. f, 293H cells stably expressing Flag-tagged NRAS(Q61K) were treated with indicated RAF inhibitors for 1 h. The mutant RAS-bound BRAF and CRAF proteins were pulled down by immunoprecipitation with anti-Flag antibody and examined by western blot with indicated antibodies. For gel source data, see Supplementary Fig. 9. g, V5 tagging identified BRAF or CRAF proteins that were expressed in 293H cells stably expressing Flag-tagged NRAS(Q61K). BRAF(T529M) and CRAF(T421N) gatekeeper mutants that do not bind inhibitors were used as controls. NRAS-bound mutant BRAF and CRAF proteins were pulled down by anti-FLAG antibody from cells treated with or without dabrafenib (1 μM, 1 h). The immunoprecipitated proteins were assayed by western blot. For gel source data, see Supplementary Fig. 9.
Extended Data Figure 3
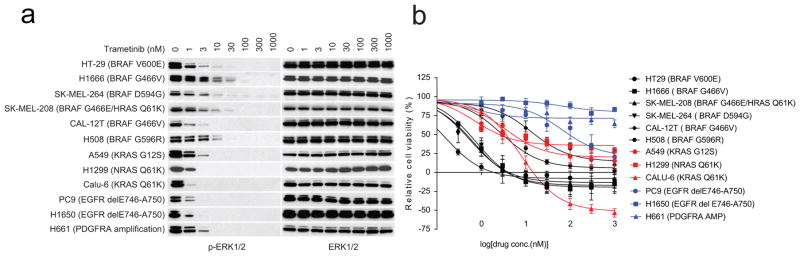
a, Inhibition of ERK signalling in a panel of cell lines exposed trametinib for 1 h at indicated doses. For gel source data, see Supplementary Fig. 10. b, The cell lines as indicated in a were treated with different concentrations of trametinib for 3 days. Cell viability was determined by ATP-Glo assay. Dose-dependent inhibition curves were generated using Prism 6 (mean ± s.d., n =8).
Extended Data Figure 4
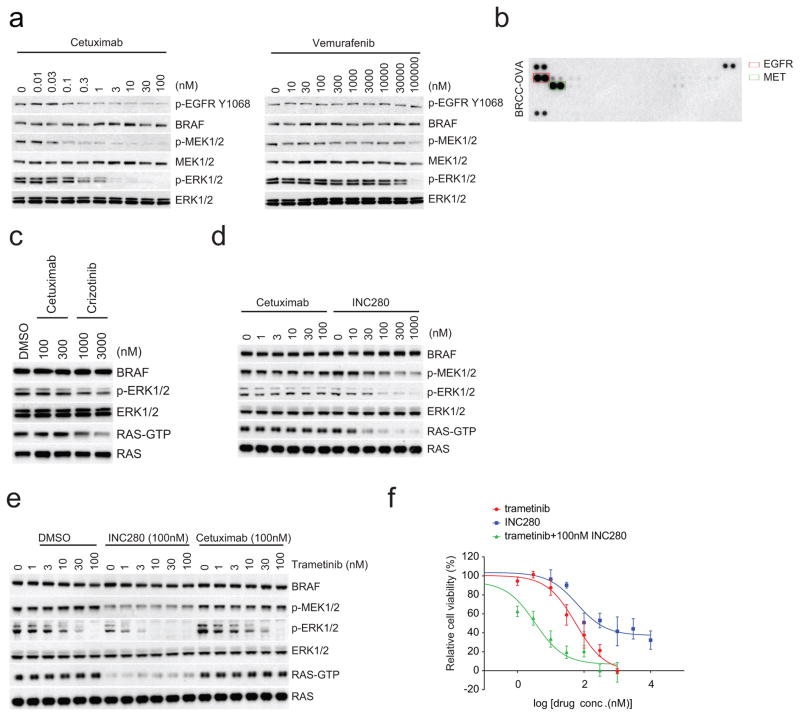
a, Cells isolated from the patient-derived xenograft (PDX) models as described (Fig. 4b) were treated with increasing doses of vemurafenib for 1 h or cetuximab for 4 h. Levels of indicated proteins were examined by western blot. For gel source data, see Supplementary Fig. 10. b, RTK phosphorylation profile of the cells isolated from the ovarian metastatisis derived PDX (BRCC-OVA) was assessed with Human Phospho-RTK arrays. The elevated p-EGFR and p-MET bands are within the red and green rectangles, respectively. c, Cells isolated from BRCC-OVA were treated with indicated drugs for 4 h and cell lysates were then analysed. For gel source data, see Supplementary Fig. 11. d, e, The BRCC-OVA cells were treated with indicated drugs or combinations thereof over a range of drug concentrations for 4 h. INC280 is a selective inhibitor of MET activity. The ERK signalling was assayed by western blot. The cellular RAS–GTP level was determined using the active RAS pull-down assay. For gel source data, see Supplementary Fig. 11. f, The growth inhibition of BRCC-OVA cells with indicated drugs at 0, 1, 3, 10, 30, 100, 300 and 1,000 nM of trametinib or 0, 10, 30, 100, 300, 1,000, 3,000 and 10,000 nM of INC280 or combination of increasing dose of trametinib with 100 nM INC280 was determined by ATP-Glo assay after 3 days of treatment. Graphs were generated using Prism 6 (mean ±s.d., n = 8).
Extended Data Table 1
Genetic alterations of the CRC patient sample
| Alterations | Gene Name | DNA change | Protein change |
|---|---|---|---|
| Missense Mutation | BRAF (NM_004333) | c.1397G>T | p.G466V |
| TP53 (NM_000546) | c.481G>T | p.A161S | |
| APC (NM_000038) | c.4461_4464delTATT | p.L1488fs | |
| c.2379_2380delAA | p.S794fs | ||
| BARD1 (NM_000465) | c.709C>T | p.Q237X | |
| FLCN (NM_144997) | c.323G>T | p.S108I | |
| LATS1 (NM_004690) | c.860G>A | p.R287Q | |
| MITF (NM_198159) | c.5A>C | p.Q2P | |
| MYCL1 (NM_001033082) | c.128A>G | p.Y43C | |
| Gene Deletion | TGFBR2 (NM_001024847) | deletion | Reduced expression |
| MAP2K4 (NM_003010) | Intragenic deletion | Reduced expression | |
| PARK2 (NM_004562) | Intragenic deletion | Reduced expression | |
| PIK3R1 (NM_181523) | Intragenic deletion | Reduced expression |
Extended Data Table 2
Mutations of the BRCC tumour samples
| Gene | Alteration | Primary Kidney (Foundation Medicine) | Ovary Metastasis (IMPACT) | Ovary Metastasis PDX (IMPACT) |
|---|---|---|---|---|
| FBXW7 | R505C | X | X | X |
| BRAF | D594N | X | X | |
| BRAF | G466A | X | X | X |
| FLT3 | V194M | X | X | |
| JAK3 | V722I | X | X | |
| CDH1 | R749L | X | X | X |
| TBX3 | H106Y | X | X | X |
| ATM | A2893P | X | X | |
| ARID1AP | P703T | X | X | X |
| PTPRD | S1554R | X | X | |
| ARID2 | V1649L | X | X | X |
| EPHA3 | R447W | X | X | X |
| BLM | fmshift | X | X | X |
| RECQL4 | V1020M | X | X | |
| CARD11 | V659M | X | X | |
| RB1 | I517S | X | X | |
| NUP93 | I583M | X | X | X |
| HGF | A46V | X | X | |
| SMARCB1 | fmshift | X | X | |
| RAD52 | R396C | X | X | |
| FOXL2 | fmshift | X | X | |
| RAD 50 | I94T | X | ||
| PREX 2 | P963S | X | ||
| BCL2L2 | R161H | X | ||
| SNCAIP | R33Q | X |
Supplementary Material
Supp Info
Supplementary Figure 1. Uncropped blots from Fig. 1a, 1b and 1d.
Supplementary Figure 2. Uncropped blots from Fig. 2a and 2b.
Supplementary Figure 3. Uncropped blots from Fig. 3b and 3c.
Supplementary Figure 4. Uncropped blots from Extended Data Fig. 1a, 1b, 1c and 1d.
Supplementary Figure 5. Uncropped blots from Extended Data Fig. 1e and 1d.
Supplementary Figure 6. Uncropped blots from Extended Data Fig. 1j.
Supplementary Figure 7. Uncropped blots from Extended Data Fig. 1l and 1n.
Supplementary Figure 8. Uncropped blots from Extended Data Fig. 2b and 2d.
Supplementary Figure 9. Uncropped blots from Extended Data Fig. 2e, 2f and 2g.
Supplementary Figure 10. Uncropped blots from Fig. 3a and and4a4a.
Supplementary Figure 11. Uncropped blots from Fig. 4c, 4d and 4e.
Acknowledgments
We are grateful to P. Lito, Y. Gao, W. Su, L. Desrochers, D. Santamaria and O. Abdel-Wahab for useful discussions. We thank S. Lowe for the vectors of the retrovirus-based inducible expression system and M. Baccarini for the Raf1-knockout MEFs. We thank Novartis for supplying INC280. We also thank I. Maruani Ryan for her help on this work. This research was supported by grants to N.R. from the National Institutes of Health (NIH) (P01 CA129243; R35 CA210085); the Melanoma Research Alliance (237059 and 348724); The Commonwealth Foundation for Cancer Research and The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center; and the Stand Up To Cancer – American Cancer Society Lung Cancer Dream Team Translational Research Grant (SU2C-AACR-DT17-15). Support was also received from the NIH MSKCC Cancer Center Support Grant P30 CA008748. Additional funding was provided by a Career Development Award from the Conquer Cancer Foundation of the American Society of Clinical Oncology (R.Y.); and from the NIH (R01 CA204749 and R01 CA180037), the Sontag and Josie Robertson Foundations and the Cycle For Survival (B.S.T.). Additional NIH funding was received by E.d.S. (U54 ODSS020355). We would like to acknowledge the support of the Arlene and Joseph Taub Foundation and of Paula and Thomas McInerney, without which this work would not have been possible. M.T.C. was supported in part by the NIH training grant T32 GM007175. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by grants from the European Research Council (ERC-AG/250297-RAS AHEAD), EU-Framework Programme (HEALTH-F2-2010-259770/LUNGTARGET and HEALTH-2010-260791/EUROCANPLATFORM) and Spanish Ministry of Economy and Competitiveness (SAF2011-30173 and SAF2014-59864-R) to M.B. M.B. is the recipient of an Endowed Chair from the AXA Research Fund.
Footnotes
The authors declare competing financial interests: details are available in the online version of the paper.
Readers are welcome to comment on the online version of the paper. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author Contributions Z.Y., R.Y., B.S.T. and N.R. conceived the project, designed the experiments. Z.Y. and N.R. wrote the manuscript. R.Y., D.B.S., V.S.R.-O., B.S.T. and M.B. provided critical revisions of the manuscript. Z.Y., R.Y., A.T., N.M.T., M.D., H.Z., V.S.R.-O., F.C., T.H., L.M., E.D.S., J.M., H.B. and M.B. established the in vitro and in vivo experimental systems, performed the laboratory experiments and analysed the results. M.T.C., D.B.S. and B.S.T. acquired and analysed the genetic data. N.M.C. reviewed and interpreted the clinical computerized axial tomography scans.
Reviewer Information Nature thanks L. Garraway and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature23291
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc5648058?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature23291
Article citations
Expert consensus on the diagnosis and treatment of solid tumors with BRAF mutations.
Innovation (Camb), 5(6):100661, 18 Oct 2024
Cited by: 0 articles | PMID: 39529955
Review
Multiplexed profiling of intracellular protein abundance, activity, interactions and druggability with LABEL-seq.
Nat Methods, 21(11):2094-2106, 21 Oct 2024
Cited by: 0 articles | PMID: 39433876
Features and efficacy of triple-targeted therapy for patients with EGFR-mutant non-small-cell lung cancer with acquired BRAF alterations who are resistant to epidermal growth factor receptor tyrosine kinase inhibitors.
ESMO Open, 9(10):103935, 09 Oct 2024
Cited by: 0 articles | PMID: 39389004 | PMCID: PMC11490925
Review Free full text in Europe PMC
Molecular markers for pediatric low-grade glioma.
Childs Nerv Syst, 40(10):3223-3228, 08 Oct 2024
Cited by: 0 articles | PMID: 39379532
Review
Therapeutic vulnerabilities and pan-cancer landscape of BRAF class III mutations in epithelial solid tumors.
BJC Rep, 2(1):77, 08 Oct 2024
Cited by: 0 articles | PMID: 39516363 | PMCID: PMC11524077
Go to all (278) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF.
Nature, 464(7287):427-430, 01 Mar 2010
Cited by: 1138 articles | PMID: 20179705 | PMCID: PMC3178447
RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E).
Nature, 480(7377):387-390, 23 Nov 2011
Cited by: 929 articles | PMID: 22113612 | PMCID: PMC3266695
RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth.
Nature, 464(7287):431-435, 03 Feb 2010
Cited by: 996 articles | PMID: 20130576
Efficacy of Dabrafenib and Trametinib in a Patient with Squamous-Cell Carcinoma, with Mutation p.D594G in BRAF and p.R461* in NF1 Genes-A Case Report with Literature Review.
Int J Mol Sci, 24(2):1195, 07 Jan 2023
Cited by: 1 article | PMID: 36674722 | PMCID: PMC9864135
Review Free full text in Europe PMC
Funding
Funders who supported this work.
European Research Council (3)
Grant ID: 259770
Grant ID: 260791
Ras Genes in Health and Disease (RAS AHEAD)
Dr Mariano Barbacid Montalban, Spanish National Cancer Research Centre
Grant ID: 250297
NCI NIH HHS (6)
Grant ID: P30 CA008748
Grant ID: R01 CA169351
Grant ID: P01 CA129243
Grant ID: R35 CA210085
Grant ID: R01 CA180037
Grant ID: R01 CA204749
NIGMS NIH HHS (1)
Grant ID: T32 GM007175
NIH HHS (2)
Grant ID: U54 ODSS020355
Grant ID: U54 OD020355





