Abstract
Free full text

Senataxin Mutation Reveals How R-loops Promote Transcription by Blocking DNA Methylation at Gene Promoters
Summary
R-loops are three-stranded nucleic acid structures found abundantly, and yet often viewed as by-products of transcription. Studying cells from patients with a motor neuron disease (amyotrophic lateral sclerosis 4, ALS4) due to a mutation in senataxin, we uncovered how R-loops promote transcription. In ALS4 patients, the senataxin mutation depletes R-loops with a consequent effect on gene expression. With fewer R-loops in ALS4 cells, the expression of BAMBI, a negative regulator of TGFβ, is reduced that then leads to the activation of the TGFβ pathway. We uncovered that genomewide R-loops influence promoter methylation of over 1,200 human genes. DNA methyl-transferase 1 favors binding to double-stranded DNA over R-loops. Thus, in forming R-loops, nascent RNA blocks DNA methylation and promotes further transcription. Hence, our results show that nucleic acid structures in addition to sequences influence the binding and activity of regulatory proteins.
Introduction
R-loops are three-stranded nucleic acid structures comprised of an RNA-DNA hybrid and a single-stranded DNA. After exiting RNA polymerase, nascent RNA is in close proximity to its negatively supercoiled DNA template; this promotes the formation of an RNA-DNA hybrid leading to the displacement of a DNA strand. In the 1990s, Crouch and others began to detail transcription-dependent R-loops (Drolet et al., 1995). Since then, studies have shown that R-loops are stable and abundant structures (Chan et al., 2014; El Hage et al., 2014; Wahba et al., 2016).
However, information on whether R-loops are merely by-products or an integral part of transcription remains limited. Much of the focus has been on the single-stranded DNAs in the three-stranded structures, as unpaired DNA strands are prone to damage (reviewed in (Aguilera and Gómez-González, 2008; Chaudhuri and Alt, 2004; Sollier and Cimprich, 2015; Stork et al., 2016)). R-loops have been associated with human diseases, including disorders due to repeat expansions such as Friedreich ataxia, fragile X syndrome (Groh et al., 2014; Lin et al., 2010; Reddy et al., 2011), and C9orf72-mediated amyotrophic lateral sclerosis (ALS)(Reddy et al., 2011; Zhang et al., 2015). R-loops are found in regions such as CpG islands (Ginno et al., 2013; Skourti-Stathaki and Proudfoot, 2014) where interactions with regulatory proteins take place. But, there is inadequate information on how R-loops interact with proteins and the functional consequences of those interactions. Many regulatory proteins act by specific binding to different nucleic acids. Some interact with double-stranded DNA, others with single-stranded DNA or even RNA-DNA hybrids. For example, the telomeric protein, POT1, binds to and protects G-rich DNA, and facilitates telomerase activity (Baumann and Cech, 2001; Nandakumar et al., 2010). The specificity of protein-nucleic acid interaction is critical for cellular functions. We posit that different sets of proteins interact with R-loops than with their single-stranded RNA or double-stranded DNA counterparts, and the specificity of these interactions confers a regulatory role on R-loops.
R-loops are not “fixed” structures; instead their formation and/or resolution are dynamic. Organisms from Escherichia coli to yeast and human have proteins that resolve R-loops. These include DNA topoisomerase (Pruss et al., 1982), RNase H (Cerritelli and Crouch, 2009; Drolet et al., 1995) and senataxin (Skourti-Stathaki et al., 2011). SEN1 was first identified by Culbertson and colleagues in saccharomyces cerevisiae as an tRNA splicing endonuclease (thus named as SEN1) (DeMarini et al., 1992; Winey and Culbertson, 1988). In mapping the gene for an autosomal recessive disorder, ataxia-ocular apraxia, Koenig and colleagues found mutations in the human homologue of SEN1, they then named the human gene as senataxin, SETX (Moreira et al., 2004). Soon after, mutation in senataxin was also identified in amyotrophic lateral sclerosis 4, ALS4, an autosomal dominant motor neuron disorder (Chen et al., 2004). In saccharomyces cerevisiae to schizosaccharomyces pombe and mammalian cells, senataxin plays multiples roles in RNA processing including as an RNA/DNA helicase that resolves R-loops (Kim et al., 1999; Martin-Tumasz and Brow, 2015; Skourti-Stathaki et al., 2011). However, whether SETX mutations affect R-loop abundance, and if so how they contribute to neurodegeneration remains largely unknown (Groh et al., 2017; Yüce and West, 2013).
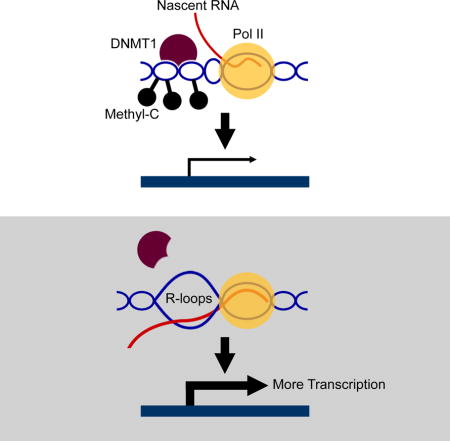
Here, cells from ALS4 patients with a senataxin mutation (Chen et al., 2004) allowed us to uncover how R-loops regulate transcription. ALS4 is characterized by early onset weakness and slow progression of symptoms. Although the causative genetic mutation was identified more than 10 years ago (Chen et al., 2004), how the mutation affects senataxin function is yet to be determined. In this study, we found that the senataxin mutation in ALS4 leads to a decrease in R-loops. This provided us with an opportunity to assess the function of R-loops. We show that ALS4 patients have fewer R-loops at the promoter of BAMBI, a pseudoreceptor which negatively regulates TGFβ (Onichtchouk et al., 1999). The decrease in R-loops unmasks the BAMBI promoter, and facilitates DNA methylation by DNA methyltransferase 1 (DNMT1). As a result, BAMBI is silenced which leads to the activation of TGFβ signaling. Target genes such as BMP2 and helix-loop-helix transcription factors ID1 and ID2 that play a key role in neuronal differentiation (Shah et al., 1996; Vierbuchen et al., 2010) are upregulated. To assess whether R-loops regulate transcription of other genes besides BAMBI, we carried out genome-wide analyses and found that R-loops decrease DNA methylation at over 1,200 gene promoters. Then to determine how R-loops regulate transcription, we uncovered that DNA methyltransferase binds to dsDNA more avidly than to RNA/DNA hybrid; thus, genes with R-loops in their promoters are less methylated, and have higher gene expression levels. Together, our results show that R-loops provide positive feedback to promote transcription. By forming R-loops, nascent RNA facilitates its own transcription by blocking methylation-mediated gene silencing.
Results
Clinical manifestations of ALS4
Our study subjects are members of a multigenerational ALS4 family with a known senataxin mutation, L389S (Chen et al., 2004). We obtained blood samples and skin biopsies from subjects with and without the senataxin mutation. Spinal cord and motor cortex from autopsies were also included.
Key clinical characteristics of ALS4 include hyperreflexia and progressive distal-predominant weakness consistent with upper and lower motor neuron disease, as reported previously (Chen et al., 2004). Features that distinguish ALS4 from sporadic ALS include early onset, slow progression, and lack of bulbar involvement (Figure 1A). Denervation was found on EMG (Figure 1A). The average creatine kinase level was 394 U/L, higher than the control group (106 U/L, P=0.03). The alpha fetoprotein (AFP) level was also higher on average in the ALS4 subjects (2.4 ng/ml) compared to the controls (1.6 ng/ml, P=0.03). Ten patients had thigh muscle MRIs, which showed a reduction in myofiber volume with disease duration (R2=0.54, P= 0.02) (Figure 1B), consistent with progressive denervation and muscle atrophy.
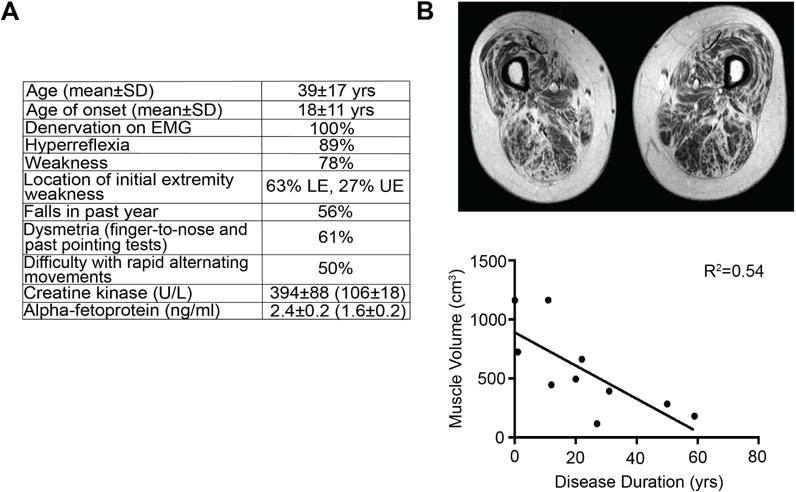
(A) Age of onset is defined as the age of initial muscle weakness. All patients had electromyography (EMG) showing denervation in at least two different muscle groups. Hyperreflexia was scored as increased reflexes in either the upper or lower extremities; diminution of reflexes was observed in patients with severe weakness. Strength was evaluated using the Medical Research Council (MRC) scale (https://www.mrc.ac.uk/research/facilities-and-resources-for-researchers/mrc-scales/mrc-muscle-scale/) with weakness determined by MRC scale less than 5 in any muscle group. Falls were assessed retrospectively; (+) = at least one fall in the past year. LE = lower extremity, UE = upper extremity. Dysmetria identified as positive findings on finger-to-nose or past pointing tests. Creatine kinase and alpha-fetoprotein levels were measured in 15 patients (average age 41) compared to 8 related, unaffected controls in parentheses (average age 50); values shown are mean ± SEM). (B) T1 MRI measurement of thigh muscle volume. Muscle volume correlates significantly with disease duration (defined as years since diagnosis) (R2=0.54, P=0.02).
R-loops are reduced in ALS4
We began by assessing the molecular effects of the senataxin mutation. Expression data from public databases (Figure S1A) and experimental results in this study (Figure S1B) show that senataxin is expressed in most if not all cell types. There are three major isoforms of senataxin, all of which include the L389S mutation in exon 10. Expression levels of senataxin mRNA and protein are not altered by the mutation (Figure S1C, D). This prompted us to ask whether the mutation affects senataxin function; we began by measuring the abundance of R-loops since as an RNA-DNA helicase, a key function of senataxin is to resolve R-loops (Kim et al., 1999; Martin-Tumasz and Brow, 2015; Skourti-Stathaki et al., 2011).
To detect R-loops, we used the well-established S9.6 antibody along with RNase H1 treatment to ensure specificity of the signals (Boguslawski et al., 1986). RNase H hydrolyzes the RNA in R-loops (Cerritelli and Crouch, 2009; Drolet et al., 1995), and is used in many studies to validate the specificity of the S9.6 antibody. We showed that our S9.6 signal is abolished by treatment with RNase H (Figure S2). Following that, we assessed R-loops in our patients’ cells. The results show a decreased S9.6 dot-blot staining, indicating an overall decrease in R-loop abundance in fibroblasts from ALS4 patients compared to controls (Figure 2A). By immunostaining, we also measured the abundance of R-loops in primary fibroblasts, spinal cord, and motor cortex from patients and controls. In all three types of samples, there were significantly fewer R-loops (P<0.001) in ALS4 patients compared to controls (Figure 2B–D). To check that the lower R-loop abundance in ALS4 cells is due to the senataxin mutation, we overexpressed wildtype and L389S mutant forms of senataxin in fibroblasts. The overexpression of the mutant L389S senataxin led to fewer R-loops, thus recapitulating the feature in patient cells (Figure 2E, ,2F).2F). Together, these results show that across different cell types, the senataxin mutation results in fewer R-loops, perhaps from a gain of helicase function which is consistent with an autosomal dominant mode of inheritance.
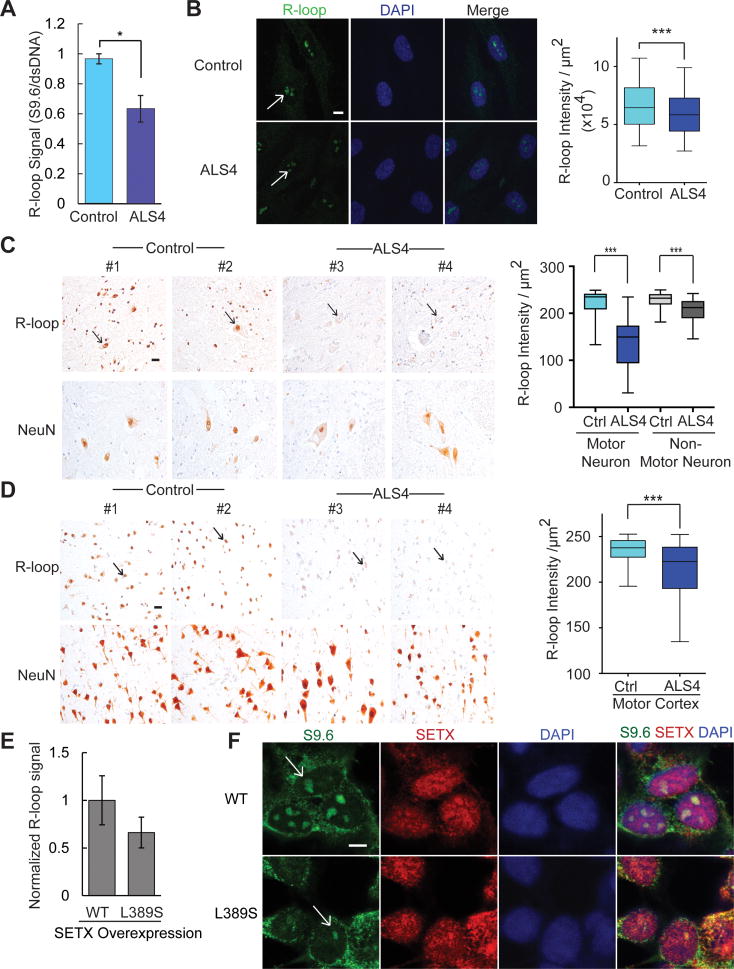
(A) Dot-blot probed with S9.6 antibody shows fewer R-loops in ALS4 fibroblasts compared to control. A duplicate blot was probed with anti-dsDNA antibody as loading control. S9.6 intensity was normalized to that of the dsDNA and average was shown. (n= 3 control & 3 patients; error bars are SEM). * represents P<0.05 (t-test). (B) Immunofluorescence staining of ALS4 fibroblasts shows a significant reduction in nuclear S9.6 signal that co-localizes with DAPI (scale bar = 7.5μm) as compared to controls. n = 3 controls & 3 patients, 10 fields of view were counted per sample and quantified using ImageJ. Arrows indicate nuclear R-loops. R-loop staining with the S9.6 antibody shows a significant reduction of R-loop signal in cell nuclei in the (C) lumbar spinal cord and (D) the motor cortex. NeuN is shown as a control of neuronal nuclei staining. Neuronal nuclei indicated by arrows. n = 2 controls & 2 patients; scale bar = 25 m, 10 fields per sample, error bars are SEM. *** represents P<0.001 (t-test). (E) Fewer R-loops in cells transfected with L389S form of SETX compared to WT (t-test, P=0.06). R-loops were analyzed by dot blot using S9.6 antibody. Error bars are SEM of triplicates. S9.6 signal was first normalized to loading control (dsDNA) and then to SETX expression as detected by western blot. (F) Immunofluorescence staining showing reduced nuclear S9.6 signal (green) in cells transfected with L389S form of SETX compared to WT. Note in both L389S and WT SETX transfected cells, S9.6 staining is reduced in those cells with higher SETX expression (red); scale bar = 5 μm, merged image with staining of S9.6, SETX, and DAPI (blue), R-loops in nuclei indicated by arrows.
ALS4 cells have low BAMBI expression and high TGFβ signaling
R-loops form co-transcriptionally; however, whether these R-loops have functional consequences or are merely unwanted products has been unclear. The ALS4 cells with fewer R-loops provide an opportunity to address this question. Initially, we compared gene expression by RNA-sequencing of fibroblasts from patients and controls. The differentially-expressed genes are highly enriched for activation of the TGFβ pathway (P<<10−10; Figure S3A, Table S1). Figure 3A shows a scatter plot of the differentially expressed genes; genes that point to activation of the TGFβ pathway in ALS4 are highlighted. Since this pathway is known to be important in motor neurons (Colavita et al., 1998; Martinou et al., 1990) and previous studies have implicated TGFβ in ALS (Endo et al., 2015; Houi et al., 2002; Nakamura et al., 2008; Phatnani et al., 2013), we focused on this pathway, and measured gene expression from additional individuals. Of the 77 genes that we examined, the expression of 47 genes differed by 20% or more between ALS4 patients and controls (Figure 3B). Among them, 36 genes including BMPs, SMADs and inhibins were expressed at higher levels in the patients than the controls. Consistent with activation of the TGFβ signaling, BAMBI which encodes a key negative regulator is expressed at a lower level in the patients. BAMBI is a pseudoreceptor that lacks an intracellular kinase domain, and thus cannot initiate signaling (Onichtchouk et al., 1999). Binding of TGFβ to the TGFβ receptors activates the signaling cascade by phosphorylation of receptor-regulated SMAD proteins such as SMAD2/3 (Eppert et al., 1996; Macías-Silva et al., 1996; Wrana et al., 1994). BAMBI interacts with the TGFβ receptors and negatively modulates signaling by deterring the formation of functional receptors (Onichtchouk et al., 1999). Lower BAMBI expression results in more activated TGFβ pathway, as the TGFβ receptors are free to initiate signaling. To check that BAMBI is indeed acting as a negative regulator in our cells, we knocked down BAMBI (Figure S3B) and confirmed this results in activation of the TGFβ pathway. Upon silencing of BAMBI, as expected we found an increase in phosphorylation of SMAD2/3 (Figure S3C) and expression of the downstream targets of the TGFβ pathway (Figure S3D). Thus, knockdown of BAMBI recapitulated the expression pattern in ALS4 cells for the TGFβ pathway (Figure S3D). Given this critical role, we examined the expression of BAMBI in additional samples, and confirmed that its expression level in ALS4 patients is lower (by 65%) than that of controls (Figure S3E).
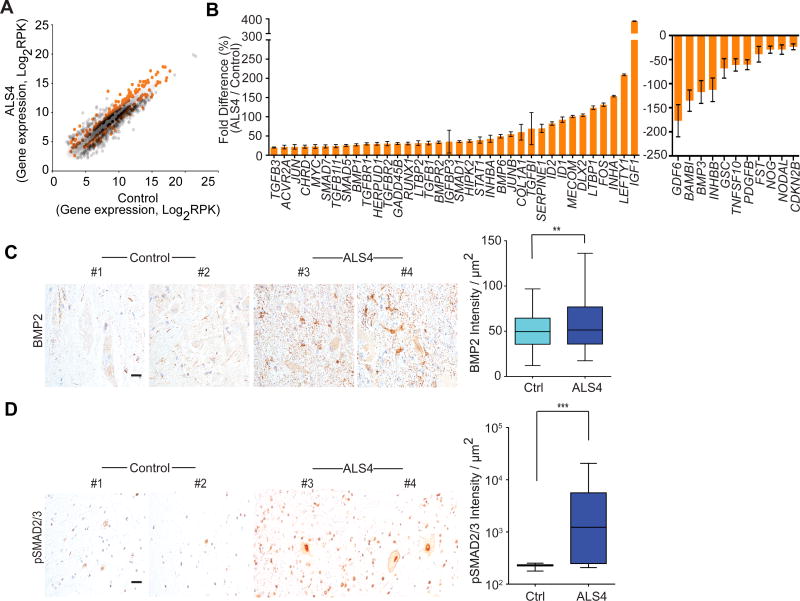
(A) Scatter plot of gene expression levels from RNA-seq of fibroblasts (n = 3 controls & 3 ALS4 patients); differentially expressed genes that point to activation of TGFβ pathway in the ALS4 patients are shown in orange; genes shown are those with ≥50% differences. (B) Expression levels of genes (RT-PCR) in TGFβ pathway from fibroblasts (n = 5 controls & 5 ALS4 patients; 2 replicates from each sample), error bars are SEM. Genes shown are those with ≥20% differences. (C) & (D) Increased staining of BMP2 and pSMAD2/3 in patient lumbar spinal cords; n = 2 controls & 2 ALS4 patients; scale bar = 25μm, 10 fields per sample, error bars are SEM. ** represents P<0.01, *** represents P<0.001 (t-test).
The TGFβ pathway is activated in neurons of ALS4 patients
We next sought to evaluate whether the changes in TGFβ signaling identified in patient fibroblasts are similarly disrupted in the relevant tissues for a motor neuron disease. We measured the expression levels of BMP2 and phosphorylated SMAD2/3 which were previously studied in the spinal cord (Katsuno et al., 2010; Nakamura et al., 2008). The expression levels of phosphorylated SMAD2/3 are important since they are the first proteins to be activated in TGFβ signaling (Eppert et al., 1996; Macías-Silva et al., 1996). We found significantly higher levels of BMP2 (P<0.01) and pSMAD2/3 (P<0.001) in the anterior horn of the ALS4 patient spinal cord by immunofluorescence, consistent with an activation of TGFβ signaling in motor neurons and neighboring cells (Figure 3C, ,3D).3D). We also found ID1 transcript levels to be ~50% higher in a patient’s spinal cord relative to controls (Figure S3F). Together, our results show an overactive TGFβ pathway in ALS4 patients.
ALS4 cells have fewer promoter R-loops and lower BAMBI expression
Next, we asked whether the low BAMBI expression can be explained by altered R-loops in ALS4. In Figure 2, we show fewer R-loops globally in cells from ALS4 patients. Here we examined R-loops at the BAMBI gene. The promoter region of BAMBI is particularly GC rich and shows high GC skewing (Figure 4A), which results in G-rich non-template DNA strand that can promote R-loop formation (Ginno et al., 2012; Hamperl and Cimprich, 2014; Yu et al., 2003). To investigate if there are R-loops in the promoter region of BAMBI, we carried out DNA-RNA immunoprecipitation with the S9.6 antibody (S9.6 DRIP). The results confirmed the known R-loops in β-actin (Skourti-Stathaki et al., 2011), and uncovered R-loops in the promoter regions of BAMBI. Figure 4B shows that at both loci, ALS4 patients have significantly (P<0.03) fewer R-loops compared to controls. Then by S9.6 DRIP-seq, we confirmed that the R-loops in the promoters are the only ones in BAMBI gene that differ between patients and controls (Figure 4C).
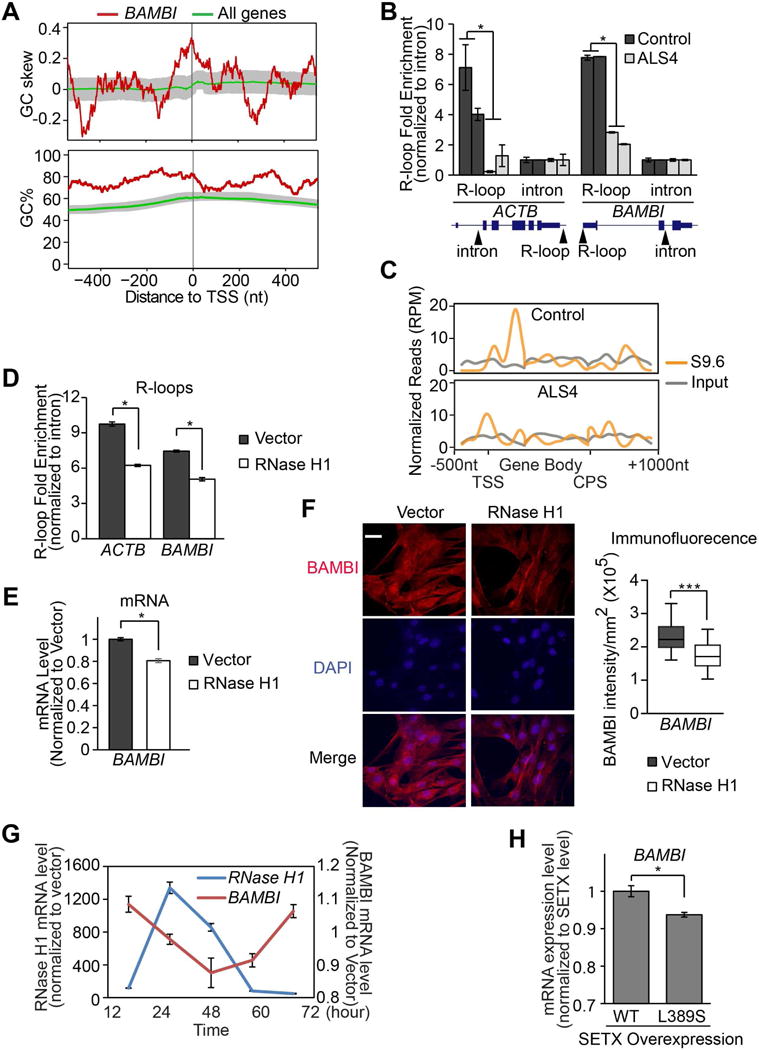
(A) BAMBI promoter is GC rich and G-skewed in comparison to median levels of all genes annotated in RefSeq. 50-bp sliding windows were used to compute GC% = [(G+C)/(G+C+A+T)]%, and GC skew = (G−C)/(G+C). The GC% and GC skew at the BAMBI promoter (in red), and their median level for all 23,682 promoters (in green) are shown. Shaded area represents the range between 40th and 60th percentile. (B) S9.6 DNA-RNA IP followed by quantitative PCR with primers specific to a region containing known R-loops at β-actin or BAMBI promoters (n = 2 controls and 2 patients). Primers specific to intronic regions free of R-loops were used as negative controls. Arrows indicate location of primers. (C) S9.6 DNA-RNA IP followed by sequencing in control and ALS4 cells. Normalized read counts across BAMBI gene are plotted. Transcription start site (TSS), Cleavage and polyadenylation site (CPS). (D) Overexpression of human RNase H1 in fibroblasts led to fewer R-loops β-actin or BAMBI promoters (same PCR primers used in 4B), and lower BAMBI transcript (E) and protein (F) expressions. 8 fields of view with 15-20 cells per field were quantified. P<0.001, scale bar = 15μm. (G) A 72-hour time course showed BAMBI gene expression correlates negatively with the expression of RNase H1. (H) Overexpression of L389S form of senataxin led to lower BAMBI gene expression (t-test, P<0.01). Error bars are SEM.
To assess whether having fewer R-loops at the BAMBI promoter region accounts for the lower BAMBI expression in ALS4, we perturbed R-loops with a protein other than senataxin. We overexpressed RNase H1 (Figure S4A), which cleaves RNA in R-loops (Cerritelli and Crouch, 2009; Drolet et al., 1995). As expected, this leads to fewer R-loops in β-actin and the BAMBI promoter region (Figure 4D). We then measured BAMBI expression, and found that with reduced R-loops from RNase H1 overexpression (Figure 4D, Figure S4B), BAMBI transcript and protein levels were significantly lower (P<0.03; Figure 4E, ,4F;4F; Figure S4C). We assessed cells up to 72 hours following RNase H1 overexpression; as the RHaseH1 level increases, we saw a corresponding decrease in BAMBI expression (Figure 4G). These results support that reduced R-loop formation at the gene promoter accounts for the lower expression of BAMBI in ALS4 patients. Additionally, we measured BAMBI expression in cells where we overexpressed the mutant L389S senataxin, and found that the overexpression of mutant senataxin resulted in significantly lower BAMBI expression (Figure 4H). As noted above, the overexpression of this mutant form of senataxin resulted in fewer R-loops (Figure S4B). Together, the results show by lowering R-loops with either RNase H or senataxin, we lowered BAMBI expression.
R-loops repel DNMT1
Next, we sought to determine how R-loops affect gene expression. Previous studies demonstrated that DNA methylation silences BAMBI in cancers (Khin et al., 2009; Marwitz et al., 2016). Chédin and colleagues showed that R-loops are correlated to de novo methylation in development (Ginno et al., 2012). However, if and how R-loops affect DNA methylation in human cells more generally remain unknown. This prompted us to ask whether R-loop abundance could affect promoter methylation and thus BAMBI expression. To address this, first we compared promoter methylation in patients and controls by MethylC-seq (Urich et al., 2015). Results showed methylated cytosines in the same region of the BAMBI promoter as found by the ENCODE consortium (ENCODE Project Consortium, 2011). With that confirmation, we compared BAMBI promoter methylation in ALS4 cells to controls, and found that the ALS4 patient cells have more methylated cytosines at the BAMBI promoter (Figure 5A).
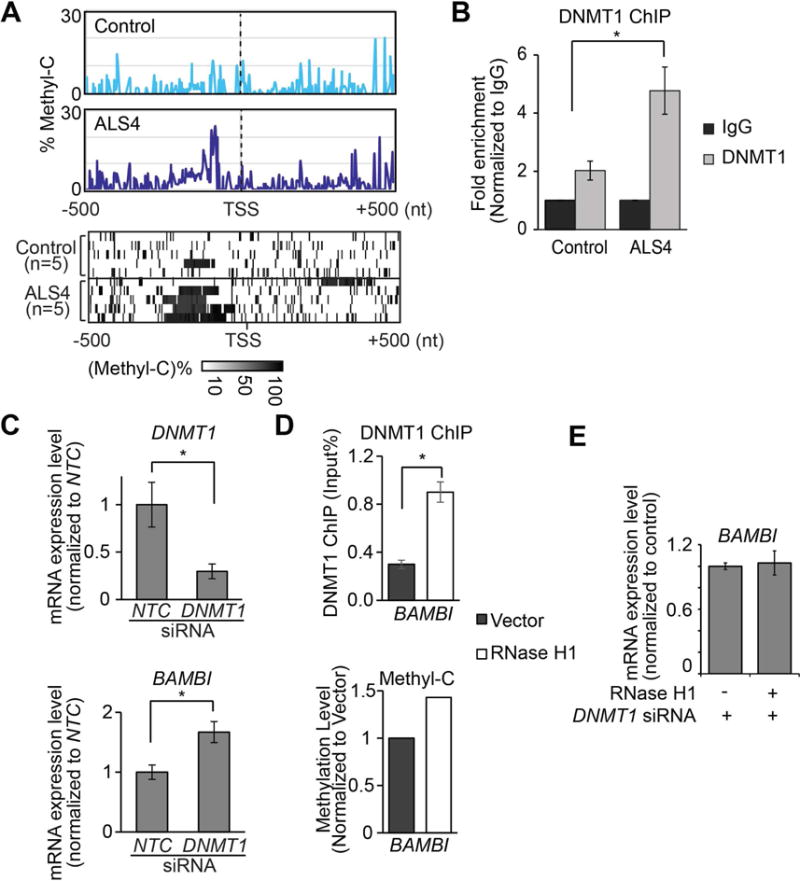
(A) Cytosine methylation is higher in ALS4 patients than in controls. From methyl-C sequencing reads, at each genomic position, level of cytosine methylation was calculated as percentage of methylated C. Upper panel: average levels of methylated cytosine in fibroblasts at BAMBI promoter are shown (n = 5 controls & 5 ALS4 patients). Lower panel: heatmap showing methylated cytosine levels from each subject. Each row shows the average methyl-C level across the BAMBI promoter region (TSS±500bp). (B) DNMT1 ChIP-qPCR shows significantly more DNMT1 binds to BAMBI promoter in ALS4 patient compared to control (P<0.05; error bar: SEM of quadruplicates.). Arrow indicates location of primers. (C) DNMT1 knockdown results in lower DNMT1 expression and higher BAMBI expression; error bars represent SEM of triplicates. (D) RNase H1 overexpression which resolves R-loops leads to more DNMT1 binding (top) and consequently higher methylation (bottom) of BAMBI promoter. (P<0.05, error bars represent SEM of triplicates). Average methylation levels of cytosines within BAMBI promoter are shown. (E) R-loop-mediated repression of BAMBI expression is dependent on DNMT1. BAMBI expression did not change following RNase H1 overexpression when DNMT1 was knocked down (error bars represent SEM of triplicates).
To examine methylation of the BAMBI promoter region more closely, using several methods, we confirmed that BAMBI expression is repressed by DNA methyltransferase 1 (DNMT1). We treated fibroblasts with 5-Aza-C, which inhibits DNMT activity (Robert et al., 2003). 5-Aza-C treatment resulted in reduced methylation at the BAMBI promoter, and increased gene expression level (Figure S4D) thus providing evidence for methylation-dependent expression of BAMBI. Then, we asked which enzyme methylates BAMBI in fibroblasts. Given that these are cells from adults, DNMT1 should be the predominant methyltransferase; we confirmed this by measuring DNMT expression levels. While DNMT1 is expressed, DNMT3 proteins are not expressed in the fibroblast (Figure S4E). Next, we showed by chromatin immunoprecipitation that DNMT1 binds to the BAMBI promoter region (Figure S4F). There is significantly more DNMT1 binding to BAMBI promoter in ALS4 cells compared to control (Figure 5B), consistent with higher methylation levels in ALS4 cells (Figure 5A). To be certain that DNMT1-mediated methylation influences BAMBI expression, we knocked down DNMT1 by RNA interference (Figure 5C, top). The expression of BAMBI was significantly higher after DNMT1 knockdown (t-test, P<0.05) (Figure 5C, bottom). Together, these results showed that DNA methylation mediated by DNMT1 silences BAMBI expression.
Then, to check that R-loop affects DNMT1-mediated methylation, we overexpressed RNase H1 and assessed binding of DNMT1 by DNM1-ChIP (O’Hagan et al., 2011). With fewer R-loops (Figure 4D), we found there was an increased in DNMT1 binding to the BAMBI promoter (Figure 5D, top) and a corresponding increase in methylation (Figure 5D, bottom). The resulting increase in promoter methylation accounts for the lower BAMBI expression (Figure 4E). In addition, we knocked down DNMT1 and then overexpressed RNase H1. When DNMT1 was knocked down, overexpression of RNase H1 did not repress BAMBI expression (Figure 5E), thus confirming that the effect of R-loops on BAMBI expression is mostly through DNMT1-mediated methylation.
Next, we asked how R-loops influence methylation of the BAMBI promoter region. To address this, we first examined whether DNA methyltransferase activity differs based on nucleic acid substrates, in particular, double-stranded DNA versus RNA/DNA hybrids in R-loops. We used a competitive immunoassay to assess the activity of recombinant DNMT1 on immobilized CpG-rich sequences, with double-stranded DNA or corresponding RNA/DNA hybrids as competitors. Binding of the methyl-C binding protein to immobilized substrates provides a colorimetric readout. We used the same oligomers as those used to characterize R-loops in previous studies as competitors to the CpG substrate (Haruki et al., 1997). The results showed that DNMT1 activity is about 2.5 times higher on double-stranded DNA than on the RNA-DNA hybrids (Figure 6A). Second, by electrophoretic mobility gel shift assay (EMSA), we asked if there is a difference in DNMT1 binding to double-stranded DNA versus RNA/DNA hybrids. Probes corresponding to the BAMBI promoter were used. The results show that DNMT1 binds more avidly to dsDNA than RNA-DNA hybrids (Figure 6B). To confirm this finding, we used biolayer interferometry to evaluate DNMT1 binding and confirmed that DNMT1 has higher affinity for double-stranded DNA than its corresponding RNA/DNA hybrids (Figure 6C and Figure S4G). Together, results from these in vitro assays (Figure 6C and Figure S4G) show that the preferred substrate of DNMT1 is double-stranded DNA. Along with our genetic knockdown of DNMT1, RNase H1 overexpression, and in vivo DNMT1 chromatin immunoprecipitation assays, these studies show that in cells with more R-loops, there is less DNMT1-mediated gene silencing of BAMBI. Since our patients in contrast have fewer R-loops, their BAMBI promoter is mostly double-stranded DNA, the preferred substrate of DNMT1, which facilitates DNMT-mediated methylation-induced silencing. To confirm that promoter methylation silences BAMBI expression, we studied nascent transcript of BAMBI by nuclear run-on assay followed by PCR. We compared the abundance of nascent transcripts of BAMBI in cells from patients and controls, and found that ALS4 cells have significantly (P<0.01) fewer nascent BAMBI transcripts (Figure 6D).
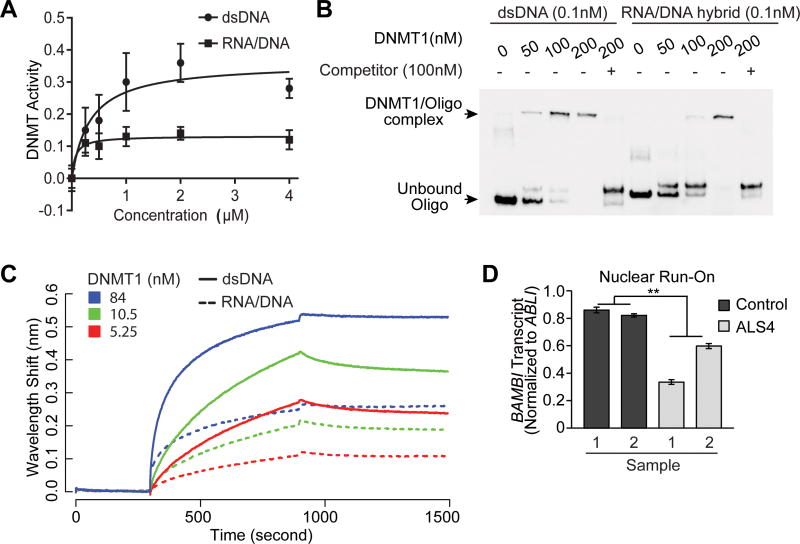
(A) DNMT1 activity is higher with dsDNA than RNA-DNA hybrid as substrates; error bars represent SEM of triplicates. (B) Binding affinity of DNMT1 for dsDNA is higher than for RNA/DNA hybrid. EMSA was used to measure binding of DNMT1 to BAMBI promoter represented as biotinylated dsDNA or corresponding RNA/DNA hybrid. Unlabeled dsDNA was added as competitor to show specificity of binding. (C) More binding of DNMT1 to dsDNA than RNA/DNA hybrid. Binding with a dilution series of DNMT1 to 2.5nM dsDNA (solid lines) or RNA/DNA hybrid (dotted lines) assayed by biolayer interferometry. Baseline was recorded from 0 to 300 seconds, association of DNMT1 with dsDNA or RNA/DNA hybrid from 300-900 seconds, followed by dissociation. (D) ALS4 patients have less nascent transcripts of BAMBI. In nuclear run-on (NRO) of primary fibroblasts (n = 2 controls & 2 ALS4 patients), nascent transcript level was measured by quantitative PCR using primers specific to promoter of BAMBI, and normalized to promoter of a housekeeping gene ABLI that has no R-loops in S9.6 pull-down assay.
Together, these results indicate that R-loops facilitate transcription by preventing DNA methylation. The reduction of R-loops in ALS4 allows DNA methylation and silencing of BAMBI, which promotes TGFβ signaling.
R-loops reduce promoter methylation of more than 1,200 human genes
Next, we asked whether R-loops regulate gene expression through DNA methylation at other genes. We searched globally for genes like BAMBI that have R-loops and high GC content in their promoter regions. In normal human primary fibroblasts, we carried out DNA-RNA immunoprecipitation with the S9.6 antibody followed by sequencing (DRIP-seq). The results identified 3,257 genes with R-loops at their promoter regions. Among these gene promoters with R-loops, 1,386 are GC-rich (GC% ≥ 0.6) like the BAMBI promoter. Figure 7A (top) is a metagene plot of R-loops at these 1,386 GC-rich gene promoter regions. Some representative examples of genes are shown in Figure S5A. We also carried out DRIP-seq with ALS4 fibroblasts. Comparison of the metagene plots from normal and ALS4 cells confirms that ALS4 cells have fewer R-loops in these gene promoters (Figure 7A, bottom).
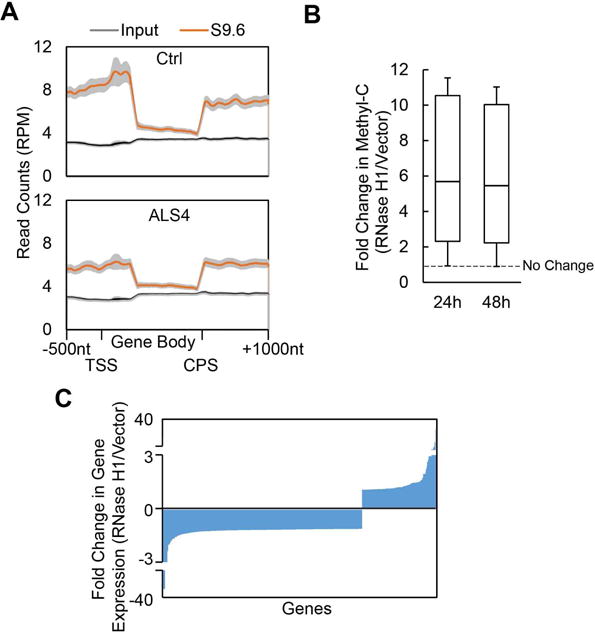
(A) -1,386 gene promoters that are GC-rich (GC%>0.6) and have R-loops. Metagene plot of DRIP-Seq data shows enrichment of R-loops near promoters (top). Metagene plots of DRIP-seq also show genome-wide decrease of R-loops in ALS4 (bottom) compared to the control (top). Average ± 95% CI was plotted. (B) Among the 1, 386 genes with GC-rich promoters, 1,240 genes showed an increase in DNA methylation following RNaseH1 overexpression; fold changes at two time points are shown. (C) Among the genes that showed R-loop dependent methylation in (B), 697 gene showed changes in gene expression following overexpression of RNase H1, of which 508 showed decreased expression.
We then asked whether as with BAMBI, R-loops prevent DNA methylation at these gene promoters. We abolished R-loops by overexpression of RNase H1, and measured the effects on DNA methylation and gene expression. Overexpression of RNase H1 led to a significant increase (t-test, P<0.001) in promoter methylation, specifically 89% (1,240) of the genes showed an increase (≥ 2.5 fold) in methylation levels (Table S2). Results from methylC-seq at 24 and 48 hours following RNase H1 overexpression show significantly higher methylation levels at both time-points (Figure 7B). Thus, like in BAMBI, R-loops decreases promoter methylation of these genes. The R-loops, and methyl-C patterns on these genes can be viewed and downloaded on the UCSC genome browser, https://goo.gl/O9psQi. Some representative examples are shown in Figure S5B. To assess the consequence of R-loop dependent methylation, we measured expression levels of these genes before and following RNase H overexpression. Promoter methylation decreases transcription; even though we do not expect RNase H1 overexpression to decrease the expression of all genes as steady-state gene expression levels are determined by transcription and degradation, we expect a trend towards lower gene expression. Indeed, we found the expression levels of most (73%) of the genes with promoter R-loops that showed changes in gene expression were reduced following RNase H1 overexpression (Figure 7C, Figure S5C). Together, these results uncover that R-loops promote transcription of over 1,200 genes by preventing DNA methylation-directed gene silencing.
Discussion
In conclusion, our study of an autosomal dominant form of ALS uncovered a function of R-loops. These three-stranded nucleic acid structures trail the RNA polymerase as negative helical torsion created by the transcribing polymerase complex leads to DNA strand separation, thus allowing the nascent RNA and its DNA template to form an RNA-DNA hybrid. R-loops have generally been considered as by-products of transcription without a defined functional role. However, studies have suggested that they may affect transcription (Finkel et al., 2010; Ginno et al., 2012; Steinmetz et al., 2006). Here we provide an example of R-loop-mediated gene regulation, where R-loops protect gene promoters such as that of BAMBI from DNA methylation. This unveils yet another regulatory role of RNA. We show that in human cells, nascent RNA positively regulates its own transcription by forming R-loops to protect its DNA template from methylation-mediated gene silencing. DNA methyltransferases bind less avidly to RNA/DNA hybrids than double-stranded DNA; thus, when R-loops accumulate at gene promoters, there is less DNA methylation. Other forms of RNA-mediated post-transcriptional gene regulation such as RNA-directed gene silencing in plants and mammalian cells are known (Hammond et al., 2000; Jones et al., 1999; Tuschl et al., 1999; Wassenegger et al., 1994). The regulation by R-loops uncovered here is different. We show that R-loops promote transcription, in contrast to RNA-mediated silencing which regulates transcription negatively. In addition, regulation by R-loops uses just the nascent RNA strands while the RNA-directed DNA silencing requires RNA interference to generate the regulatory RNAs.
Our finding illustrates the importance of nucleic acid structure in gene regulation. R-loops are common; we found them at the promoters of over 3,000 genes. Our results show that they influence gene expression by limiting DNA methylation. We expect that DNMT1 is not the only protein whose activity is influenced by R-loops; thus, there is much to uncover on R-loops’ regulatory function. Recent methods, such as those by Fu and colleagues (Chen et al., 2017) offer an alternate technique for detection of R-loops in addition to the S9.6 antibody, are important steps to facilitate further studies. Most gene regulation studies focus on sequences as sites for binding of regulatory proteins. To obtain a more complete understanding of gene regulation, we must move beyond sequences to include structures and topologies in determining nucleic acid-protein interactions.
Our results also provide mechanistic insights into the role of TGFβ in motor neuron diseases. There is accumulating literature that associates TGFβ with motor neuron function and diseases. However, it has not been clear whether the disruption of TGFβ signaling is the cause or the consequence of motor neuron dysfunction. Here, we showed that the mutation in ALS4 allows methylation of the BAMBI promoter, which alters expression of genes in the TGFβ pathway. The activated genes include ID1, ID2 and IGF1, which play critical roles in neurons (Shah et al., 1996; Vierbuchen et al., 2010). By showing that excessive activation of TGFβ is associated with motor neuron degeneration, our results have implications beyond ALS4 to more common forms of ALS and other motor neuron diseases. Our results illustrate that by altering R-loop levels, the senataxin mutation in ALS4 leads to disruption of transcription and abnormal TGFβ signaling, which might result in motor neuron dysfunction and death.
STAR Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-NeuN | Millipore | Cat# MAB377 |
| Rabbit monoclonal anti p-SMAD 2/3 | Cell Signaling Technology | Cat# 8828 |
| Rabbit polyclonal anti-BMP2 | LSBio | Cat# LS-B12549 |
| Mouse monoclonal S9.6 | A gift from Dr. Stephen Leppla | N/A |
| Rabbit polyclonal anti-RNase H1 | Novus Biologicals | Cat# NBP2-38501 |
| Rabbit polyclonal anti-SETX | Bethyl Laboratories | Cat# A301-104A-M |
| Mouse monoclonal anti-dsDNA | Abcam | Cat# ab27156 |
| Mouse IgG (For DRIP IP) | Santa Cruz | Cat# SC2025 |
| Mouse monoclonal anti-Halo | Promega | Cat# G9211 |
| Rabbit polyclonal anti-BAMBI | ThermoFisher | Cat#PA5-38027 |
| Rabbit polyclonal anti-pSMAD2 | Cell Signaling Tech. | Cat# 3108 |
| Rat monoclonal anti-DNMT1 | Sigma-Aldrich | Cat# MABE306 |
| Mouse monoclonal anti-DNMT3A | Novus | Cat# NB120-13888 |
| Rabbit polyclonal anti-DNMT3B | Sigma-Aldrich | Cat# HPA001595 |
| Biological Samples | ||
| ALS4 patient spinal cord | Johns Hopkins Pathology Department | N/A |
| Control patient spinal cord and brain | Brain endowment Bank at the University of Miami | Miamibrainbank.org |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM medium | Thermo Fisher Scientific | Cat# 11965-084 |
| Fetal Bovine Serum | Atlantic Biologicals | Cat# S11150 |
| Trypsin-EDTA (0.25%) | Thermo Fisher Scientific | Cat# 25200056 |
| Penicillin-Streptomycin-Glutamine (100X) | Thermo Fisher Scientific | Cat# 10378016 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596026 |
| 3M Sodium Acetate | Thermo Fisher Scientific | Cat# 351035721 |
| DAPI | Sigma | Cat# D9546 |
| Lipofectamine 3000 Transfection | Thermo Fisher | Cat# L3000015 |
| Reagent | Scientific | |
| Purified RNase H1 | A gift from Dr. Robert Crouch | N/A |
| Pooled human tissue cDNA | Clontech | Cat# 636742, 636743 |
| Recombinant DNMT1 protein | Active Motif | Cat# 31404 |
| Recombinant DNMT3A protein | Active Motif | Cat# 31406 |
| Recombinant DNMT3B protein | Active Motif | Cat# 31413 |
| RNaseOUT | Thermo Fisher Scientific | Cat# 10777019 |
| biotin-11-ATP | Perkin-Elmer | Cat# NEL544001EA |
| biotin-11-UTP | Perkin-Elmer | Cat# NEL543001EA |
| biotin-11-CTP | Perkin-Elmer | Cat# NEL542001EA |
| biotin-11-GTP | Perkin-Elmer | Cat# NEL545001EA |
| Dynabeads M-280 Streptavidin | Thermo Fisher Scientific | Cat# 11206D |
| 5-Azacytidine (5-Aza-C) | Sigma-Aldrich | Cat# A1287 |
| MEGAscript T7 Transcription kit | Invitrogen | Cat#AM1333 |
| SfcI restriction enzyme | NEB | Cat#R0561S |
| HpaII restriction enzyme | NEB | Cat#R0171S |
| Critical commercial Assays | ||
| RNeasy MinElute Cleanup Kit | Qiagen | Cat# 74204 |
| High Capacity cDNA Reverse Transcriptase kit | Thermo Fisher Scientific | Cat# 4368814 |
| RT2 SYBR Green ROX qPCR Mastermix | Qiagen | Cat# 330521 |
| RT2 First Strand Kit | Qiagen | Cat# 330404 |
| Human TGF-ß/BMP Signaling Pathway RT2 Profiler PCR Array | Qiagen | Cat# PAHS-035ZE-4 |
| TaqMan 2x PCR master mix | Thermo Fisher Scientific | Cat# 4304437 |
| SYBR Safe DNA Gel Stain | Thermo Fisher Scientific | Cat# S33102 |
| FastStart PCR Master | Sigma-Aldrich | Cat# 4710444001 |
| RNase-Free DNase Set | Qiagen | Cat# 79254 |
| DNMT Activity/Inhibition Assay | Active Motif | Cat# 55006 |
| Ovation Ultralow Library system V2 | NuGEN | Cat# 0344 |
| EZ DNA Methylation-Gold™ Kit | Zymo Research | Cat# D5005 |
| TruSeq DNA Methylation Kit | Illumina | Cat# EGMK81312 |
| TruSeq Stranded Total RNA Library Prep Kit | Illumina | RS-122-2201 |
| QuikChange Lighting Site-directed Mutagenesis Kit | Agilent | Cat# 210518 |
| MEGAscript T7 Transcription kit | Thermo Fisher Scientific | Cat# AM1333 |
| Deposited Data | ||
| Deep Sequencing Data | dbGaP | phs001322.v1.p1 |
| Experimental Models: Human cell lines | ||
| Control Primary Fibroblasts | This paper | N/A |
| ALS4 Primary Fibroblasts | This paper | N/A |
| HEK293 cell line | ATCC | Cat#: CRL1573 |
| Oligonucleotides | ||
| TaqMan qPCR assay human BAMBI | Thermo Fisher Scientific | Cat# Hs03044164_m1 |
| TaqMan qPCR assay human HPRT1 | Thermo Fisher Scientific | Cat# Hs02800695_m1 |
| TaqMan qPCR assay human SETX | Thermo Fisher Scientific | Cat# Hs00981128_m1 |
| DNMT1 siRNA | Dharmacon | Cat# A-004605-14; A-004605-16 |
| Non-targeting control siRNA | Dharmacon | Cat# D-001910-10-20 |
| Recombinant DNA | ||
| RNase H1-GFP plasmid | A gift from Dr. Robert Crouch | N/A |
| Halo-SETX plasmid | Promega | Cat# FHC00111 |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Prism 7 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| FASTX-toolkit | Hannon Lab | http://hannonlab.cshl.edu/fastx_toolkit/index.html |
| GSNAP (version 2013-10-28) | Wu and Nacu, 2010 | http://research-pub.gene.com/gmap/ |
| Bismark | Krueger and Andrews, 2011 | https://www.bioinformatics.babraham.ac.uk/projects/bismark/ |
| SAMtools | Li, H. et al 2009 | http://www.htslib.org/ |
| Ingenuity Pathway Analysis | Qiagen | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| Other | ||
| Illumina HiSeq 2500 Platform | Illumina | Cat# SY-401-2501 |
| Avanto 1.5T MRI scanner | Siemens | N/A |
| QuantStudieo 6 Flex Real-Time | Thermo Fisher | Cat# 4485691 |
| PCR system | Scientific | |
| Bioruptor | Diagenode | Cat# B01020001 |
| UV Stratalinker 2400 | Stratagene | Cat# 53274-1 |
| Octet RED96 | ForteBio | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vivian G. Cheung (ude.hcimu@gnuehcgv).
Participant recruitment
Eighteen ALS4 patients were identified by genetic testing and confirmed to have the SETX mutation at codon L389S. 14 related and unaffected control subjects were similarly tested. All subjects received clinical evaluations at the National Institutes of Health in Bethesda, MD under IRB-approved protocol 00-N-0043 “Clinical and Molecular Manifestations of Inherited Neurological Disorders.” Written informed consent was received from all participants prior to inclusion in the study.
Sample collection and processing
All subjects completing the biopsy received 1% lidocaine local anesthesia. Dermal skin fibroblasts were obtained by 3mm punch biopsy of the anterior forearm and subsequently expanded in media containing DMEM with 10% fetal bovine serum and passaged at a 1:3 splitting ratio using Trypsin-EDTA (0.25%) and grown at 37°C with 5% CO2. All experiments were performed on patient cells with less than 10 passages. ALS4 patient post-mortem spinal cords were obtained from a 68 y/o female, 88 y/o female, and 60 y/o male subject affected with typical features of the disease. Control samples were obtained from the Brain Endowment Bank at the University of Miami from healthy controls; 48 y/o female and a 58 y/o male.
MRI Imaging
The patients completed brain and leg imaging in a 1.5-T MR scanner. T1 images of the thigh were reconstructed with a slice thickness of 10 mm. The processing of images was performed as previously described (Kovacs et al., 2016). Briefly, a fuzzy c-means algorithm was used to define muscle and adipose tissue. Cortical bone and bone marrow were segmented out, and adipose tissue was divided into subcutaneous adipose and intermuscular adipose tissue (IMAT). Muscle, fat, and total thigh volumes were calculated from images collected at distance of 6 cm proximal and distal to the midpoint of the thigh.
Immunohistochemistry
Lumbar spinal cord and Brodmann area 4 motor cortex samples were fixed in formalin and paraffin embedded (FFPE). FFPE blocks were then sectioned at 5 μm thickness and dried at room temperature. Sections from each block were stained with Hematoxylin and Eosin (H&E). Subsequently, deparaffinization, antigen retrieval specific to each antibody and immunostaining was performed with a Leica Bond Max IHC automated stainer in the Neuropathology Unit of the Surgical Neurology Branch, NINDS. Tissue slides were stained with 1:500 S9.6 antibody (gift of Dr. Stephen Leppla), 1:100 NeuN (Millipore), 1:1000 pSMAD2/3 (Cell Signaling Technologies), and 1:50 BMP2 (LSBio). RNase H1 treatment was performed overnight in a humidified chamber at 37°C using 40 nM human RNase H1 (gift of R. Crouch Lab, NICHD/NIH) in reaction buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 10 mM MgCl2, 10 mM DTT). Mock digestion was performed in parallel using the same reaction and buffer conditions without RNase H1. Imaging was performed with a Nikon Eclipse Ti inverted microscope. Image quantification analysis using ImageJ was performed blinded with a sampling of 10 fields per sample, and 10 to 15 cells analyzed per field. S9.6 and pSMAD2 were quantified within the nuclei of cells, and total cell area quantified for BMP2 staining.
Expression levels of SETX Isoforms
Primers specific for various isoforms of SETX were designed and ordered from Integrated DNA Technologies. PCR reactions were carried out using FastStart PCR Master Mix (Roche) and analyzed by agarose electrophoresis. Sequence of each PCR product was validated by Sanger Sequencing. Senataxin primers used are as follows: Total SETX (exon 13,14) (F-5′-AAGCCATGTCTCTGTTGGGT 3′, R-5′ GGCAACTGATGGTGAGTGTT-3′), exon 23,25 (F-5′-AGGCCCAGAAGACGATGATT-3′, R-5′-TACTTGGCTCGTGTGATGGT-3′), exon 26,27 (F-5′-TTCAGCCCAGAACCCAAGTA-3′, R-5′-CTCTGGGGCTATGGTAGGAG-3′), and HPRT1 (F-5′-CCTGGCGTCGTGATTAGTGA-3′, R-5′-TCTCGAGCAAGACGTTCAGT-3′). Protein level of total SETX in fibroblasts was measured by western blot using specific antibody (Bethyl).
S9.6 Dot Blot
Genomic DNA containing R-loops was incubated with 1 μl of RNase H1 (gift from Crouch lab) or mock digestion in 1X RNase H reaction buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 10 mM MgCl2, 10 mM DTT) at 37°C for 12 hours. DNA was phenol extracted, ethanol precipitated, and reconstituted in 10 μl TE buffer. 5 μl DNA solution was loaded to Hybond N+ nylon membrane (GE Life Sciences) presoaked with PBS, and crosslinked in UV Stratalinker 2400 (Stratagene) at the “Auto Crosslink” setting (1200 μJoulesX100). The membrane was blocked in 5% milk in PBS-0.1% Tween-20 for one hour and incubated with 1:1000 S9.6 antibody overnight at 4°C to detect RNA-DNA hybrids. A duplicate blot was incubated with anti-dsDNA antibody (ABCAM) as loading control. Double-stranded DNA, double-stranded RNA and RNA-DNA hybrid controls were annealed using synthetic oligos reported previously (Haruki et al, 1997). ssRNA, top strand: 5′- UGGGGGCUCGUCCGGGAUAUGGGAACCACUGAUCCC-3′; ssDNA, top strand: 5′-TGGGGGCTCGTCCGGGATATGGGAACCACTGATCCC-3′; ssDNA, bottom strand: 5′-GGGATCAGTGGTTCCCATATCCCGGACGAGCCCCCA-3′; ssRNA, bottom strand: 5′-GGGAUCAGUGGUUCCCAUAUCCCGGACGAGCCCCCA-3′.
Immunofluorescence
Fibroblasts were fixed with 4% paraformaldehyde for 15 minutes at room temperature, then washed three times with phosphate-buffered saline (PBS). Slides were then placed in blocking solution (5% normal goat serum, 0.3% Triton X-100 in PBS) for one hour at room temperature. Primary antibody staining was done at 4°C overnight in PBS with 1% bovine serum albumin and 0.3% Triton X-100 using 1:500 S9.6 antibody, 1:500 pSMAD2 antibody, or 20 μg/ml of BAMBI polyclonal antibody. Slides were then washed three times with PBS for 5 minutes each, incubated with 1:500 secondary antibody (Invitrogen) for two hours at room temperature in the dark, and then washed three times with PBS for 5 minutes each before DAPI nuclear staining. Imaging was performed with a Leica DMI 6000CS laser confocal microscope with a Leica HCX PL APO 40X oil objective. Quantification of staining was performed blinded using ImageJ quantification. For S9.6 and pSMAD2 staining, only nuclear signal that co-localizes with the DAPI was quantified. For BAMBI staining, total cell area was quantified. 10 fields of view were analyzed for each sample with 15-20 cells per field for S9.6 and 8 fields of view with 15-20 cells per field for pSMAD2 and BAMBI.
mRNA expression analysis
Primary fibroblasts were homogenized in 500 μl TRIzol reagent (Invitrogen) and centrifuged for 10 min at 4°C. 100 μl chloroform was added to the supernatant, and the mixture was vortexed and spun down again. The aqueous phase was transferred to a separate tube with 250 μl isopropanol and 50 μl 3 M sodium acetate. The RNA was precipitated, washed with 70% ethanol, and resuspended in water. The sample was then purified using the RNA-clean up kit (Qiagen). 1 μg total RNA was converted to cDNA using the High Capacity cDNA Reverse Transcriptase kit (Applied Biosystems).
Human TGFβ/BMP Signaling Pathway RT2 Profiler PCR Array (PAHS-035Z, Qiagen) was used to measure gene expression. RNA was purified from fibroblasts of five controls and five ALS4 patients. 500 ng purified RNA was reverse transcribed using RT2 First Strand Kit (Qiagen) protocol. cDNA was measured on the array according to manufacturer protocol for RT2 Profiler PCR Array. Each sample was measured in duplicates and the average Ct value was normalized to that of the housekeeping gene RPLP0. Fold change was calculated based on the average of the normalized Ct values. To further validate gene expression level of BAMBI, TaqMan probe (Hs03044164_m1) was used to determine the mRNA levels of BAMBI. HPRT1 TaqMan probe (Hs02800695_m1) was used as a housekeeping gene control. The qPCR was run on QuantStudio 6 Flex Real-Time PCR System (ThermoFisher Scientific). Each sample was measured on same plate in triplicates, and the average Ct value of the three technical replicates was determined for each sample. Pooled human tissue RNA for SETX expression characterization was obtained from Clontech and evaluated using TaqMan probe (Hs00981128_m1).
Sequencing libraries were prepared from total RNA from 3 patients and 3 controls using TruSeq Stranded Total RNA Library Prep Kit (Illumina). Sequencing was performed on Illumina HiSeq 2500 and >150 million 100-nt reads were generated from each sample. Low-quality bases were trimmed from the 3′ end of reads and 3′ adapter was trimmed using FASTQ/A Clipper with default settings (Hannon lab). Reads shorter than 35 bp were excluded from analysis. Sequencing reads were aligned to human reference (HG18) using GSNAP (Version 2013-10-28) (Wu and Nacu, 2010) using the following parameters: Mismatches ≤ [(read length+2)/12-2]; Mapping score ≥ 20; Soft-clipping on (-trim-mismatch-score=−3). Reads counts from each samples were normalized to total number of mapped reads. Gene expression levels (RPK) were then calculated using the RUV (remove unwanted variation) method (Risso et al., 2014). Genes with RPK>5 were defined as “expressed genes”. Expressed genes with fold differences (ALS4/control) ≥1.5 or ≤0.67 were identified as differentially expressed genes. Gene ontology analysis was carried out using Ingenuity Pathway Analysis (IPA) software (Qiagen).
GC content analysis
GC content at promoters was analyzed similarly to Ginno et al, 2012. Promoter sequence (TSS±500bp) of 23,682 genes (including BAMBI) was downloaded from RefSeq, excluding genes that are within 1kb of neighboring genes. GC content in a 50bp sliding window for each promoter was computed: GC% = [(G+C)/(G+C+A+T)]%; GC skew = (G−C)/(G+C). The GC% and GC skew at the BAMBI promoter, and their median levels (+/− 10 %) for 23,682 gene promoters, are shown in Figure 4A.
RNase H1 and SETX overexpression
Primary human fibroblasts were trypsinized and seeded at 80% confluency for 18 hours before transfection. Overexpression plasmid of GFP-tagged human RNase H1 (gift from Crouch lab) or vector expressing GFP alone was transfected into human primary fibroblasts using Lipofectamine 3000 reagent (Invitrogen). Cells were harvested 24~72 hours post transfection. Overexpression of RNase H1 was validated by quantitative PCR and western blot (anti-RNase H1 from Novus). Wildtype SETX was cloned in overexpression plasmids with N-terminal or C-terminal Halo tag (Promega). The L389S mutation of SETX was generated by site-directed mutagenesis (QuikChange kit, Agilent). Sequence of all constructs was confirmed by Sanger sequencing. Overexpression plasmids of WT and L389S senataxin were transfected to HEK293 cells. Protein expression level of SETX was quantified by western blot using Halo antibody (Promega). Cells harvested 48 hours post transfection were analyzed by immunofluorescence and dot blot using S9.6 antibody. Overexpression plasmids with N-terminal or C-terminal Halo tag showed similar results. Data from C-terminal Halo tag plasmids were shown.
DNMT1 and BAMBI siRNA knockdown
Primary fibroblasts were seeded at 2 × 105 per well in 6-well dishes. Cells were transfected with 20 nM siRNA against DNMT1 (pool of Dharmacon A-004605-14 and A-004605-16) or non-targeting control siRNA (Dharmacon D-001910-10-20) using Lipofectamine RNAiMax. Cells were harvested for expression analysis 72 hrs post-transfection. For experiments with RNase H1 overexpression, cells were transfected with GFP-RNase H1 24 hrs after siRNA transfection, and grown for an additional 48 hrs prior to harvest. For BAMBI knockdown, primary fibroblasts were seeded at the density above and transfected with siRNA against BAMBI (pool of Dharmacon A-019596-25, A-019596-26, A-019596-27, and A-019596-28) or non-targeting control siRNA using Lipofectamine RNAiMax. Cells were transfected with 10 nM siRNA at time 0 and 24 hours, and then collected at 48 hours. Fibroblasts were fixed with 4% paraformaldehyde for immunofluorescence analysis or collected in TRIzol for RNA extraction and RT-PCR analysis.
DNA-RNA hybrid immunoprecipitation (DRIP)
Immunoprecipitation procedure was adapted from previous studies (Skourti-Stathaki et al., 2011). 5×106 primary fibroblasts were lysed in 600 μl cell lysis buffer (50 mM PIPES, pH 8.0, 100 mM KCl, 0.5% NP-40) and nuclei were collected by centrifugation. Nuclei pellet was resuspended in 300 μl nuclear lysis buffer (25 mM Tris-HCl, pH 8.0, 1% SDS, 5mM EDTA). Genomic DNA, along with R-loops was then extracted by phenol:chloroform and precipitated by ethanol. Purified DNA was resuspended in IP dilution buffer (16.7 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.01% SDS, 1% Triton-X100, 167 mM NaCl) and sonicated 15 minutes in Bioruptor (Hi setting, 30sec on/30sec off) to fragments with average size of 500 bp. 3 μg of S9.6 monoclonal antibody (gift from Dr. Stephen H. Leppla at NIH) or non-specific mouse IgG (Santa Cruz) was used for each immunoprecipitation. Input and precipitates were analyzed by quantitative PCR using primers. BAMBI promoter: F-5′ CCCTGGGGGTTCCAGAAAAC 3′; R-5′ CTCGGGTGGTCAGCGTTC 3′ BAMBI intron: F-5′ TTACAGAGGGCTGCACGATG 3′; R-5′ AGAAACGGCTTCCACCTACC 3′. Control primers for known R-loops: ACTB R-loops: F-5′ TTACCCAGAGTGCAGGTGTG 3′; F-5′ CCCCAATAAGCAGGAACAGA 3′; ACTB intron: F-5′ CGGGGTCTTTGTCTGAGC; R-5′ CAGTTAGCGCCCAAAGGAC.
Sequencing libraries were prepared from input and DRIP DNA using Ovation Ultralow System (NuGen) and sequenced on Illumina HiSeq 2500 platform. An average of 100 million 100 nt reads per sample were generated. Sequencing reads were pre-processed to remove adapter sequences from the end of reads using the program fastx_clipper from FASTX-Toolkit (Hannon Lab). Low-quality sequences at ends of reads represented by stretches of “#” in the quality score string in FASTQ file were also removed. Reads shorter than 35 nt after trimming were excluded from analysis. Sequencing reads were then aligned to human reference HG18 using GSNAP (Version 2013-10-28) (Wu and Nacu, 2010) using the following parameters: Mismatches ≤ [(read length+2)/12−2]; Mapping score ≥ 20; Soft-clipping on (-trim-mismatch-score=−3). Reads with identical sequences were compressed into one unique sequence. We focused on R-loop signal at promoter (500 nt upstream to 500 nt downstream of transcription start site). Read counts were normalized as number of reads per million of uniquely mapped reads (RPM). Fold enrichment = (RPM in DRIP/RPM in input). An R-loop peak was determined as a region larger than 100 nt and ≥2 fold enrichment.
To obtain distribution profile across the whole gene, metagene analysis was carried out as previously described with modification (Kwak et al., 2013). Specifically, each gene was scaled to a “3kb-long metagene”, with promoter region defined as TSS-500bp to TSS+500bp, 3′ UTR defined as 3′end CPS+1kb, and gene body of all genes with various length scaled to 1kb. Genes that are less than 1kb in length or are closer than 1kb to nearby genes were excluded from analysis. Each gene is divided into 300 bins (100 bins each for promoter, gene body and 3′UTR). Number of sequencing reads mapped within each bin was normalized by total million number of mapped reads (RPM). Sequencing data from IP and input were computed by same method and were shown in parallel. For the 1,386 genes with GC-rich promoters, RPM within each bin was computed and plotted in Figure 7A. For metagene profile in Figure 4C, RPM value of BAMBI was shown. A smoothing window of 200 bp was used in plotting.
DNMT1 Chromatin Immunoprecipitation (ChIP)
ChIP was performed in skin fibroblasts with chromatin crosslinked in 1% formaldehyde in PBS for 10 min at room temperature. Nuclei were isolated by rotating crosslinked cells for 10 min at 4°C in 5 ml lysis buffer 1 (50 mM Hepes pH 7.6, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) followed by 10 min rotating in 5 ml lysis buffer 2 (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris, pH 8). Nuclei swelled in lysis buffer 3 (10 mM Tris, pH 8, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 0.1% deoxycholic acid, 10% N-lauryl sarcosine) for 10 min, then were sonicated to shear chromatin to < 500 bp in a Diagenode Bioruptor. 50 μg sheared chromatin was immunoprecipitated in RIPA buffer (50 mM Tris, pH8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with 5 μg of antibody against DNMT1 (Millipore) or IgG (Biolegend). Bound DNA was recovered with 40 μl protein G beads, then washed in low salt RIPA (150 mM NaCl) and high salt (500 mM NaCl) RIPA, then eluted twice in 50 μl elution buffer (1% SDS, 0.1 M sodium bicarbonate). Eluates were combined and crosslinks reversed overnight at 65°C. Samples were treated with proteinase K (0.5 μg/μl) at 55°C for 2 hours, then purified over Qiagen PCR purification columns. Isolated DNA was quantified by qPCR with values reported as average ±SEM of triplicate or quadruplicate quantification. Primers. BAMBI promoter: F-5′ CCCTGGGGGTTCCAGAAAAC 3′; R-5′ CTCGGGTGGTCAGCGTTC 3′; BAMBI intron: F-5′ TTACAGAGGGCTGCACGATG 3′; R-5′ AGAAACGGCTTCCACCTACC 3′.
Nuclear Run-On Assay (NRO)
Nuclear Run-On was carried out as previously reported (Kwak et al., 2013). Briefly, 2×107 primary fibroblasts were washed with 20 ml of ice-cold PBS and resuspended in 10 ml lysis buffer [20 mM Tris-HCl pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.5% Igepal CA-630, 1X Complete Protease Inhibitor Cocktail (Roche) and 4 units/ml RNAseOUT (Invitrogen)]. Cell suspension was incubated on ice for 10 minutes before nuclei were centrifuged by 500 × g for 1 min at 4°C. Pellets containing nuclei were washed carefully with 10 ml ice-cold lysis buffer, collected by centrifugation (500 × g, 1 min, 4°C). 5×106 nuclei were added to the same volume of 2X Nuclear Run-On reaction mixture (10 mM Tris-HCl pH 8.0, 300 mM KCl, 1% Sarkosyl, 5 mM MgCl2, 1 mM DTT, 0.03 mM each of biotin-11-A/C/G/UTP (Perkin-Elmer), 0.4 U/μl RNase inhibitor) and incubated for 3 min at 37°C. Nascent RNA was extracted using Trizol and precipitated in ethanol. Extracted nascent RNA was fragmented by base hydrolysis in 0.2 N NaOH on ice for 15 min, and neutralized by 0.68X volume of 1 M Tris-HCl pH 6.8. Fragmented nascent RNA was purified using 30 μl of Streptavidin M-280 magnetic beads (Invitrogen) following the manufacturer’s instructions three times. NRO transcripts were quantified by RT-quantitative PCR using the following primers. BAMBI: F-5′-GCGCGGAGCCGGAGAGAC-3′; R-5′-CAGCTCCCGCCCGCGCCA 3′. ABL1: F-5′-CGAGGGCGTGTGGAAGAA-3′; R-5′ GTCTTCACGGCCACCGTC-3′.
MethylC-Seq
100 ng genomic DNA purified from primary fibroblasts was incubated with 130 μl CT conversion reagent (Zymo Research, EZ DNA Methylation-Gold Kit) at 98°C for 10 min and 64°C for 2.5 hours. Bisulfite-treated DNA was purified by Zymo-Spin IC Column and eluted in 10 μl elution buffer. Bisulfite-treated DNA was ligated to adapters and amplified following Illumina protocol (Tru-seq DNA Methylation Kit). To analyze effect of 5-Aza-C, primary human fibroblasts were treated with 10 μM 5-Aza-C (Sigma) or mock buffer for 24 hours before harvest. Treated cells were then analyzed by MethylC-Seq.
MethylC-seq libraries were sequenced on Illumina HiSeq 2500 platform. Low-quality bases were trimmed from the 3′ end of reads and 3′ adapter was trimmed using FASTQ/A Clipper with default settings (Hannon lab). Reads shorter than 35 bp were discarded. Sequencing reads were aligned to human reference (HG18) by Bismark (Krueger and Andrews, 2011) using default parameters. For each samples from the 5 patient cells and 5 normal controls, 120 ~ 150 million reads were uniquely aligned. We focused analysis on gene promoters (500 nt upstream to 500 nt downstream of TSS) and counted reads that were aligned to the promoter regions using SAMtools. Average sequencing depth of cytosines in the promoter regions are is 5.8×. The sequence coverage for the samples in RNaseH time course experiment is very similar, and the same analysis method was used. Methylation level at each cytosine was calculated as the ratio between the number of reads with methylated cytosine and total number of reads i.e. those with methylated and non-methylated cytosine: Methyl-C level (%) = (#methylated Cytosine)/(#all Cytosine). Average methyl-C levels of 5 controls and 5 ALS4 patients at each position within BAMBI promoter were calculated and plotted (Figure 5A, top panel). Methyl-C levels of each individual were also shown in a heatmap (Figure 5A, bottom panel): the BAMBI promoter region (TSS±500bp) was divided into 100 bins of 10-bp size; then, the average methyl-C level within each bin was computed, and shown in a heatmap; each row represents an individual.
dsDNA and RNA/DNA hybrids representing BAMBI promoter
The biotinylated and non-biotinylated BAMBI dsDNA 600-mers were prepared using the following primers:
BAMBI_1T7DNMT_F: GAATTTAATACGACTCACTATAGGGGCACCCAAAAGAAGCCCC
BAMBI_1DNMT_R: CCCAGGAGCCCAGAAAAGTT
BAMBI_1DNMT_bio_R: biotin-CCCAGGAGCCCAGAAAAGTT
The 600 nt RNA transcript was generated from an aliquot of the BAMBI dsDNA using the MEGAscript T7 Transcription kit (ThermoFisher Scientific). In another aliquot of the dsDNA, the ~20 bp T7 RNA polymerase promoter was removed by SfcI digestion (NEB, catalog # R0561S), and this dsDNA and the transcribed RNA were denatured, and then annealed in 10 mM Tris-HCl pH 8.0, 1 mM EDTA, 50 mM NaCl by holding at 95°C for 5 min, followed by slow cooling. Any reannealed dsDNA was removed by HpaII digestion (NEB, catalog # R0171S) and the RNA/DNA duplex was isolated using agarose gel electrophoresis.
DNMT1 Assay and EMSA
A commercial DNMT Activity/Inhibition Assay (Active Motif, #55006) was used to quantify DNMT1 binding and activity. In this assay, we pre-incubated dsDNA or RNA/DNA hybrid with recombinant DNMT1, and then measured the methylation of CpG-rich sequences that were immobilized on 96-well plates. DNMT1 transfers the methyl group from S-Adenosyl methionine (SAM) supplied in the reaction to the immobilized DNA, and the methylated cytosine was quantified by ELISA with a specific methyl-binding protein and HRP-conjugated antibody. 500 ng of recombinant DNMT1 (Active Motif) was incubated with a titration of dsDNA or RNA/DNA hybrid at 25°C for 30 min in 10 μl binding buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl). Methylation level of CpG nucleotides by DNMT1 was measured on a spectrophotometer at 450 nm following manufacturer’s manual. Each reaction was carried out in triplicate. DNMT1 activity on dsDNA or RNA/DNA hybrid was calculated as Activity = 1-(OD450 of sample with competitor - OD450 background)/(OD450 sample without competitor – OD450 background). Average and SEM of triplicated assay were reported.
Electrophoretic mobility shift assay was carried out using biotinylated dsDNA or RNA/DNA hybrids with the sequence described above. Serial titrations of recombinant DNMT1 were incubated with 0.1 nM of biotinylated nucleotide substrates in DNMT1 binding buffer (5 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 1 mM DTT, 3% (v/v) glycerol, 100 mM NaCl) in a 10 μl reaction at 25°C for 30 min. For the competition assay, 100 nM of non-biotinylated dsDNA was added to the appropriate tubes and incubated for 15 min before biotinylated substrates were added. The 10 uL samples were mixed with 1.1 uL of Novex Hi-Density TBE Sample buffer (5X) and loaded to 6% DNA retardation gel (ThermoFisher, #EC6365BOX) and run at 100V in 0.5X TBE at 4°C for 1.5 hours. The gel was blotted to Hybond N+ nylon membrane in 0.5X TBE at 4°C at 15V for 17 hours. Biotin signal was detected using Chemiluminescent Nucleic Acid Detection Kit (ThermoFisher, #89880).
Biolayer Interferometry
Analysis of dsDNA or RNA/DNA hybrid binding to DNMT1 was carried out using the Octet RED96 system (ForteBio) with sensor detection of the change in wavelength (nm shift). Biotinylated dsDNA or RNA/DNA hybrid at concentrations of 0.625, 1.25, 2.5, and 5nM were immobilized onto a Streptavidin-SA biosensor. Double-stranded DNA, double-stranded RNA and RNA-DNA hybrid controls were annealed using synthetic oligos reported previously (Haruki et al, 1997). ssRNA, top strand: 5′-UGGGGGCUCGUCCGGGAUAUGGGAACCACUGAUCCC-3′; ssDNA, top strand: 5′-TGGGGGCTCGTCCGGGATATGGGAACCACTGATCCC-3′; ssDNA, bottom strand: 5′-GGGATCAGTGGTTCCCATATCCCGGACGAGCCCCCA-3′; ssRNA, bottom strand: 5′-GGGAUCAGUGGUUCCCAUAUCCCGGACGAGCCCCCA-3′. dsDNA and RNA/DNA hybrid were loaded onto the sensors until saturation. The nucleotide-labeled sensors were then washed with buffer, followed by addition of DNMT1 at concentrations of 5.25, 10.5, 42, and 84 nM. All reactions were tested in TBS buffer (10 mM Tris, pH 7.4, 68 mM NaCl, 0.02% Tween-20). A reference sample of buffer and DNMT1 alone did not show any signal drift. Association and dissociation were monitored for 10 minutes each. All experiments were conducted in the Octet instrument with agitation at 1,000 rpm.
Quantification and Statistical Analysis
Analysis of data from deep sequencing and other experiments was carried out using programs described in “Key resources table” and corresponding sections in STAR methods. Experimental results from replicates are shown as mean ± SEM. In each of the box-whisker plots, middle line of the box represents median value, and upper and lower boundaries of the box represent first and third quartiles. Length of whiskers represents 1.5 times of interquartile range. Student’s t-test was used to compare means. In figures where significance is represented by asterisks, * stands for p < 0.05; ** for p < 0.01, and *** for p < 0.001.
Data Availability
The deep sequencing data reported in this paper are deposited in dbGaP via accession number phs001322.v1.p1.
Supplementary Material
1
Table S1. Genes in TGFβ pathway that are differentially expressed between ALS4 patients and controls (Related to Figure 3A).
2
Table S2. Genes with R-loops in GC-rich promoter regions (Related to Figure 7A).
Acknowledgments
We thank Dr. David Cornblath for patient referrals, insightful discussions and suggestions on ALS4, Dr. Susana Cerritelli & Dr. Robert Crouch for the expression clone of human RNase H1, their expert advice and enthusiastic discussions of R-loops, Dr. Stephen Leppla for the S9.6 antibody and advice on R-loop detection. We thank Ms. Yaojuan Liu and Ms. Jennifer Fox for technical help. We thank Dr. Story Landis for review of and comments on the manuscript. The authors thank Ms. Elizabeth Hartnett for her help in scheduling subjects during the study, the clinical staff of the outpatient NIH neurology clinic for their assistance during the subjects’ visits. Lastly, the authors thank the research subjects who inspire and encourage us, and their participation made this study possible. This work is supported by funds from the Howard Hughes Medical Institute and the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
C.G., I.X.W., R.D.G., J.A.W, A.B., and V.G.C. designed the experiments. C.G., I.X.W., R.D.G., J.A.W., T.L., A.B.S., N.E., A.R.C., A.B., T.L., B.C. performed the experiments and collected the patient data. C.G., I.X.W., R.D.G., J.A.W., Z.Z., J.T.B., K.C., A.R.C., J.Y., K.H.F., and V.G.C. analyzed the data. C.G., I.X.W., K.H.F. and V.G.C., wrote the first draft of the paper; all authors edited the manuscript.Competing Financial Interests: The authors declare no competing interests.
References
- Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. [Abstract] [Google Scholar]
- Baumann P, Cech TR. Pot1, the Putative Telomere End-Binding Protein in Fission Yeast and Humans. Science. 2001;292:1171–1175. [Abstract] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA. RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986;89:123–130. [Abstract] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. [Europe PMC free article] [Abstract] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PYT, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10:e1004288. [Europe PMC free article] [Abstract] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. [Abstract] [Google Scholar]
- Chen L, Chen JY, Zhang X, Gu Y, Xiao R, Shao C, Tang P, Qian H, Luo D, Li H, et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol Cell. 2017;68:745–757.e5. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. [Europe PMC free article] [Abstract] [Google Scholar]
- Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science. 1998;281:706–709. [Abstract] [Google Scholar]
- DeMarini DJ, Winey M, Ursic D, Webb F, Culbertson MR. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2154–2164. [Europe PMC free article] [Abstract] [Google Scholar]
- Drolet M, Phoenix P, Menzel R, Massé E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A. 1995;92:3526–3530. [Europe PMC free article] [Abstract] [Google Scholar]
- El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10:e1004716. [Europe PMC free article] [Abstract] [Google Scholar]
- ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. [Europe PMC free article] [Abstract] [Google Scholar]
- Endo F, Komine O, Fujimori-Tonou N, Katsuno M, Jin S, Watanabe S, Sobue G, Dezawa M, Wyss-Coray T, Yamanaka K. Astrocyte-derived TGF-β1 accelerates disease progression in ALS mice by interfering with the neuroprotective functions of microglia and T cells. Cell Rep. 2015;11:592–604. [Abstract] [Google Scholar]
- Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, et al. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. [Abstract] [Google Scholar]
- Finkel JS, Chinchilla K, Ursic D, Culbertson MR. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics. 2010;184:107–118. [Europe PMC free article] [Abstract] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. [Europe PMC free article] [Abstract] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, Chédin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–1600. [Europe PMC free article] [Abstract] [Google Scholar]
- Groh M, Lufino MMP, Wade-Martins R, Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 2014;10:e1004318. [Europe PMC free article] [Abstract] [Google Scholar]
- Groh M, Albulescu LO, Cristini A, Gromak N. Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. J Mol Biol. 2017;429:3181–3195. [Abstract] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. [Abstract] [Google Scholar]
- Hamperl S, Cimprich KA. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair. 2014;19:84–94. [Europe PMC free article] [Abstract] [Google Scholar]
- Haruki M, Noguchi E, Kanaya S, Crouch RJ. Kinetic and stoichiometric analysis for the binding of Escherichia coli ribonuclease HI to RNA-DNA hybrids using surface plasmon resonance. J Biol Chem. 1997;272:22015–22022. [Abstract] [Google Scholar]
- Houi K, Kobayashi T, Kato S, Mochio S, Inoue K. Increased plasma TGF-beta1 in patients with amyotrophic lateral sclerosis. Acta Neurol Scand. 2002;106:299–301. [Abstract] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. [Abstract] [Google Scholar]
- Katsuno M, Adachi H, Minamiyama M, Waza M, Doi H, Kondo N, Mizoguchi H, Nitta A, Yamada K, Banno H, et al. Disrupted transforming growth factor-beta signaling in spinal and bulbar muscular atrophy. J Neurosci Off J Soc Neurosci. 2010;30:5702–5712. [Europe PMC free article] [Abstract] [Google Scholar]
- Khin SS, Kitazawa R, Win N, Aye TT, Mori K, Kondo T, Kitazawa S. BAMBI gene is epigenetically silenced in subset of high-grade bladder cancer. Int J Cancer. 2009;125:328–338. [Abstract] [Google Scholar]
- Kim HD, Choe J, Seo YS. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry (Mosc) 1999;38:14697–14710. [Abstract] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinforma Oxf Engl. 2011;27:1571–1572. [Europe PMC free article] [Abstract] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. [Europe PMC free article] [Abstract] [Google Scholar]
- Lin Y, Dent SYR, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A. 2010;107:692–697. [Europe PMC free article] [Abstract] [Google Scholar]
- Macías-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. [Abstract] [Google Scholar]
- Martinou JC, Le Van Thai A, Valette A, Weber MJ. Transforming growth factor beta 1 is a potent survival factor for rat embryo motoneurons in culture. Brain Res Dev Brain Res. 1990;52:175–181. [Abstract] [Google Scholar]
- Martin-Tumasz S, Brow DA. Saccharomyces cerevisiae Sen1 Helicase Domain Exhibits 5′- to 3′-Helicase Activity with a Preference for Translocation on DNA Rather than RNA. J Biol Chem. 2015;290:22880–22889. [Europe PMC free article] [Abstract] [Google Scholar]
- Marwitz S, Depner S, Dvornikov D, Merkle R, Szczygieł M, Müller-Decker K, Lucarelli P, Wäsch M, Mairbäurl H, Rabe KF, et al. Downregulation of the TGFβ Pseudoreceptor BAMBI in Non-Small Cell Lung Cancer Enhances TGFβ Signaling and Invasion. Cancer Res. 2016;76:3785–3801. [Abstract] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Németh AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schöls L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–227. [Abstract] [Google Scholar]
- Nakamura M, Ito H, Wate R, Nakano S, Hirano A, Kusaka H. Phosphorylated Smad2/3 immunoreactivity in sporadic and familial amyotrophic lateral sclerosis and its mouse model. Acta Neuropathol (Berl) 2008;115:327–334. [Abstract] [Google Scholar]
- Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci U S A. 2010;107:651–656. [Europe PMC free article] [Abstract] [Google Scholar]
- O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. [Europe PMC free article] [Abstract] [Google Scholar]
- Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massagué J, Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. [Abstract] [Google Scholar]
- Phatnani HP, Guarnieri P, Friedman BA, Carrasco MA, Muratet M, O’Keeffe S, Nwakeze C, Pauli-Behn F, Newberry KM, Meadows SK, et al. Intricate interplay between astrocytes and motor neurons in ALS. Proc Natl Acad Sci U S A. 2013;110:E756–765. [Europe PMC free article] [Abstract] [Google Scholar]
- Pruss GJ, Manes SH, Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982;31:35–42. [Abstract] [Google Scholar]
- Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Nichol Edamura K, Wang YH, Pearson CE. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39:1749–1762. [Europe PMC free article] [Abstract] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. [Europe PMC free article] [Abstract] [Google Scholar]
- Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. [Abstract] [Google Scholar]
- Shah NM, Groves AK, Anderson DJ. Alternative neural crest cell fates are instructively promoted by TGFbeta superfamily members. Cell. 1996;85:331–343. [Abstract] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. [Europe PMC free article] [Abstract] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. [Europe PMC free article] [Abstract] [Google Scholar]
- Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–522. [Europe PMC free article] [Abstract] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. [Abstract] [Google Scholar]
- Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chédin F, Swigut T, Cimprich KA. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. ELife. 2016;5 [Europe PMC free article] [Abstract] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. [Europe PMC free article] [Abstract] [Google Scholar]
- Urich MA, Nery JR, Lister R, Schmitz RJ, Ecker JR. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nat Protoc. 2015;10:475–483. [Europe PMC free article] [Abstract] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. [Europe PMC free article] [Abstract] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016;30:1327–1338. [Europe PMC free article] [Abstract] [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sänger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. [Abstract] [Google Scholar]
- Winey M, Culbertson MR. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988;118:609–617. [Europe PMC free article] [Abstract] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. [Abstract] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinforma Oxf Engl. 2010;26:873–881. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. [Abstract] [Google Scholar]
- Yüce Ö, West SC. Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol. 2013;33:406–417. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.molcel.2017.12.030
Read article for free, from open access legal sources, via Unpaywall:
https://www.cell.com/article/S1097276517310092/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The hidden architects of the genome: a comprehensive review of R-loops.
Mol Biol Rep, 51(1):1095, 26 Oct 2024
Cited by: 0 articles | PMID: 39460836
Review
ALKBH5 modulation of ferroptosis in recurrent miscarriage: implications in cytotrophoblast dysfunction.
PeerJ, 12:e18227, 18 Oct 2024
Cited by: 0 articles | PMID: 39434797 | PMCID: PMC11493020
The Thermodynamic and Kinetic Properties of the dA-rU DNA-RNA Hybrid Base Pair Investigated via Molecular Dynamics Simulations.
Molecules, 29(20):4920, 17 Oct 2024
Cited by: 0 articles | PMID: 39459288 | PMCID: PMC11510705
R-loops' m6A modification and its roles in cancers.
Mol Cancer, 23(1):232, 18 Oct 2024
Cited by: 0 articles | PMID: 39425197 | PMCID: PMC11487993
Review Free full text in Europe PMC
Human SKI component SKIV2L regulates telomeric DNA-RNA hybrids and prevents telomere fragility.
iScience, 27(11):111096, 04 Oct 2024
Cited by: 0 articles | PMID: 39493885 | PMCID: PMC11530851
Go to all (115) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
HPA - The Human Protein Atlas
- (1 citation) HPA - HPA001595
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mutation in senataxin alters the mechanism of R-loop resolution in amyotrophic lateral sclerosis 4.
Brain, 145(9):3072-3094, 01 Sep 2022
Cited by: 22 articles | PMID: 35045161 | PMCID: PMC9536298
SETX (senataxin), the helicase mutated in AOA2 and ALS4, functions in autophagy regulation.
Autophagy, 17(8):1889-1906, 07 Aug 2020
Cited by: 29 articles | PMID: 32686621 | PMCID: PMC8386630
Protein interaction analysis of senataxin and the ALS4 L389S mutant yields insights into senataxin post-translational modification and uncovers mutant-specific binding with a brain cytoplasmic RNA-encoded peptide.
PLoS One, 8(11):e78837, 11 Nov 2013
Cited by: 13 articles | PMID: 24244371 | PMCID: PMC3823977
Role of Senataxin in Amyotrophic Lateral Sclerosis.
J Mol Neurosci, 73(11-12):996-1009, 20 Nov 2023
Cited by: 2 articles | PMID: 37982993
Review
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIDDK NIH HHS (1)
Grant ID: T32 DK007378