Abstract
Free full text

Unravelling the molecular basis for regulatory T‐cell plasticity and loss of function in disease
Abstract
Regulatory T cells (Treg) are critical for preventing autoimmunity and curtailing responses of conventional effector T cells (Tconv). The reprogramming of T‐cell fate and function to generate Treg requires switching on and off of key gene regulatory networks, which may be initiated by a subtle shift in expression levels of specific genes. This can be achieved by intermediary regulatory processes that include microRNA and long noncoding RNA‐based regulation of gene expression. There are well‐documented microRNA profiles in Treg and Tconv, and these can operate to either reinforce or reduce expression of a specific set of target genes, including FOXP3 itself. This type of feedforward/feedback regulatory loop is normally stable in the steady state, but can alter in response to local cues or genetic risk. This may go some way to explaining T‐cell plasticity. In addition, in chronic inflammation or autoimmunity, altered Treg/Tconv function may be influenced by changes in enhancer–promoter interactions, which are highly cell type‐specific. These interactions are impacted by genetic risk based on genome‐wide association studies and may cause subtle alterations to the gene regulatory networks controlled by or controlling FOXP3 and its target genes. Recent insights into the 3D organisation of chromatin and the mapping of noncoding regulatory regions to the genes they control are shedding new light on the direct impact of genetic risk on T‐cell function and susceptibility to inflammatory and autoimmune conditions.
Introduction: The Treg phenotype
A healthy immune system must balance robust reactivity towards pathogens with limited reactivity, or tolerance, to self‐antigens, commensal bacteria and food/environmental antigens.1 The effector arm of the adaptive immune system comprises CD8 and CD4 cells, defined by their function and ability to respond to antigens presented on either MHC class 1 or 2, respectively. Within the CD4+ T‐cell compartment, there are specific effector subsets that are capable of responding to defined antigens in specific locations, and antigen specificity is determined by the TCR repertoire. This TCR repertoire is generated during CD4+ commitment and differentiation in the thymus in a process that includes deletion of (strongly) self‐reactive TCR bearing T cells. Despite this, there is the possibility of self‐reactive T cells being released into the periphery. To manage this, special subsets of CD4+ T cells called regulatory T cells (Treg) are also generated, both in the thymus and in the periphery. While both natural thymic Treg or nTreg, and induced or peripheral Treg are similar in function, they have different origins and roles in regulating other immune cells.2, 3 In a general sense, Treg act as ‘policemen’ of the immune system to limit rogue immune activity, and this role in immune homeostasis is critical. In addition to regulating antigen‐specific immune responses, Treg are capable of regulating cell function in an antigen‐independent manner.4
Treg play a central role in immune regulation by actively controlling the proliferation and activation of cells of both the adaptive and innate immune systems using multiple mechanisms.1 Treg use multiple suppressor mechanisms, and the mechanism is likely to differ according to the physiological and inflammatory states encountered.1, 5 Failure of tolerance can lead to autoimmunity and chronic inflammation, and the loss of tolerance can be caused by defects in Treg function or insufficient Treg numbers, or by unresponsive or overactivated effector cells.
The regulatory T‐cell first gained widespread recognition as a T‐cell lineage upon the demonstration by Sakaguchi et al.6 that stable expression of the IL2 receptor (CD25) tracks with suppressor function in CD4+ T cells. Many groups have since investigated this lineage, and it is now clear that while CD25 expression is strongly upregulated on Treg, it is also a transient activation marker for CD4+ T‐cells without regulatory function, so is not an exclusive Treg marker. The second major discovery in this field was that the Treg lineage is dependent on the expression of the transcription factor FOXP3, which controls a gene regulatory network essential for suppressor function. A third significant discovery was the existence of two Treg populations of different origin: natural or thymic Treg emerge from the thymus fully mature and stably expressing FOXP3. In contrast, peripheral or induced Treg arise from naïve T‐cells which do not express FOXP3 until stimulated in the presence of cytokines and transcriptional activators which turn on the FOXP3 gene. This reveals the ability of the immune system to acquire tolerance to antigens from both inherited repertoires (thymic Treg) and to de novo antigen exposure, such as commensal bacteria, food, chemical compounds and alloantigens in pregnancy (induced Treg). More recently, a search for cell surface surrogates of FOXP3 (FOXP3 cannot be used to isolate viable Treg as it requires intracellular staining) has revealed that reduced expression of the IL7 receptor (CD127) is also a hallmark of the human Treg phenotype.7, 8 It has been reported that CD127 is not selective for mouse Treg, as activated murine Treg express CD127 strongly.9 The differential expression of cytokine receptors on Treg may be important in restricting survival or function as, for example, mature human Treg are dependent on exogenous IL2, but not on IL7. This also allows for the possibility that the expression of a specific cytokine receptor on Treg may enable them to act as a biological sink to sequester cytokine in a tissue microenvironment.10
Regulatory T cells are dependent on the expression of FOXP3 for both their formation and function, and FOXP3 controls a gene regulatory network essential for suppressor function. Each lineage in the helper arm of the CD4 pool has a defining transcription factor, and it is the expression of this transcription factor that shapes function. T‐cell transcription factors can be induced by specific external stimuli, and as a result, a transcriptional programme is established which enables the cell to express pathogen‐specific effector molecules. This raises the possibility that function may not be predetermined and fixed in a given lineage, but is plastic. The need for plasticity in T‐cell responses may be twofold; it may be part of a mechanism to quell the active immune response once the pathogen has been cleared. In contrast, functional plasticity may enable tailored responses which are tuned to the challenge type and site. A logical consequence is that different defects in a Treg may be implicated in different autoimmune and inflammatory disease settings.
It is now clear that much complexity in the Treg phenotype exists than originally appreciated, and a growing number of other cell surface markers are found on specific Treg subsets, for example TIGIT,11, 12 GARP,13, 14, 15, 16 CD73 and CD39.17, 18 These markers are frequently also found on effector cell populations, and an emerging theme is that either Treg are able to transition between functional states or that the Treg compartment is paired with the Tconv effector compartment so that any immune response mediated by a T‐cell can be controlled by a matching Treg.19 This is supported by the detection of lineage‐specific transcription factors (e.g. Tbet,20 IRF421) being co‐expressed with FOXP3 in Treg subsets, using various mouse models, but this has yet to be confirmed in humans. It is possible that the balance of Treg to effector lineages may be altered under specific circumstances or that the Treg themselves may switch fates. As more signature molecules for each functional subset are confirmed, and highly purified cell populations are subjected to expression profiling and functional assay, the question of altered committed lineage proportions vs plasticity will be better understood.
Treg and disease
Decreased Treg numbers or impaired function in adult mice leads to autoimmune diseases.1 Adoptive transfer of Treg ameliorates many diseases, including the nonobese diabetic (NOD) and inflammatory bowel disease mouse models,1 as well as mouse models of pregnancy disorders which mimic autoimmune disease in many regards.22 These studies strongly indicate that a threshold of Treg function is required throughout life to restrain autoreactive T cells and/or inflammatory responses, and this prevents autoimmune disease onset. The early development of autoimmune disease in IPEX patients, who lack FOXP3 and Treg,23 confirms that Treg are also essential in humans.24 Functional defects in Treg in type 1 diabetes and inflammatory bowel disease have been reported,25, 26 but there are conflicting reports in the literature about reduced Treg numbers in autoimmune cohorts. These conflicts arise in part because of methods for enumerating Treg, the need to consider the amount of FOXP3, as well as the absolute presence or absence of staining for FOXP3 in flow cytometric data, and the impact of activation and source of sample on analysis.
There are likely to be many points in relevant gene regulatory networks that alter Treg or Tconv function, if affected by genetic or environmental risk factors. It is relatively rare to find mutations in FOXP3 itself, so many Treg‐specific defects in autoimmune disease are likely to result from reduced FOXP3 function or alterations in expression of downstream targets. There are also genes involved in Treg function that are FOXP3‐independent. Type 1 diabetes is an example of a disease which arises as a result of complex interactions between genetic and environmental factors conspiring to drive disease progression. A meta‐analysis of six genome‐wide association studies to identify single nucleotide polymorphisms that track with type 1 diabetes identified 45 loci,27 and using our FOXP3 ChIP data,28 we observed that 34 (75%) contain a FOXP3 binding site. A more recent fine mapping of genetic risk of type 1 diabetes identified 51 distinct loci, of which 34 mapped to a gene.29 Using the filter that a gene is a potential target of FOXP3 if there is a FOXP3 binding site within 20Kb of the transcription start site, 37 (72%) of these loci are putative targets of FOXP3. This suggests the potential for there to be a Treg‐specific defect directly as a result of polymorphism in these genes or their regulatory elements, but the functional link between them is currently unknown. While many cell types contribute to immune homeostasis, Treg/Tconv defects play a major role in the pathology of human autoimmune disease, and epigenetic profiling is now providing compelling evidence that much of the autoimmune disease‐associated risk impacts Treg‐ and Tconv‐specific transcriptional programmes. This suggests that susceptibility to disease is causally linked to either Treg plasticity, altered Treg development or altered Treg function, and for example, a Treg defect which is responsive to TNF antagonism has been demonstrated in rheumatoid arthritis.30
Transcriptional control of Treg formation and function
Since its identification as the key transcription factor for the formation and function of Treg in mouse and humans,23, 31, 32 FOXP3 has been the subject of much investigation. Molecular mechanisms for the action of FOXP3 in shaping regulatory T cells and their critical role in maintaining lifelong tolerance are now being better understood. It is increasingly clear that loss of function is implicated in a wide variety of autoimmune and chronic inflammation settings; however, there remains the possibility that loss of function is a consequence rather than the cause of autoimmunity and chronic inflammation. High‐resolution functional genomics using genotyped patient samples, when combined with mouse models, will undoubtedly resolve this.
Important insights into the transcriptional control by FOXP3 have been defined by chromatin immunoprecipitation (ChIP) experiments, which trap transcription factors bound to DNA. Studies carried out in mouse Treg33 and in human Treg28, 34 have defined the target regions in chromatin bound by FOXP3. As well as identifying a significant number of loci both directly bound by FOXP3 and associated with differentially expressed genes in Treg, many loci were identified that are either too far from a transcription start site to annotate to a target gene easily, or do not appear to be associated with differentially expressed genes in Treg. For example, in our human FOXP3 ChIP data set, of almost 3000 FOXP3‐bound regions, only 750 were also annotated to genes differentially expressed in human Treg28 (Figure (Figure1).1). This has revealed a core network of genes that are tightly regulated by FOXP3 and has also identified significant bioinformatic limitations in annotation of FOXP3‐bound regions. It is now clear that noncoding regulatory regions are critical for T‐cell‐specific gene regulation, may be far from their target gene and that they can exert their function via DNA looping.35 This DNA looping brings specific genes and regulatory elements together into transcriptionally active hubs36 and this may be different in Tconv or Treg. However, since DNA looping cannot currently easily be predicted using bioinformatics approaches, proximity‐based annotation of FOXP3 targets based on linear DNA organisation under‐ascribes transcriptional targets to FOXP3 binding sites in chromatin. Hence, alterations to the binding of FOXP3 to its target genes may contribute to plasticity or lineage commitment and function.

Functional annotation of FOXP3‐bound mRNA regions of the human genome intersected with differential mRNA expression (left Venn diagram), and FOXP3‐bound microRNA regions of the human genome intersected with differential miR expression (right Venn diagram), showing that a subset of mRNAs and miRs are both differentially expressed and FOXP3 targets.
FOXP3 epigenetic regulation
FOXP3 is itself subject to tight regulation, and this is both at the level of transcription and post‐transcription. The modifiable mediators of this regulation include methylation,37, 38, 39, 40, 41, 42, 43, 44 acetylation45, 46, 47 and microRNA‐mediated control of transcript translation.28, 48, 49, 50, 51, 52, 53, 54, 55 The global regulation of FOXP3 is influenced by chromatin organises, including SATB1 acts both during development and in mature cells.48, 58 The regulation of FOXP3 by distinct modules including the Treg‐specific demethylated region (TSDR)37, 38, 59 has revealed marks for FOXP3 expression control and can discriminate between thymic Treg FOXP3 expression and activation‐dependent expression of FOXP3 in naive T cells in the periphery37, 43 (gene annotation and species conservation depicted in Figure Figure2).2). The methylation or demethylation of the TSDR is controlled by DMT3 or TET, respectively, and the regulation of this process is associated with thymic induction of FOXP3 as well as induction of FOXP3 in the periphery.60, 61, 62 Additional analysis of the regulatory elements in the FOXP3 locus has defined specific regions near to the TSDR named conserved noncoding sequence (CNS) 1, 2 and 3.63 CNS1 is responsible for expression of FOXP3 specifically in iTreg. CNS2 contains the TSDR and is responsible for maintenance of FOXP3 in all dividing Treg and CNS3 is the pioneer site for FOXP3 expression in thymic origin nTreg.63 Occupancy by specific transcription factors at each of these loci is now characterised, and this includes AP1 and NFAT at CNS1,59 Runx1 and CBFβ at CNS264 and cREl at CNS3.63 The observation that naïve human CD4+ T cells induce FOXP3 upon activation,65, 66, 67 and that in the presence of TGFβ and all‐trans retinoic acid (ATRA)63 the expression of FOXP3 is stabilised to some degree, was defined transcriptionally by the finding that the activation‐induced expression of FOXP3 results from partial but not complete demethylation of the FOXP3 locus in iTreg.37 This suggests that the relative methylation state of the FOXP3 regulatory elements encoded by CNS1, 2 and 3 are an axis for Treg plasticity. In addition to gain of FOXP3, there is also the consequence of loss of FOXP3 on Treg function due to exhaustion or chronic overstimulation, which might occur during a potent immune response. This has led to the concept of the ex‐Treg. Fate mapping studies in mice elegantly demonstrate that Treg can lose expression of FOXP3, but are of thymic Treg origin based on genetic marking. These studies link ex‐Treg to susceptibility to multiple sclerosis68 and RA,69 and suggest that high levels of IL6 can induce this loss of FOXP3 expression in vivo. A contributing factor may be reduced IL2 signalling, which is in part mediated by SOCS1.70 Hence, a second axis on plasticity may be the impact of local pro‐inflammatory cytokines including IL‐6.

(a) Schematic representation of the human FOXP3 gene, annotating the three conserved noncoding sequences involved in regulating gene expression (CNS1, 2, 3). (b) A VISTA alignment of mouse and human FOXP3 showing regulatory regions which are conserved between species in red, conserved untranslated regions in light blue and conserved exons in lilac.
Tissue and metabolic cues can alter transcriptional programming of T cells
The recruitment of effector cells to sites of infection is driven by local tissue cues as well as pathogen signals, and the pro‐inflammatory milieu at these sites may contribute to the shaping of the phenotype of the cells once they arrive.19, 71 It is now emerging that in addition to these classic cytokine/cytokine receptor‐mediated signals, T cells are also highly responsive to other metabolic cues, and these are also sensed by surface receptors and biochemical pathways.
There are a wide range of molecules that can be detected via cell surface receptors or transporters, and these include sugars, amino acids, energy molecules, vitamin metabolites and food metabolites from the microbiome, as well as oxygen tension and environmental toxins.72 Well‐characterised sensors of metabolic stimuli include mTor and HIF,73, 74 which can influence the differentiation of Treg vs effector T‐cells including Th17. These exert a network effect as they in turn regulate gene networks influencing biochemical responses including glycolysis. The balance of oxidative phosphorylation is linked to FOXP3 expression and regulatory phenotype,75, 76 and this in part enables Treg to function in environments that are under oxidative stress.77
Metabolites which can alter transcriptional programming have also been identified. The role of the vitamin A metabolite retinoic acid ATRA has been well characterised in the context of T‐cell differentiation, and it is evident that either alone or in combination with other factors ATRA can induce either induce Treg or Th17 phenotypes by upregulating FOXP3 expression or ROR gamma expression, respectively.78 This axis can also be influenced by sensing toxins and pollutants via the aryl hydrocarbon receptor (AHR).79, 80 Induction of FOXP3 in response to food metabolites including short‐chain fatty acids is now well characterised at the molecular levels and is particularly relevant for induced Treg generation in the gut. Butyrate generated by commensals in the colon is able to activate the FOXP3 locus and promote a tolerogenic bias.81, 82 These mediators play a key role in differentiation of naïve T cells, but it is less clear whether they drive fate change in committed T‐cell subsets. Nonetheless, all of these pathways and inducer molecules are implicated in altered Treg function or numbers in disease, and are the targets for interventions to restore balance.
Tissue and metabolic cues can alter Treg function
Given that the Treg have to home to the same locations to regulate the effector response after pathogen has been cleared, they are also exposed to the same environment. The gene regulatory network controlled by FOXP3 has to be robust enough to resist this environment, and this is achieved by the combination of repression of expression of key inflammatory cytokine receptors, rendering the Treg unresponsive, and induction of key suppressor genes. However, it is also possible that these tissue‐specific cues can shape Treg function and limit their potency to enable pathogen clearance. Given that activation can induce FOXP3 in effector cells, this may also be the mechanism by which the effector cells themselves return to a resting state, but this requires further validation.
MicroRNA‐mediated tuning of regulatory phenotype
The regulatory network of genes controlled by FOXP3 is essential for the Treg phenotype. Of the 2000‐3000 target genes identified by FOXP3 ChIP,28, 83 a subset are directly differentially expressed or repressed in Treg at any given time, including SATB148 (Figure (Figure1).1). These genes combine with FOXP3 to form the FOXP3 gene regulatory network and shape the function of Treg. It is important to note that there are also a number of differentially expressed genes in Treg that are either indirect targets of FOXP3 or are downstream of FOXP3 direct regulation, but which may be controlled by FOXP3‐induced miRs. Transcriptomics analysis reveals that microRNAs (miRs) are both direct and indirect FOXP3 targets in Treg, suggesting a key role for miRs in Treg function. MicroRNAs (miRNAs) are 21‐22nt, noncoding RNAs commonly found in the introns of protein‐coding genes. Their primary function is to post‐transcriptionally regulate gene expression by binding to a complementary target sequence, typically found in the 3′ or 5′ UTR of a gene. Depending on the affinity, this interaction leads to cleavage of the mRNA transcript or inhibition of translation. A single miRNA has the ability to target and regulate multiple genes. Likewise, multiple miRNAs are able to target a single mRNA transcript, which gives rise to complex regulatory networks and fine‐tuning of regulation.
A key discovery demonstrating that miRNA are critical to T‐cell function was revealed by conditional null mutation in Dicer or Drosha. Cobb et al. selectively inactivated miRNA formation in Treg at a late stage in T‐cell development.49 This induced a fatal inflammatory disease in mice due to a significant reduction in Treg.49 Liston et al. also showed that miRNAs are crucial to Treg suppressive function. In comparison with their healthy counterparts, the Dicer knockout mice showed significantly reduced suppressive function.51 Under similar conditions, Zhou et al. demonstrated that depletion of mature miRNAs leads to uncontrolled autoimmunity and skewing of iTreg to a Th1/Th2‐like effector phenotype, such that iTreg exhibited altered surface markers and cytokine expression profile. Interestingly, no effect was seen in tTreg.55
FOXP3 cooperates with microRNAs via regulatory loops which tightly control the FOXP3 transcriptome
These results suggest that miRNAs are critical in the development and function of Tregs and that miRNAs may confer a rheostat‐like function in T‐cell lineage differentiation and plasticity. Therefore, there is interest in the cooperation of miRNAs and FOXP3 and how they may control Treg phenotype and function. Genome‐wide molecular approaches are being utilised to identify the key genes and gene regulation interactions in human Treg, including ChIP, iCLIP and expression profiling, and this aides understanding Treg function and stability. We have previously demonstrated that miRNAs, such as miR‐155, and FOXP3 cooperate to coordinately regulate other key genes in Treg, including SATB148 and ZEB228 (Figure (Figure3),3), and have identified a number of other candidate miRNAs involved in the Treg genotype. It is interesting to observe that a common miRNA/FOXP3‐mediated molecular switch is able to regulate several key genes, which forms a core part of the FOXP3 gene regulatory network (GRN).
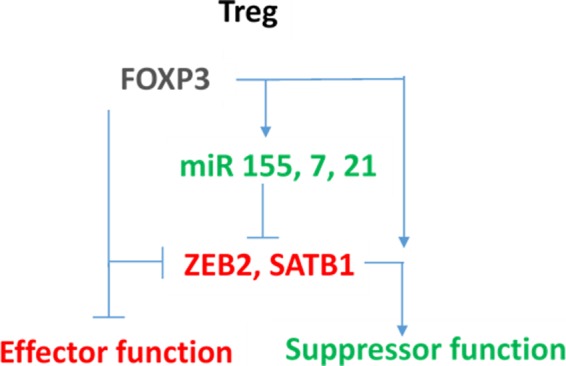
FOXP3‐ and FOXP3‐induced microRNAs (green) cooperate to tightly repress (red) target genes in Treg, to reinforce the suppressor phenotype. These genes are actively expressed in effector T cells.
MicroRNAs are hence implicated as key regulators of Treg function. A differential miRNA signature for human nTreg comprising positively regulated miRNAs includes miR‐21, 155, 125a, 146a, 181c and 374. These miRNAs form part of the network of feedforward regulators in Treg (Figure (Figure4).4). However, less is known about the miRNAs which form part of a negative regulatory loop. Recent analysis reveals a miR‐31 target sequence in the 3′ UTR of FOXP3, suggesting that miR‐31 may be able to negatively regulate FOXP3. Lentiviral expression of miR‐31 in cord blood‐isolated nTreg showed a significant reduction in FOXP3, measured by quantitative RT‐PCR. Likewise, antagonism of miR‐31 expression leads to increased expression of FOXP3, suggesting that miR‐31 regulates FOXP3. A luciferase reporter assay was performed in HEK293 and HeLa to determine whether this regulation was by direct, rather than indirect targeting. The results suggest that FOXP3 is a direct target of miR‐31.53

Molecular model of the FOXP3 gene regulatory network feedforward loops. FOXP3 represses key effector function genes both by direct binding to regulatory elements associated with the gene (e.g. SATB1) and by inducing miRs that themselves target the same gene 3′ UTR. In addition, FOXP3 induces Treg functional genes to reinforce the Treg lineage and phenotype, while suppressing inducers of effector lineage commitment.
MicroRNA‐31 was not identified in the mouse miRNA Treg signature.49 However, in mouse Zhang et al. identified a potential FOXP3 binding site within the promoter region of the gene encoding murine miR‐31,84 suggesting FOXP3 itself may directly target miR‐31. Analysis of miRNA expression by qRT‐PCR revealed miR‐31 was approximately 90‐fold higher in CD4+FOXP3‐ cells in comparison with CD4+FOXP3+ (Treg) cells. Furthermore, a FOXP3 ChIP assay performed in mouse Treg showed significant recruitment of FOXP3 to the miR‐31 promoter.9 Combined, these experiments suggest that FOXP3 binds and downregulates miR‐31 expression in mouse Treg, and alignment with the human miR‐31 host gene suggests that there is also a FOXP3 consensus region in the human miR‐31 locus.
Given the potential relationship between FOXP3 and miR‐31 (Figure (Figure5),5), it is unsurprising to find that miR‐31 is dysregulated in several autoimmune diseases such as IBD or Crohn's disease85, 86 and Kawasaki disease.87 Therefore, understanding the molecular mechanisms by which FOXP3 and miR‐31 regulate each other, and identifying the target genes within this regulatory network, could contribute to the development of novel treatments for autoimmune disease.
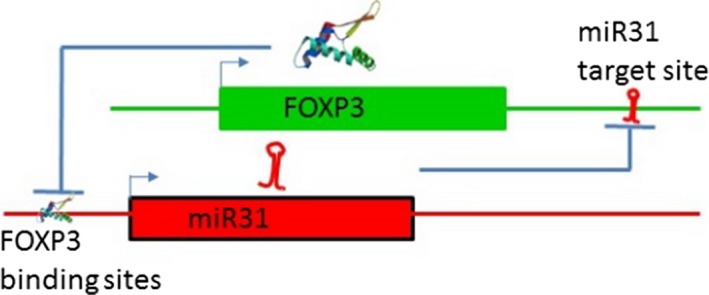
Molecular model of the FOXP3 gene regulatory network feedback loop controlling FOXP3 expression. In a stable Treg, FOXP3 represses miR31 by direct binding to regulatory elements associated with the gene, and in stable effector T cells, miR31 expression prevents the expression of FOXP3 by targeting FOXP3 mRNA for degradation. Subtle alterations in levels of either miR31 or FOXP3 will have reciprocal effects on the expression of each, providing a threshold for transition between FOXP3 +ve and −ve states.
The combined impact of the sentinel transcription factors in each lineage and the miRNAs controlled by them shapes CD4+ T‐cell phenotypes. As modelled in Figure Figure66 , the commitment to the Treg lineage is dependent on FOXP3 and a set of miRNAs controlled by it. In effector cells such as Th17 cells, an opposing transcription factor orchestrates the gene regulatory network, and external signals can influence the expression of these.88 This may enable switching between functional phenotypes by enabling one transcription factor to decline and another to dominate, which is a potential molecular mechanism for plasticity. In addition, it is interesting to speculate that the transient expression of FOXP3 in activated conventional T cells then induces miRs which repress effector function gene networks, and this enables the effector cells to return to a resting state (Figure (Figure7).7). These models require further evaluation in human cells.

Integrated model of the plasticity of phenotypes based on molecular switching driven by induction of miRs which regulate the sentinel transcription factor for one phenotype and allow expression of the transcription factor driving the other phenotype. In regulatory T cells, a suite of FOXP3‐induced miRs are expressed in the steady state, but as a result of external cues such as cytokine signalling, these miRs are reduced, and competing miRs including miR31 are increased, reducing FOXP3 expression and releasing other transcription factors from repression, facilitating a transition and reversible functional switching.
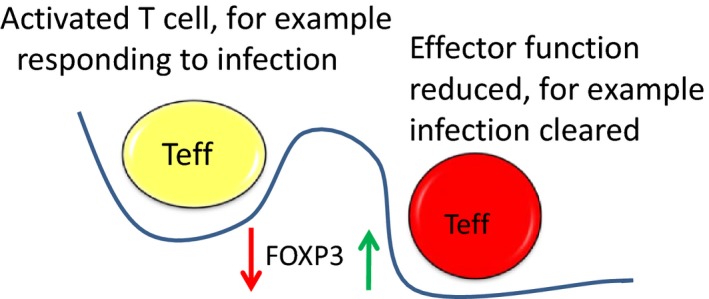
Integrated model of regulation of effector function in effector T cells based on molecular switching driven by transient repression of miR(s) which de‐represses FOXP3, therefore transiently repressing effector function genes. This enables shut down of an immune response upon clearance of the pathogen.
Long noncoding RNAs and Treg
In addition to short noncoding RNAs such as miRs, an emerging theme from high‐resolution functional annotation of the human and mouse genomes is the prevalence and importance of long noncoding RNAs (lncRNAs) as subtle regulators of gene expression. These are transcripts of greater than 200bp that are not translated, but that can interact with mRNA and chromatin to regulate gene expression. There is a evidence that they function either as stabilisers of DNA looping to form transcriptional hubs and as part of the RNA binding protein complex to regulate transcription.89 In addition to a role in shaping the development of many tissues and cells, there is a defined role for lncRNAs in differentiation of T‐cell subsets.90 Evidence of the impact of long noncoding RNAs in immune function is becoming clear,91, 92 and alterations in function can be identified in autoimmune and chronic inflammation samples which are linked to lncRNAs.93 The importance of lncRNA in Treg has been elegantly demonstrated for a lncRNA (Flicr) which subtly controls the expression of FOXP3 in mouse and human Treg.94 Interestingly, the impact of Flicr on FOXP3 expression is via interactions with conserved noncoding elements and loss of Flickr results in reduced expression of FOXP3. This may be the prerequisite for transcriptional reprogramming of Treg under external cues such as from reduced IL2 signalling, reinforcing the model that plasticity can be induced by relatively small changes in transcriptional networks. It is interesting that lncRNA plays a role in stabilising DNA looping and is often encoded within enhancer regions, which shape the expression of multiple genes. The observation that autoimmune genetic risk is found in enhancers and the enhancers loop to form the regulatory hubs on key immune function genes suggests that there could be a complex network effect on multiple targets from genetic risk at a single lnc/enhancer module.95
Genetic risk of autoimmune disease alters Treg function
Genome‐wide mapping of epigenetic variation in Treg and conventional T‐cells reveals cell type‐specific transcriptional activity,83, 96 and large‐scale cataloguing of the marks associated with active, open or closed chromatin has opened the way for understanding cell type‐specific gene regulation (e.g. epigenomics roadmap,97, 98 FANTOM99, 100, 101, 102). In the post‐GWAS era, there are very strong data linking genetic variation (single nucleotide polymorphism or snp) in a wide variety of immunological disease cohorts, but identification of individual causal target genes has been difficult, as it is now apparent that the majority of this genetic variation does not disrupt the coding region of genes, but is located in noncoding regions.103 Numerous independent approaches combining these genotyping data with the functional annotation of the human genome indicate that the majority of this noncoding genetic variation overlaps, and thus likely influences, transcriptional regulatory elements and long noncoding RNAs (lncRNAs).104, 105, 106
In the case of autoimmune diseases data sets, there is also significant enrichment of genetic variation in T‐cell‐specific regulatory elements, including promoters and enhancers,106, 107 suggesting that dysregulation of multiple separate pathways in the immune system can result in the same phenotype from unrelated genotypes. In the case of type 1 diabetes, a recent Bayesian analysis of the intersection between genetic risk and regulatory motifs found significant enrichment of snps within enhancer regions,29 suggesting that the consequence of single nucleotide polymorphism may be to alter the expression of multiple target genes. This is inferred from the observation that genetic risk alters the function of regulatory elements, and the targets of those regulatory elements are dependent on either short‐ or long‐range DNA looping in many cases. While bioinformatics is powerful in identifying enrichment of sequence motifs, and this can be combined with other data such as transcription factor binding sites, epigenetic modifications and genetic variation, it does not yet have the ability to efficiently predict the targets of regulatory regions. This is partly because the conformational networks formed in any cell are exquisitely cell type‐dependent and also functionally dependent on the expression of cell type‐restricted transcription factors such as FOXP3.
Enhancer–promoter interactions shape the function of Treg
Gene coding regions comprise as little as 1.5% of the human genome, encoding approximately 21 000 genes. Genome‐wide functional annotation endeavours including ENCODE, FANTOM and the Roadmap have enabled a far deeper understanding of the architecture of the human genome, and it has become clear that interactions between coding and noncoding elements are essential for normal gene regulation and maintenance of stable phenotype. This suggests that the much of the genome is involved in controlling the expression of those protein‐coding genes.
Chromatin structure is known to have a major influence on gene expression by controlling transcription factor access to binding sites in enhancers and promoters.108 Localised patterns of chromatin modification, such as DNA methylation, histone modification and nucleosome remodelling, correlate with enhancer activity, transcription factor binding and initiation or repression of transcription.99, 109, 110 More recently, these approaches have enabled the identification of a rare subset of enhancers, termed super‐enhancers.103, 107 Super‐enhancers are characterised by a relative enrichment of active enhancer‐associated epigenetic marks and transcriptional cofactor binding, and they appear to regulate key genes involved in cell type‐specific function.103, 107
Determining the genes that enhancers control will complete the picture of Treg gene regulation. DNA looping plays a critical role in transcriptional regulation by allowing widely separated enhancers to interact with promoters at gene hubs.111, 112 Hence, DNA looping facilitates gene network formation as a single enhancer may interact with more than one promoter, and a single promoter may be contacted by more than one enhancer. Many of these interactions occur in a tissue‐specific manner and appear to be the major determinants of cell type‐specific responses.111, 113, 114, 115 Critically, many enhancers do not interact with the nearest active gene but contact gene promoters many hundreds to thousands of kilobases away through long‐range DNA looping (modelled for T cells in Figure Figure88).111, 112
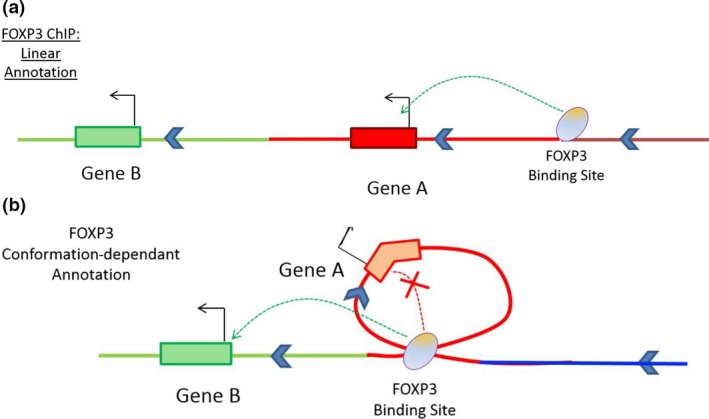
Conformation‐dependent interactions contribute to gene regulation networks, and these are not readily predicted by bioinformatics approaches. (a) Linear annotation of TF binding site data to nearest neighbour is only accurate if within a short distance from the TSS. (b) unidentified FOXP3 binding sites, and distal regulatory interactions are annotated after incorporation of DNA looping data from conformation capture experiments.
These findings highlight the difficulty in annotating regulatory regions to specific target genes using ChIP and chromatin accessibility data alone. Likewise, while genome‐wide association studies have identified potential disease‐associated variation in regulatory regions, the genes they affect remain largely unknown116 (mutation impact modelled in Figure Figure9).9). FOXP3 itself may actively drive intrachromosomal interactions in Treg. Structural data indicate FOXP3 multimers can bind widely separate recognition sites, as demonstrated by 4C assay of the Ptpn22 locus following FOXP3 overexpression in murine Tconv.34 New approaches combining chromatin state profiles, transcription factor binding and autoimmune disease loci with enhancer–promoter contact maps generated by chromosome conformation capture are beginning to unravel the functional consequences of disease‐associated noncoding DNA variation.

Functional annotation of genetic risk from the GWAS data set which maps to FOXP3 ChIP binding regions. The addition of chromatin conformation data allows identification of the target genes, and this enables analysis of the impact of genetic variation. In a Tconv, the gene is expressed because there is no FOXP3 (a). In a normal Treg, the gene is repressed (b), but in a Treg from the disease cohort, the sequence variation results in loss of a FOXP3 binding site, as a result the target gene is no longer repressed (c). The functional impact is at a distal regulatory element, not in the target gene itself.
Mapping the 3D genome
Chromosome conformation capture (3C) assays are now the method of choice to study the role of DNA looping in transcriptional regulation. These assays directly identify genomic loci that are in sufficiently close proximity in living cells to be cross‐linked.117 Several versions of 3C such as 4Cseq, ChIA‐PET,112 5C111 HiChIP118 and Hi‐C113 allow for mapping of chromatin interactions on a whole genome level. Recently, to improve the resolution and sensitivity of Hi‐C assays, in situ protocols have been developed. In addition, for focussed interaction mapping oligonucleotide capture technology has been used to enrich for regions of interest (such as promoters) in the Hi‐C library prior to high‐throughput sequencing119, 120, 121 or chromatin immunoprecipitation combined with Hi‐C to interrogate individual protein‐centric conformation maps122 The use of oligo‐capture technology significantly improves sensitivity and resolution without the associated increase in sequencing costs required for high‐resolution analysis of traditional Hi‐C libraries.113 Importantly, Chromosome conformation capture‐based assays have been used to successfully identify targets of disease‐associated variation in other cell types111, 112, 120, 123 including human CD4 and CD8 populations,124 and data from a genome‐wide H3K27ac Hi CHIP (acetylated histone CHIP and conformation capture) performed in naïve T cells, Th17 and Treg, have annotated accessible chromatin interacting with regulatory elements,125 shedding new light on the lineage‐specific interactome in these cells. However, intersection with FOXP3 binding site data, genetic risk and functional validation still needs to be performed on Treg.
Conclusion and future directions
There are complex dynamic regulatory processes controlling Treg generation and stability that involve microRNA and long noncoding RNA‐based regulation of gene expression. These interact with transcription factors which themselves define function. In addition, enhancers and super‐enhancers establish the level of gene expression and response to T‐cell activation. This gives three layers of reinforcement of Treg phenotype, and each may be a point of disruption in either disease, altering the functional Treg pool, or in response to tissue and inflammatory cues, altering fate. It is evident that in the steady state, FOXP3 establishes and maintains a strong regulatory phenotype, and this is resistant to reprogramming; however, under specific circumstances this can be changed, and this either results in formation of so‐called ex‐Treg, or in a cell with effector‐like function that expresses less or no FOXP3. The definition of the process as either plasticity or exhaustion is hard to define in man. By combining genomics and high‐resolution cell phenotyping, followed by functional validation by methods including gene editing, the answer to the question of mechanisms underpinning plasticity in human Treg will likely soon be provided. There is no doubt that microRNAs have the capacity to tune the alterations in genotype required to alter phenotype, and perhaps these are the rheostat of fate. New insights into the contribution of genetic risk to gene expression in Treg in diseases including autoimmunity will also reveal whether dysregulation of microRNAs is a tipping point for altered phenotype. While elegant mouse models provide mechanistic insights and have considerable power, for example in fate mapping of Treg subsets, there is still a need to demonstrate the plasticity or paired generation of Treg subsets in man.
References
Articles from Clinical & Translational Immunology are provided here courtesy of Australasian Society of Immunology
Full text links
Read article at publisher's site: https://doi.org/10.1002/cti2.1011
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/cti2.1011
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1002/cti2.1011
Article citations
Infiltrating treg reprogramming in the tumor immune microenvironment and its optimization for immunotherapy.
Biomark Res, 12(1):97, 04 Sep 2024
Cited by: 0 articles | PMID: 39227959 | PMCID: PMC11373505
Review Free full text in Europe PMC
Beyond Immune Balance: The Pivotal Role of Decidual Regulatory T Cells in Unexplained Recurrent Spontaneous Abortion.
J Inflamm Res, 17:2697-2710, 01 May 2024
Cited by: 1 article | PMID: 38707955 | PMCID: PMC11070170
Review Free full text in Europe PMC
A disrupted FOXP3 transcriptional signature underpins systemic regulatory T cell insufficiency in early pregnancy failure.
iScience, 27(2):108994, 23 Jan 2024
Cited by: 1 article | PMID: 38327801 | PMCID: PMC10847744
OX40 (CD134) Expression on T Regulatory Cells Is Related to Serious Hypertensive Disorders in Pregnancy.
J Cardiovasc Dev Dis, 10(10):431, 17 Oct 2023
Cited by: 0 articles | PMID: 37887878 | PMCID: PMC10607140
The Role of Gut Microbiota-Derived Lithocholic Acid, Deoxycholic Acid and Their Derivatives on the Function and Differentiation of Immune Cells.
Microorganisms, 11(11):2730, 08 Nov 2023
Cited by: 2 articles | PMID: 38004742
Review
Go to all (15) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Molecular Insights Into Regulatory T-Cell Adaptation to Self, Environment, and Host Tissues: Plasticity or Loss of Function in Autoimmune Disease.
Front Immunol, 11:1269, 15 Sep 2020
Cited by: 12 articles | PMID: 33072063 | PMCID: PMC7533603
Review Free full text in Europe PMC
Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection.
Curr Top Microbiol Immunol, 293:43-71, 01 Jan 2005
Cited by: 47 articles | PMID: 15981475
Review
Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity.
Proc Natl Acad Sci U S A, 114(17):E3472-E3480, 10 Apr 2017
Cited by: 96 articles | PMID: 28396406 | PMCID: PMC5410798
Genetic and epigenetic basis of Treg cell development and function: from a FoxP3-centered view to an epigenome-defined view of natural Treg cells.
Immunol Rev, 259(1):192-205, 01 May 2014
Cited by: 104 articles | PMID: 24712467
Review

 2
2




