Abstract
Free full text

The Cancer Genomics Cloud: Collaborative, reproducible, and democratized—a new paradigm in large-scale computational research
Abstract
The Seven Bridges Cancer Genomics Cloud (CGC; www.cancergenomicscloud.org) enables researchers to rapidly access and collaborate on massive public cancer genomic datasets, including The Cancer Genome Atlas. It provides secure on-demand access to data, analysis tools and computing resources. Researchers from diverse backgrounds can easily visualize, query and explore cancer genomic datasets visually or programmatically. Data of interest can be immediately analyzed in the cloud using more than 200 pre-installed, curated bioinformatics tools and workflows. Researchers can also extend the functionality of the platform by adding their own data and tools via an intuitive software development kit. By colocalizing these resources in the cloud, the CGC enables scalable, reproducible analyses. Researchers worldwide can use the CGC to investigate key questions in cancer genomics.
Introduction
As the size and complexity of cancer genomic datasets continue to grow, the availability of scalable compute resources (i.e. the ‘cloud’) facilitates rapid and cost effective data analysis (1). The Seven Bridges Cancer Genomics Cloud (CGC; www.cancergenomicscloud.org) was funded as a pilot project by the US National Cancer Institute (NCI) to explore novel approaches to democratize access to massive cancer genomic datasets alongside the tools and computational resources to analyze them. The CGC was publicly launched in February 2016 and is open to all cancer researchers worldwide, who can create a free profile online, or log in via their eRA Commons or NIH Center for Information Technology account.
The CGC enables researchers to quickly access The Cancer Genome Atlas (TCGA; 2), which contains genomic, transcriptomic, and clinical data from more than 11,000 cancer patients. TCGA has contributed greatly to understanding the molecular basis of cancer and identifying novel therapeutic targets (3, 4). However, downloading and storing TCGA requires significant time and resources. Furthermore, querying and using the data can be challenging for researchers without adequate computational resources or appropriate technical knowledge. The CGC addresses these challenges to make TCGA and other large cancer genomics datasets usable by a wide range of cancer researchers.
Methods: Scalable cancer genomics analysis in the cloud
The CGC comprises an intuitive interface for rapidly accessing and using large public genomic datasets; comprehensive security controls; preloaded bioinformatics tools and workflows; access to scalable cloud-based computation; a software development kit; an application programming interface (API) for automation; data visualization and querying tools; and extensive support for collaborative, reproducible research (Figure 1A, Video 1). Built on Amazon Web Services’ enterprise level cloud services, the CGC provides researchers with secure access to public genomic datasets (including TCGA and the Cancer Cell Line Encyclopedia [CCLE; 5]), alongside the high-performance computation needed to analyze them. Because the data are stored together with cloud computing resources, analysis is readily scalable on-demand, allowing thousands of samples to be quickly analyzed (Figure 1B).
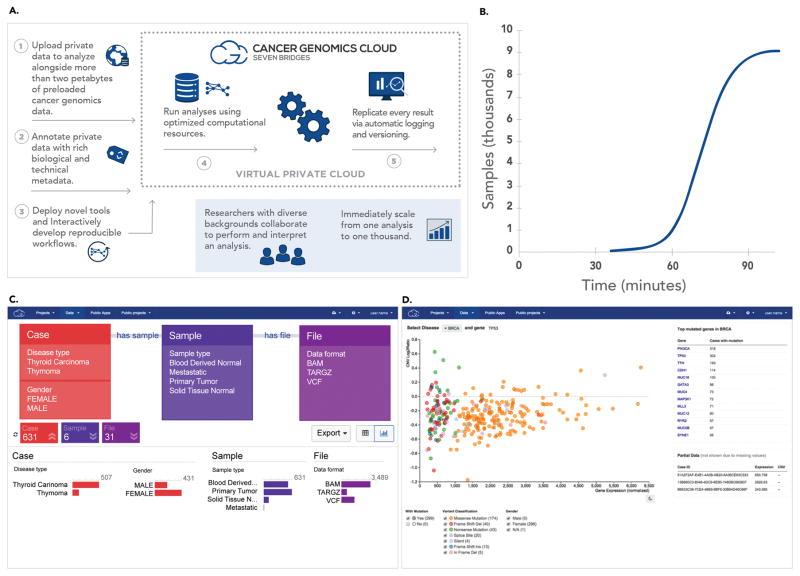
The Cancer Genomics Cloud is designed to enable scalable cancer genomics research, with features that support usability. A) In addition to hosting public cancer genomics datasets and providing curated analysis tools, the CGC enables users to upload and annotate their own data, as well as integrate their own tools. Data analyses are run using optimized computing resources and executions are recorded to ensure reproducibility. B) Time course of an RNA-Seq quantitation experiment in which more than 9,000 samples were analyzed in parallel. All samples were completed within 100 minutes. C) The visual Data Browser allows users to explore and select data by specifying properties of interest. Once selected, these files can be added to a project for analysis. D) The interactive Case Explorer allows users to visualize and select cases based on type of cancer, genes of interest, types of mutations, and more.
The hosted datasets are complemented by more than 200 biomedical data analysis tools and workflows, including pipelines for variant calling on whole genome and exome sequencing data, differential expression analysis on RNA sequencing data, and complex data visualization. These curated tools are continually revised to include updated versions, according to demand as determined by user interviews and surveys. Tools on the CGC are packaged within Docker containers, a lightweight software virtualization technology (www.docker.com). Execution instructions are described using Common Workflow Language (CWL; www.commonwl.org), an open-source, community-developed specification for describing analysis workflows and tools in a way that is portable and scalable across software and hardware environments. The CGC also offers researchers a robust software development kit, which enables them to easily describe their own tools and custom scripts in CWL for use on the platform. A visual workflow editor allows users to intuitively build reproducible workflows from individual tools.
The CGC offers a suite of technologies for visualizing, querying, and exploring to identify data of interest within complex datasets. By using a Semantic Web approach to link more than 140 clinical, biospecimen, and analysis metadata properties, the CGC enables researchers to build complex queries both visually and programmatically (Figure 1C). This allows scientists to quickly access, for example, all RNA sequencing count files from normal and tumor samples taken from patients with thyroid cancer who were treated with local radiation therapy. Importantly, this approach is readily extendable to support multi-study integration. We have made data from the CCLE available with TCGA, and we will continue to add further important public datasets. While the Data Browser allows users to explore datasets based on metadata, the Case Explorer focuses on the genetic properties of the data and allows global views of gene expression, copy number variation, and gene mutation status (Figure 1D).
Methods: Collaborative, reproducible, extensible and scalable
The CGC has been designed to support best-practice, reproducible scientific research at scale. On the basis of our engagements and collaborations with cancer researchers, and our experience in user-centric software development, we identified key principles to guide the design and implementation of the platform. We suggest that any modern biomedical ‘knowledge cloud’ should follow similar principles.
First, public cancer genomic data need to be usable by all collaborators. Few institutions have the resources to download and manage TCGA, and specialized skills are required to manipulate the data. By contrast, users of the CGC can immediately begin to explore and analyze more than a petabyte of cancer genomics data through a simple web interface. Users benefit from visual tools and guides, documentation, a dedicated user support team, and the continual development of the Seven Bridges core infrastructure upon which the CGC is built. We regularly review the usability of the CGC and incorporate feedback from researchers to create new solutions and features. In addition, collaboration between multiple researchers and institutions promotes scientific advances (6). By colocalizing data, computation, and analysis the CGC enables distributed, multidisciplinary teams to collaborate on data analysis. Shared project spaces allow approved collaborators to access the same data and workflows, and to see the same results.
Second, reproducibility is required throughout the entire research workflow, from data management to analysis. Replication studies highlight the difficulty of reproducing a number of key studies in cancer biology (7, 8). The CGC ensures reproducibility of computational analyses by recording all aspects of data analysis, including files used, tool versions, and parameter settings. Moreover, because workflows are defined using CWL, they can be readily reproduced by collaborators, reviewers, and journal editors across different computational environments. The CGC currently supports CWL 1.0 via the API, and we are in the process of updating the CGC with Rabix (9), our open-source CWL executor capable of testing and running all CWL applications in any Unix-based environment. Rabix integration will further facilitate developer testing and interoperability with external platforms.
Third, the impact of large public cancer genomics datasets is extended by new tools and data. Besides providing curated biomedical data analysis tools and workflows, we created a software development kit for users to add their own tools and workflows, and a RESTful (10) API supported in multiple languages (Python, R). Using these tools, researchers on the CGC have developed custom pipelines for a variety of scientific applications, including characterizing tumor microbiomes, epitope identification, and segmenting patient populations by gene sequence. Researchers can extend the utility of TCGA data by adding their own private datasets and analyzing them together with the same workflows. Robust upload utilities (including a visual interface, command line, and API) use all available client bandwidth while ensuring secure data transfer. Once uploaded, data can be annotated with more than 40 standard metadata properties or any number of custom properties. Integration and semantic mapping of user-defined metadata to a common ontology is an area of active development.
Finally, data analyses must be scalable to fully make use of available datasets. The elasticity of the CGC infrastructure means that as data analysis scales up, additional computational resources are allocated to enable parallelization and processing of batch jobs. Cloud computing costs have decreased by more than 80% over the last 10 years (11), making it the most cost efficient way to analyze large genomic datasets (12). As an example, one researcher was able to perform targeted variant calling across the 11,000 TCGA participants in about three hours for under $15.
Results: User uptake and impact
Within the 15 months since the launch of the CGC, over 1,900 researchers have registered on the platform, representing 150 institutions across 30 countries. In total, CGC users have deployed more than 5,000 tools or workflows and performed 80,000 executions, representing over 97 years of total computation. There is significant collaboration among users, with an average of seven members per project on the platform.
The CGC enables a diverse range of research (13, 14). For example, last year, Diermeier and colleagues (13) reported on potentially oncogenic mammary-tumor-associated RNAs (MaTARs). After identifying and characterizing MaTARs using mouse models of breast cancer, they confirmed the relevance of human MaTAR orthologs in clinical breast cancer with the CGC. By analyzing RNA-Seq data available in TCGA, the researchers found that some of the MaTARs were upregulated in breast tumors.
Discussion
The CGC makes massive cancer genomic datasets available and usable for research, while safeguarding privacy and security. This approach is readily extensible: in addition to hosting TCGA, we have made CCLE data available and empowered users to integrate their private data with these public resources. New genomics datasets, including TARGET, CGCI, and Simons diversity data as well as new types of data (imaging and proteomic data) are being added to further extend the utility of the system. The CGC represents a successful model for democratizing access to and use of massive public datasets, allowing users to maximize their research productivity. Biomedical ‘knowledge clouds’ like this can serve as the gateway to a wide ecosystem of interoperable cloud resources to support scientific discovery.
Acknowledgments
Financial support: The Cancer Genomics Cloud is powered by Seven Bridges and has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN261201400008C.
We thank the entire Seven Bridges team, the Cancer Genomics Cloud Pilot teams from the National Cancer Institute, the Broad Institute, and the Institute of Systems Biology, the Genomic Data Commons team, countless early users, and data donors. Thank you to Laura Tramontozzi for assistance with the figure. This work has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN261201400008C.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-17-0387
Read article for free, from open access legal sources, via Unpaywall:
https://cancerres.aacrjournals.org/content/canres/77/21/e3.full.pdf
Citations & impact
Impact metrics
Article citations
NCI's Proteomic Data Commons: A Cloud-Based Proteomics Repository Empowering Comprehensive Cancer Analysis through Cross-Referencing with Genomic and Imaging Data.
Cancer Res Commun, 4(9):2480-2488, 01 Sep 2024
Cited by: 0 articles | PMID: 39225545 | PMCID: PMC11413857
Robust estimation of cancer and immune cell-type proportions from bulk tumor ATAC-Seq data.
Elife, 13:RP94833, 09 Oct 2024
Cited by: 0 articles | PMID: 39383060 | PMCID: PMC11464006
A Pan-Cancer Patient-Derived Xenograft Histology Image Repository with Genomic and Pathologic Annotations Enables Deep Learning Analysis.
Cancer Res, 84(13):2060-2072, 01 Jul 2024
Cited by: 1 article | PMID: 39082680
Integrative genomic analysis of the lung tissue microenvironment in SARS-CoV-2 and NL63 patients.
Heliyon, 10(12):e32772, 10 Jun 2024
Cited by: 0 articles | PMID: 39183848 | PMCID: PMC11341340
Creating cloud platforms for supporting FAIR data management in biomedical research projects.
F1000Res, 13:8, 29 Apr 2024
Cited by: 0 articles | PMID: 38779317 | PMCID: PMC11109697
Go to all (72) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The ISB Cancer Genomics Cloud: A Flexible Cloud-Based Platform for Cancer Genomics Research.
Cancer Res, 77(21):e7-e10, 01 Nov 2017
Cited by: 33 articles | PMID: 29092928 | PMCID: PMC5780183
Using the Seven Bridges Cancer Genomics Cloud to Access and Analyze Petabytes of Cancer Data.
Curr Protoc Bioinformatics, 60:11.16.1-11.16.32, 08 Dec 2017
Cited by: 2 articles | PMID: 29220078 | PMCID: PMC5726550
Building Portable and Reproducible Cancer Informatics Workflows: An RNA Sequencing Case Study.
Methods Mol Biol, 1878:39-64, 01 Jan 2019
Cited by: 0 articles | PMID: 30378068
Using Semantic Web Technologies to Enable Cancer Genomics Discovery at Petabyte Scale.
Cancer Inform, 17:1176935118774787, 28 Sep 2018
Cited by: 2 articles | PMID: 30283230 | PMCID: PMC6166304
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH (1)
Grant ID: HHSN261201400008C
NCI NIH HHS (1)
Grant ID: HHSN261201400008C





