Abstract
Free full text

Structure determination of microbial metabolites by the crystalline sponge method†
Associated Data
Abstract
The structures of metabolites produced in microgram quantities by enzymatic reductions with baker's yeast were analyzed using the crystalline sponge method. The X-ray data provided reliable structures for trace metabolites including their relative and absolute stereochemistries that are not fully addressed by conventional NMR and LC-MS analyses. Technically, combining two or more chromatographic purification techniques is essential because, unlike abundant synthetic compounds, extracted metabolites contain many low level UV-silent impurities. The crystalline sponge method coupled with HPLC purification (LC-SCD) would thus be a useful method for metabolic analysis and drug discovery.
Introduction
The structural information of metabolites is key to understanding enigmatic cellular processes and for drug design.1 Important insights into active enzymes, gene expression of RNA sequences, and biochemical reactions can be extracted when the structures of metabolites are fully characterized. They are in most cases produced in minute quantities as low level components of a complex mixture. Combinations of spectroscopic measurements (e.g. NMR and MS) and separation techniques (e.g. GC and LC) have been frequently used and established to elucidate the structures of scarce metabolites from a mixture. However, full characterization of their structures, including their absolute stereochemistry, is still a laborious task because of limited sample supply.
The crystalline sponge method2,3 is a recently developed technique to prepare single crystal samples for X-ray crystallographic analysis of trace and non-crystalline compounds. Given the low limit of the requisite sample amount (<0.1 μg), the method can be an innovative analytical tool for trace metabolites, but has only been used to examine abundant synthetic compounds in the past.4,5 The application of the method to bio-synthetically produced scarce metabolites is challenging because, unlike synthetic samples, extracted metabolites contain many unpredictable impurities. The major advantage of the crystalline sponge method is that a microgram level quantity of samples is sufficient to be analyzed. In this paper, we demonstrate that the microbial metabolites obtained only in microgram quantities are fully characterized using the crystalline sponge method coupled with HPLC separation (LC-SCD analysis).
Results and discussion
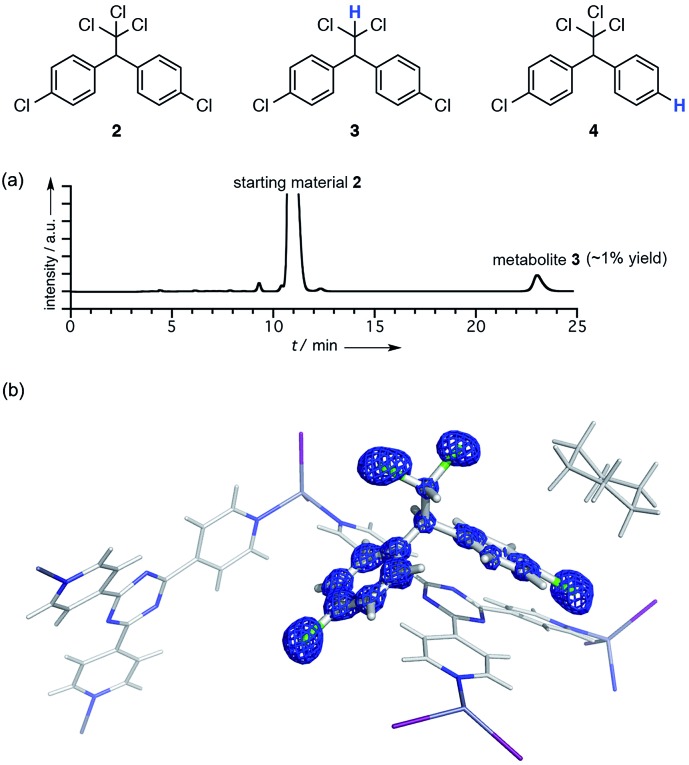
As a model case of metabolic analysis using a crystalline sponge [(ZnI2)3(tpt)2·(c-C6H12)x]n (1; tpt = tris(4-pyridyl)-1,3,5-triazine),3a,6 we conducted reductive dechlorination of 1,1-bis(4-chlorophenyl)-2,2,2-trichloroethane (DDT; 2) by reductases found in baker's yeast. When compound 2 (100 mg) was treated with fermenting baker's yeast (dry yeast (11.2 g) and sucrose (23.5 g) in 100 ml of water) at 30 °C for 1 week, a small amount of a metabolic product (~1.3% yield based on UV spectroscopy) was detected in the ether extract obtained from the mixture. LDI-TOF MS spectrometry indicated the loss of one chlorine atom (m/z = 320.040; calcd for C14H10Cl4+ [2 – Cl + H]+: 319.9507) from 2, suggesting the formation of 1,1-bis(4-chlorophenyl)-2,2-dichloroethane (DDD; 3). The formation of 3 in a low yield is consistent with the previous report.7 However, another dechlorinated compound, 4, cannot be excluded as the product based solely on MS information.
To unambiguously determine the structure of 3, the crude extract was subjected to the LC-SCD analysis. After pre-purification of the ether extract (ca. 15 μg) using PTLC, the analytical sample was further purified using HPLC with a narrow collection time window (Fig. S3†). This purification protocol is particularly important, or else the HPLC-separated sample was considerably contaminated with many UV-silent impurities. In fact, no satisfactory results were obtained in our earlier attempts in the crystalline sponge analysis when the crude extract was directly purified using HPLC with a wide collection window (Fig. S5†).
The HPLC separation system was slightly modified so that fraction collection and subsequent guest soaking could be done in the same microvial (Fig. S1†). The fraction was directly received by a microvial and, after the evaporation of the eluent (hexane), one crystal of crystalline sponge 1 and a solvent were added therein and the vial was allowed to stand at room temperature for 3 d for guest soaking.
The crystallographic analysis confirmed the structure of the metabolite to be 3 (Fig. 1). The crystal structure revealed that there are two major binding sites for guest 3 in the pores of the crystalline sponge 1.‡ One is located near a tpt ligand where guest 3 was observed with 100% occupancy (guest A; Fig. S5†). Another one is located on a crystallographic twofold axis where guest 3 was statistically disordered (guest B). A better resolution was obtained for A, and the electron density map F0 clearly shows the structure of 3 (Fig. 1b). Slightly high R1 and wR2 values (0.0780 and 0.2736, respectively) were obtained presumably due to very minor disorder in 3, which cannot be modeled yet contributes to the residual electron density significantly because of the heavy atom (Cl).
When a prochiral functional group is enzymatically reduced to a chiral one, the stereochemical issues are of major concern for the structural characterization. Moreover, with the product(s) isolated only in microgram quantities, NMR analyses (particularly, 13C or 2D NMR) become difficult even for determination of the relative stereochemistry. The absolute stereochemistry can hardly be addressed by any spectroscopic methods unless empirical rules or data for authentic compounds are available. Thus, we applied the LC-SCD analysis for the full structural characterization (including absolute stereochemistry) of a metabolite from tetralone 5. This compound exists as a racemate due to rapid enolization at C1 and can give four possible stereoisomers (including enantiomers) upon reduction of the carbonyl group at C2.
Treating tetralone 5 (100 mg) with baker's yeast (11.2 g) for 18 h gave analytically pure chiral alcohol 6 in 10 μg quantity after isolation using HPLC. The relative stereochemistry of 6 was confirmed to be cis by comparison with both an authentic cis–trans mixture obtained by the NaBH4 reduction of 5 and literature reported spectroscopic data.8 Chiral HPLC analysis indicated >98% ee for the isolated cis isomer 6. An inclusion crystal 1·6 was prepared by soaking a crystal of 1 in a cyclohexane/1,2-dichloroethane (v/v = 9 :
: 1) solution of 6. The crystal structure was solved with a non-centrosymmetric space group C2.§
A reasonable Parsons' Flack parameter value of 0.010(4) was obtained and the final R1 and wR2 values were 0.0511 and 0.1396, respectively. In an asymmetric unit, three independent guest molecules 6 (G1–G3 in Fig. 2) were clearly observed with ~100% occupancy. All of the observed molecules 6 (G1–G3) show a 1R,2S absolute configuration, consistent with a previous report,8 in which an absolute configuration was speculated based on the absolute stereochemistry of analogous compounds.8 Our crystallographic study provided unquestionable proof for the absolute configuration of 6. From this, a plausible reaction mechanism to give 6 is suggested to involve reduction from the Re face of a 1R isomer of 5 that is kinetically resolved within the enzyme pocket.
1) solution of 6. The crystal structure was solved with a non-centrosymmetric space group C2.§
A reasonable Parsons' Flack parameter value of 0.010(4) was obtained and the final R1 and wR2 values were 0.0511 and 0.1396, respectively. In an asymmetric unit, three independent guest molecules 6 (G1–G3 in Fig. 2) were clearly observed with ~100% occupancy. All of the observed molecules 6 (G1–G3) show a 1R,2S absolute configuration, consistent with a previous report,8 in which an absolute configuration was speculated based on the absolute stereochemistry of analogous compounds.8 Our crystallographic study provided unquestionable proof for the absolute configuration of 6. From this, a plausible reaction mechanism to give 6 is suggested to involve reduction from the Re face of a 1R isomer of 5 that is kinetically resolved within the enzyme pocket.
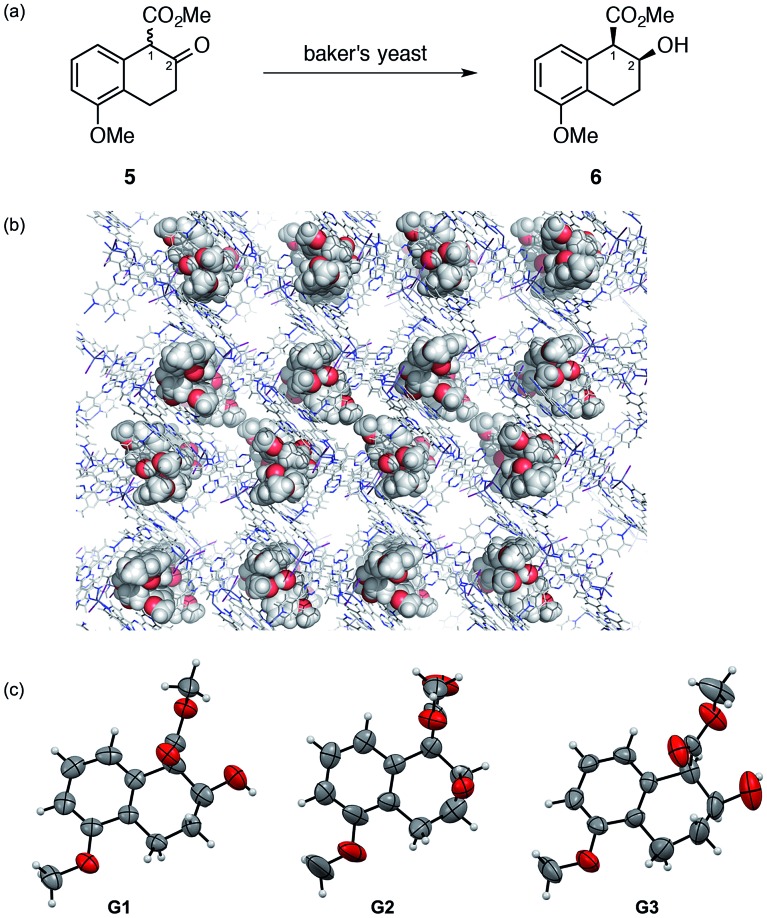
Conformational analysis of the observed guests G1–G3 is worthy of additional discussion. The methoxycarbonyl groups in G1 and G3 are located at the pseudo-axial position, while that in G2 is at the pseudo-equatorial position. For G2, intramolecular hydrogen bonding between the OH and COOMe groups was indicated by the short OO–H–OC O distance (~2.8 Å) that may stabilize this conformation. Presumably, this conformer exists in solution and the crystalline sponge traps both conformers at different binding sites, enabling the concomitant observation of both conformers.
O distance (~2.8 Å) that may stabilize this conformation. Presumably, this conformer exists in solution and the crystalline sponge traps both conformers at different binding sites, enabling the concomitant observation of both conformers.
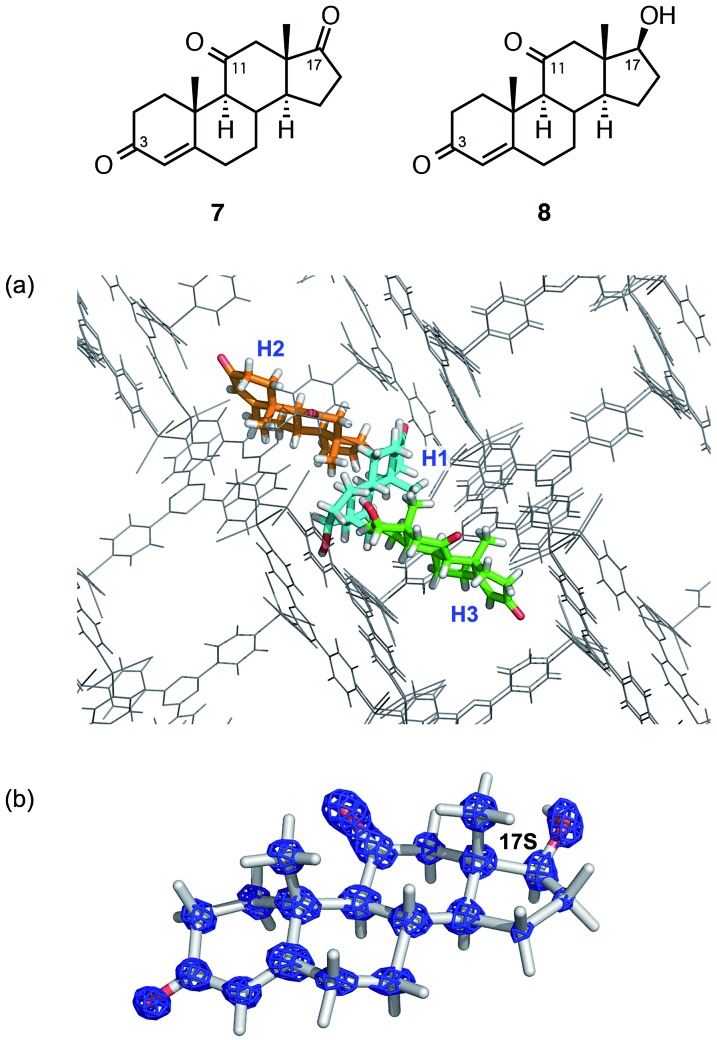
The structural and stereochemical analysis of a metabolite from adrenosterone (7) provided a more challenging task because three prochiral carbonyl carbons at C3, C11, and C17 can in principle generate 26 different compounds upon reduction of the carbonyl group(s). When steroid 7 was metabolized by baker's yeast on a 200 μg scale, HPLC analysis revealed the exclusive formation of a single metabolic product (hereafter denoted as 8).
The parent ion peak of metabolite 8 was observed at m/z = 325.1781 (calcd for C19H26NaO3+: 325.1780). The increase in mass unit by 2.0 Da from 7 suggests a mono-hydrogenation of 7 at either a carbonyl or C C group. The 1H NMR spectrum of 8 was, unfortunately, not conclusively able to determine its structure and stereochemistry because of overlapping signals. The unequivocal structure determination of 8 can be achieved, given that 8 is obtained only in microgram quantity, most reliably by the LC-SCD analysis.
C group. The 1H NMR spectrum of 8 was, unfortunately, not conclusively able to determine its structure and stereochemistry because of overlapping signals. The unequivocal structure determination of 8 can be achieved, given that 8 is obtained only in microgram quantity, most reliably by the LC-SCD analysis.
Thus, ca. 20 μg of a crude mixture obtained from the ether extract was purified using HPLC and subjected to guest soaking with a crystal of 1. After guest soaking at 50 °C for 2 d, the crystallographic analysis revealed three independent molecules of 8 (H1–H3 in Fig. 3a) in an asymmetric unit with occupancies of 100, 80.5, and 100%, respectively.¶
All the structures revealed that the carbonyl group at C17 in 7 is stereoselectively reduced to a hydroxy group with an S configuration. The two carbonyl groups at C3 and C11 remain intact: typical C O distances (1.19(3) and 1.20(3)) were observed at C3 and C11, respectively, and these carbon atoms still adopt a trigonal planar geometry. In contrast, elongation of the C–O bond length of 1.41(3) Å and a tetrahedral geometry at C17 were observed, indicating that the carbonyl reduction took place only at C17. Notably, favorable host–guest interactions were observed. For example, guest H1 is trapped by the host framework of 1 with C
O distances (1.19(3) and 1.20(3)) were observed at C3 and C11, respectively, and these carbon atoms still adopt a trigonal planar geometry. In contrast, elongation of the C–O bond length of 1.41(3) Å and a tetrahedral geometry at C17 were observed, indicating that the carbonyl reduction took place only at C17. Notably, favorable host–guest interactions were observed. For example, guest H1 is trapped by the host framework of 1 with C O···H–C or C–H···I hydrogen bond interactions.
O···H–C or C–H···I hydrogen bond interactions.
Conclusions
In conclusion, LC-SCD analysis has been demonstrated as a useful tool for structural analysis of trace microbial metabolites. Some important lessons are learned in this study working with trace microbial metabolites, which we have not noticed in the previous analysis of abundant synthetic compounds. First, a key to the success of the analysis is to obtain high purity samples of the microbial metabolites by HPLC separation. As the crystalline sponge 1 often preferentially absorbs minor components, even low-level impurities may disturb efficient guest soaking. Given that trace metabolites separated using HPLC are normally contaminated with many impurities, great care should be taken in the purification steps. Therefore, pre-purification with PTLC or LC–LC is highly recommended. Second, pre-analysis of the abundant parent compounds would be beneficial because soaking conditions optimized for the parent compounds can usually be applied to the analysis of their metabolites. Third, having successful results in this study, we are more convinced that the crystalline sponge method, coupled with HPLC separation, will innovate the structural analysis of scarce amounts of microbial products in natural product chemistry as well as in drug discovery.
Supplementary Material
Acknowledgments
This research was promoted as a part of the JST-PRESTO and JST-ACCEL projects in which Y. I. and M. F. are principle investigators, respectively. We thank Hiroyasu Sato and Akihito Yamano (Rigaku Corporation) for the support of the X-ray measurement of compound 1·8.
Footnotes
‡Crystallographic data for 1·3: C36H24N12Zn3I6·(C14H10Cl4)·(C7H4Cl2)·2.5(C3H6), M = 2230.02, colorless block, 0.23 × 0.11 × 0.09 mm3, monoclinic, space group C2/c, a = 35.7365(13) Å, b = 15.0610(5) Å, c = 30.9345(11) Å, β = 104.141(7)°, V = 16 145.2(11) Å3, Z = 8, Dc = 1.835 g cm–3, T = 93(2) K, 2.989° < θ < 27.468°, 17
145.2(11) Å3, Z = 8, Dc = 1.835 g cm–3, T = 93(2) K, 2.989° < θ < 27.468°, 17 652 unique reflections out of 88
652 unique reflections out of 88 524 with I > 2σ(I), GoF = 1.063, final R factors R1 = 0.0780, and wR2 = 0.2736 for all data, CCDC deposit number 1451761.
524 with I > 2σ(I), GoF = 1.063, final R factors R1 = 0.0780, and wR2 = 0.2736 for all data, CCDC deposit number 1451761.
§Crystallographic data for 1·6: C72H48I12N24Zn6·3.79(C13H16O4)·1.93(C6H12), M = 4220.24, colorless rod, 0.18 × 0.11 × 0.06 mm3, monoclinic, space group C2, a = 36.3783(8) Å, b = 14.6755(2) Å, c = 31.2278(6) Å, β = 103.184(2)°, V = 16 232.2(5) Å3, Z = 4, Dc = 1.727 g cm–3, T = 100(2) K, 4.1720° < θ < 74.0140°, 31
232.2(5) Å3, Z = 4, Dc = 1.727 g cm–3, T = 100(2) K, 4.1720° < θ < 74.0140°, 31 961 unique reflections out of 69
961 unique reflections out of 69 980 with I > 2σ(I), GoF = 1.031, final R factors R1 = 0.0511, and wR2 = 0.1396 for all data, Flack parameter (Parsons) = 0.010(4), CCDC deposit number 1451762.
980 with I > 2σ(I), GoF = 1.031, final R factors R1 = 0.0511, and wR2 = 0.1396 for all data, Flack parameter (Parsons) = 0.010(4), CCDC deposit number 1451762.
¶Crystallographic data for 1·8: C72H48I12N24Zn6·2.8(C19H26O3)·1.5(C6H12), M = 4147.41, colorless plate, 0.28 × 0.08 × 0.07 mm3, monoclinic, space group C2, a = 34.7707(6) Å, b = 14.8406(3) Å, c = 31.2152(5) Å, β = 102.601(2)°, V = 15 719.6(5) Å3, Z = 4, Dc = 1.749 g cm–3, T = 93(2) K, 2.9030° < θ < 73.3970°, 29
719.6(5) Å3, Z = 4, Dc = 1.749 g cm–3, T = 93(2) K, 2.9030° < θ < 73.3970°, 29 574 unique reflections out of 106
574 unique reflections out of 106 306 with I > 2σ(I), GoF = 1.005, final R factors R1 = 0.0788, and wR2 = 0.2259 for all data, Flack parameter (Parsons) = –0.008(8), CCDC deposit number 1451763.
306 with I > 2σ(I), GoF = 1.005, final R factors R1 = 0.0788, and wR2 = 0.2259 for all data, Flack parameter (Parsons) = –0.008(8), CCDC deposit number 1451763.
References
- (a)
Evans G., A Handbook of Bioanalysis and Drug Metabolism, CRC Press, 2004. [Google Scholar]
(b) Bodor N. and Buchwald P., Retrometabolic Drug Design and Targeting, Wiley, 2012. [Google Scholar] - (a) Inokuma Y., Yoshioka S., Ariyoshi J., Arai T., Hitora Y., Takada K., Matsunaga S., Rissanen K., Fujita M. Nature. 2013;495:461–466. [Abstract] [Google Scholar]
(b) Inokuma Y., Yoshioka S., Ariyoshi J., Arai T., Hitora Y., Takada K., Matsunaga S., Rissanen K., Fujita M. Nature. 2013;501:262. [Abstract] [Google Scholar] - For practical guidelines, see:
(a) Inokuma Y., Yoshioka S., Ariyoshi J., Arai T., Fujita M. Nat. Protoc. 2014;9:246–252. [Abstract] [Google Scholar]
(b) Ramadhar T. R., Zheng S., Chen Y., Clardy J. Acta Crystallogr. 2015;A71:46–58. [Europe PMC free article] [Abstract] [Google Scholar]
(c) Hoshino M., Khutia A., Xing H., Inokuma Y., Fujita M. IUCrJ. 2016;3:139–151. [Europe PMC free article] [Abstract] [Google Scholar] - For applications in organic chemistry, see:
(a) Kamimura D., Urabe D., Nagatomo M., Inoue M. Org. Lett. 2013;15:5122–5125. [Abstract] [Google Scholar]
(b) Vinogradova E. V., Müller P., Buchwald S. L. Angew. Chem., Int. Ed. 2014;126:3189–3192. [Google Scholar]
(c) O'Brien A. G., Maruyama A., Inokuma Y., Fujita M., Baran P. S., Blackmond D. G. Angew. Chem., Int. Ed. 2014;53:11868–11871. [Europe PMC free article] [Abstract] [Google Scholar]
(d) Zigon N., Hoshino M., Yoshioka S., Inokuma Y., Fujita M. Angew. Chem., Int. Ed. 2015;54:9033–9037. [Abstract] [Google Scholar]
(e) Yoshioka S., Inokuma Y., Hoshino M., Sato T., Fujita M. Chem. Sci. 2015;6:3765–3768. [Europe PMC free article] [Abstract] [Google Scholar]
(f) Takizawa S., Kishi K., Yoshida Y., Mader S., Arteaga F., Lee S., Hoshino M., Rueping M., Fujita M., Sasai H. Angew. Chem., Int. Ed. 2015;54:15511–15515. [Abstract] [Google Scholar]
(g) Urban S., Brkljača R., Hoshino M., Lee S., Fujita M. Angew. Chem., Int. Ed. 2016;55:2678–2682. [Abstract] [Google Scholar] - (a) Ramadhar T. R., Zheng S., Chen Y., Clardy J. Chem. Commun. 2015;51:11252–11255. [Europe PMC free article] [Abstract] [Google Scholar]
(b) Sanna E., Escudero-Adán E. C., Bauzá A., Ballester P., Frontera A., Rotger C., Costa A. Chem. Sci. 2015;6:5466–5472. [Europe PMC free article] [Abstract] [Google Scholar] - (a) Biradha K., Fujita M. Angew. Chem., Int. Ed. 2002;41:3392–3395. [Abstract] [Google Scholar]
(b) Ohmori O., Kawano M., Fujita M. J. Am. Chem. Soc. 2004;126:16292–16295. [Abstract] [Google Scholar] - Kallman B. J., Andrews A. K. Science. 1963;141:1050–1051. [Abstract] [Google Scholar]
- Seebach D., Roggo S., Maetzke T., Braunschweiger H., Cercus J., Krieger M. Helv. Chim. Acta. 1987;70:1605–1615. [Google Scholar]
Articles from Chemical Science are provided here courtesy of Royal Society of Chemistry
Full text links
Read article at publisher's site: https://doi.org/10.1039/c6sc00594b
Read article for free, from open access legal sources, via Unpaywall:
https://pubs.rsc.org/en/content/articlepdf/2016/sc/c6sc00594b
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Statistical optimization of guest uptake in crystalline sponges: grading structural outcomes.
IUCrJ, 11(pt 4):578-586, 01 Jul 2024
Cited by: 0 articles | PMID: 38864498 | PMCID: PMC11220878
Benchmarking Guanidinium Organosulfonate Hydrogen-Bonded Frameworks for Structure Determination of Encapsulated Guests.
ACS Mater Lett, 6(5):1906-1912, 10 Apr 2024
Cited by: 0 articles | PMID: 38726044 | PMCID: PMC11077584
Molecular-Simulation-Inspired Synthesis of [6]-Prismane via Photoisomerisation of Octafluoro[2.2]paracyclophane.
Molecules, 29(4):783, 08 Feb 2024
Cited by: 0 articles | PMID: 38398535 | PMCID: PMC10891812
The benefits of Cu Kβ radiation for the single-crystal X-ray structure determination of crystalline sponges.
IUCrJ, 9(pt 3):349-354, 10 Mar 2022
Cited by: 2 articles | PMID: 35546798 | PMCID: PMC9067116
Discovery of Cyanobacterial Natural Products Containing Fatty Acid Residues*.
Angew Chem Int Ed Engl, 60(18):10064-10072, 22 Mar 2021
Cited by: 12 articles | PMID: 33599093 | PMCID: PMC8252387
Go to all (12) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Differentiation of volatile aromatic isomers and structural elucidation of volatile compounds in essential oils by combination of HPLC separation and crystalline sponge method.
J Chromatogr A, 1474:130-137, 29 Oct 2016
Cited by: 7 articles | PMID: 27825701
Metabolic Profiling of S-praziquantel: Structure Elucidation Using the Crystalline Sponge Method in Combination with Mass Spectrometry and Nuclear Magnetic Resonance.
Drug Metab Dispos, 50(4):320-326, 03 Feb 2022
Cited by: 0 articles | PMID: 35115299
Combined Analysis Based on a Crystalline Sponge Method.
Anal Sci, 37(1):167-175, 30 Oct 2020
Cited by: 1 article | PMID: 33132236
Review
Crystalline Sponges as a Sensitive and Fast Method for Metabolite Identification: Application to Gemfibrozil and its Phase I and II Metabolites.
Drug Metab Dispos, 48(7):587-593, 20 May 2020
Cited by: 6 articles | PMID: 32434832

 a,c
a,c