Abstract
Free full text

Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy
Abstract
CD8+ T cell–dependent killing of cancer cells requires efficient presentation of tumor antigens by human leukocyte antigen class I (HLA-I) molecules. However, the extent to which patient-specific HLA-I genotype influences response to anti–programmed cell death protein 1 or anti–cytotoxic T lymphocyte–associated protein 4 is currently unknown. We determined the HLA-I genotype of 1535 advanced cancer patients treated with immune checkpoint blockade (ICB). Maximal heterozygosity at HLA-I loci (“A,” “B,” and “C”) improved overall survival after ICB compared with patients who were homozygous for at least one HLA locus. In two independent melanoma cohorts, patients with the HLA-B44 supertype had extended survival, whereas the HLA-B62 supertype (including HLA-B*15:01) or somatic loss of heterozygosity at HLA-I was associated with poor outcome. Molecular dynamics simulations of HLA-B*15:01 revealed different elements that may impair CD8+ T cell recognition of neoantigens. Our results have important implications for predicting response to ICB and for the design of neoantigen-based therapeutic vaccines.
Immune checkpoint inhibitors that target cytotoxic T lymphocyte–associated protein 4 (CTLA-4) or programmed cell death protein 1 (PD-1) or its ligand (PDL-1) have markedly improved the treatment of patients with meta-static cancer (1–3). However, tumor responses to these drugs are variable, and treatment resistance is common (4–6). To date, most research to predict clinical efficacy of immune checkpoint blockade (ICB) therapies has focused on tumor immune phenotype, somatic genomic features, or the gut microbiome (7–21), but how host germline genetics affects response is unclear.
The human leukocyte antigen class I (HLA-I) genotype has been linked with differential immune responses to infection, inflammatory conditions, and autoimmune diseases (22–30). Each HLA-I molecule binds specific peptides derived from intracellular proteins for presentation on the cell surface to CD8+ T cells (31–33). The anti-tumor activity of ICB has been shown to depend on CD8+ T cell, HLA class I–dependent immune activity (34–36). We performed survival and genetic association analyses to address two hypotheses: (i) Zygosity at HLA-I genes influences survival of cancer patients to ICB, and (ii) individual HLA-I germline alleles influence survival to ICB.
We examined two sets of cancer patients (henceforth called cohort 1 and cohort 2) treated with ICB. Cohort 1 (n = 369 patients) was treated with anti–CTLA-4 or anti–PD-1 therapy, and exome sequencing and clinical data were obtained. Within cohort 1, 269 patients had advanced melanoma [previously reported (7, 11, 12, 17)], and 100 patients had advanced non–small cell lung cancer (NSCLC) (table S1) (10). Patients with NSCLC were treated mainly with anti–PD-1 mono-therapy. Cohort 2 (n = 1166 patients) comprised different cancer types, including melanoma and NSCLC (table S1), and tumors were subjected to targeted next-generation sequencing (MSK-IMPACT) (37). These patients were treated with drugs targeting CTLA-4, PD-1/PD-L1, or a combination of both, at the Memorial Sloan Kettering Cancer Center (37). For all patients in both cohorts, we performed high-resolution HLA-I genotyping from normal DNA using DNA sequencing data or a clinically validated HLA typing assay (LabCorp).
HLA-I molecules are highly polymorphic, with variation located in the peptide-binding region; each variant binds a select repertoire of peptide ligands. As such, an individual homo-zygous in at least one HLA-I locus would be predicted to present a smaller, less diverse repertoire of tumor-derived neoantigens to cytotoxic T lymphocytes (CTLs) as compared with a person who is heterozygous at each class I locus (32). We therefore asked whether greater diversity (heterozygosity) in the repertoire of antigen-presenting HLA-I molecules could be associated with better survival after ICB therapy. We examined HLA-I variation at each of the genes (HLA-A, -B, and -C) in cohort 1 and cohort 2 by using a Cox proportional hazard regression model to examine overall survival probability. HLA-I homozygosity in at least one locus was associated with reduced survival in cohort 1 [n = 369 patients; P = 0.036, hazard ratio (HR) = 1.40, 95% confidence interval (CI) 1.02 to 1.9] (Fig. 1A) and was validated in the independent cohort of 1166 patients (cohort 2; P = 0.028, HR = 1.31, 95% CI 1.03 to 1.70) (Fig. 1B). The number of somatic mutations in tumors was not statistically different between homozygous and heterozygous patients (fig. S1, A and B). Furthermore, the association of HLA-I homozygosity with reduced survival remained significant in multivariable Cox regression modeling when analyzed for mutation load, tumor stage, age, and drug class in cohort 1 (P = 0.02, HR = 1.50, 95% CI 1.07 to 2.10) (table S2) and in cohort 2 (P = 0.028, HR = 1.31, 95% CI 1.03 to 1.67) (table S3).
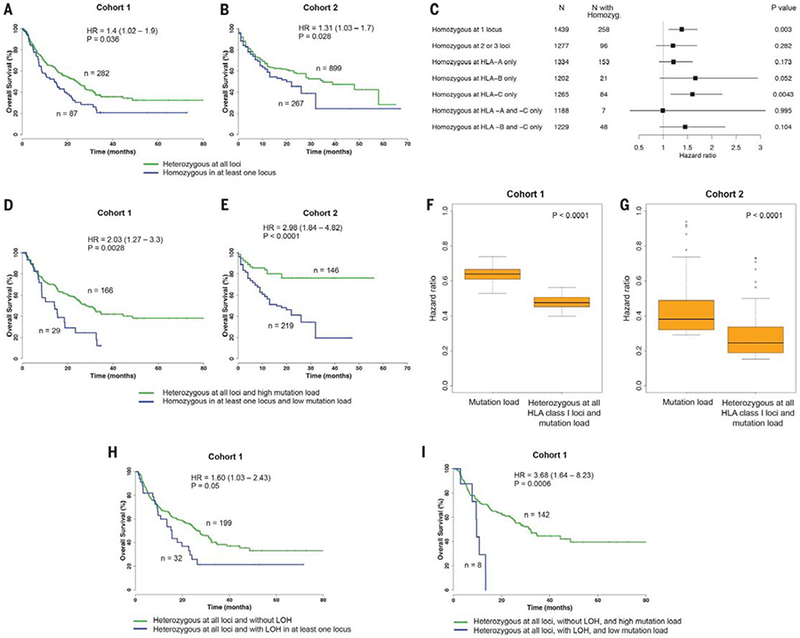
(A) Association between homozygosity in at least one HLA-I locus and reduced overall survival in cohort 1. (B) Association between homozygosity in at least one HLA-I locus and reduced survival in cohort 2. (C) Association between HLA-I homozygosity and decreased survival from all 1535 patients. Data show one or more HLA-I loci or individual loci (HLA-A, HLA-B, and HLA-C). Indicated are the number of patients and HR. Horizontal lines represent the 95% CI. P value was calculated by using the Log-rank test. (D) Patients in cohort 1 with heterozygosity at all HLA-I loci and a high mutation load (defined as >113 mutations) compared with patients that are homozygous for at least one HLA-I locus and have a low mutation load. (E) Patients in cohort 2 with heterozygosity at all HLA-I loci and a high tumor mutational load (defined as >16.72 mutations) compared with patients that are homozygous in at least one HLA-I locus and have a low mutation load. (F) Distribution of HRs to stratify cohort 1 patients based on tumor mutational load. The combined effect of HLA-I heterozygosity at all loci and mutation load on improved survival was greater as compared with mutation load alone. (G) Distribution of HRs to stratify cohort 2 patients based on mutation load. A range of cutoffs across the quartiles of mutation load was used. P values were calculated by using the Wilcoxon-rank sum test. (H) Survival analysis showing that LOH of heterozygous germline HLA-I is associated with decreased overall survival in patients treated with ICB. (I) Survival analysis showing that the effect of LOH of heterozygous germline HLA-I is greater in tumors with low mutation burden compared with tumors with high mutation load and without LOH. High mutation load is defined as >113 mutations.
We next examined all 1535 patients from cohort 1 and 2 together to determine whether the effect of homozygosity may be due to a single HLA-I locus or a combination of different loci. This analysis revealed that homozygosity at one HLA-I locus (“A,” “B,” or “C”) was associated with significant reduction of overall survival (P = 0.003, HR = 1.38, 95% CI 1.11 to 1.70) (Fig. 1C). The effect of homozygosity on survival because of specific HLA-I locus seemed mostly associated with HLA-B (P = 0.052, HR = 1.66, 95% CI 0.93 to 2.94) (Fig. 1C) and HLA-C (P = 0.004, HR = 1.60, 95% CI 1.16 to 2.21) (Fig. 1C). The number of patients available likely limited the interpretability of analyses involving combinations of loci (such as HLA-A and -B). Our findings may be explained because HLA-B is generally expressed at higher levels on the cell surface than HLA-A and HLA-C and because HLA-B alleles bind to a greater diversity of peptides (38, 39). Amino acids that bind to the B pocket of HLA-A alleles are broadly hydrophobic. By contrast, the B pocket of HLA-B alleles can accommodate a greater variety of residues (29, 39). Antigen-presenting cells express higher levels of HLA-C on the cell surface than do other cell types (40), suggesting that heterozygous HLA-C may facilitate continuous CTL priming (41).
Previous reports have shown that the total number of somatic coding mutations in a cancer genome correlates with response to ICB (7, 8, 10–12, 17). An explanation for this observation is that the number of tumor mutations presented on the cell surface increases the probability of neoantigen recognition by cytotoxic T cells (42). We found that HLA-I homozygosity and low mutation burden were strongly associated with decreased survival compared with patients who were heterozygous at each class I locus and whose tumors had high mutation burden, in cohort 1 (P = 0.003, HR = 2.03, 95% CI 1.27 to 3.30) (Fig. 1D) and in cohort 2 (P < 0.0001, HR = 2.98, 95% CI 1.84 to 4.82) (Fig. 1E). The combined effect of HLA class I heterozygosity and mutation load on improved survival was greater as compared with mutation load alone (Fig. 1, F and G).
Previous work has reported loss of heterozygosity (LOH) of HLA-I genes in cancer (43, 44). We thus analyzed all tumor exomes from cohort 1 and identified 32 patients who were heterozygous at all HLA-I loci but had LOH in at least one HLA-I locus in their tumors (table S1). Patients with LOH of HLA-I were associated with reduced survival (P = 0.05, HR = 1.60, 95% CI 1.03 to 2.43) (Fig. 1H). Furthermore, the effect of LOH of HLA-I on survival was greater in patients whose tumors contained low mutation load (P = 0.0006, HR = 3.68, 95% CI 1.64 to 8.23) (Fig. 1I). Given that only a small fraction of presented tumor mutations are immunogenic in cancer patients (45, 46), our findings suggest that relatively small differences in the number of available HLA-I molecules in a given individual can present major challenges to effective antitumor T cell responses and efficacy to ICB. Furthermore, the demonstration of a significant survival advantage to HLA-I heterozygosity in patients treated with ICB both at the germline and somatic level highlights its importance in the dynamic anti-tumor immune response and immune evasion.
As an exploratory analysis, we also found that HLA-II homozygosity at HLA-DP was associated with reduced survival (P = 0.018, HR = 1.45, 95% CI 1.06 to 2.00) (fig. S2A). Additionally, homozygosity at the HLA-DPB locus was associated with decreased survival (P = 0.04, HR = 1.37, 95% CI 1.07 to 1.87). This effect was independent of the associations of HLA-I homozygosity and mutation burden (tables S4 and S5). Mismatched HLA-DP has been shown to be associated with graft-versus-host disease (47).
Additionally, we used next-generation deep sequencing of T cell receptor CDR3 regions (TCR-seq) (48, 49) from a subset of tumor samples collected on-therapy (4 weeks after Nivolumab initiation) (17). We found significantly higher on-therapy clonality of TCR CDR3s in HLA heterozygous patients as compared with HLA homozygotes (in at least one class I locus or at HLA-DP) (Wilcoxon rank-sum test, P = 0.0093) (Fig. 3F and table S1). To refine the interpretation of this result with respect to the antigen-binding properties of the TCR repertoire, the clonality of CDR3s encoded by a single VJ cassette combination was analyzed individually (17, 50). Higher on-therapy clonality of TCR CDR3s per VJ was observed in HLA heterozygotes (Wilcoxon rank-sum test, P = 0.023) (Fig. 3G). Altogether, these results indicate that the diversity of HLA molecules in a given patient influences the selection and the resulting T cell clonal expansion reactive against neoantigens after ICB (51).
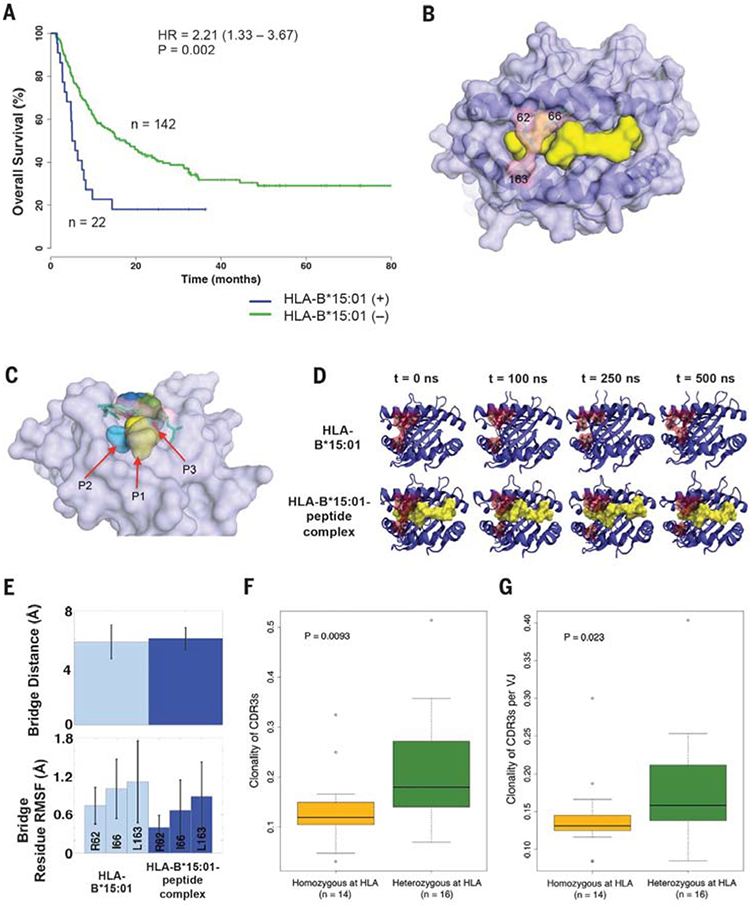
(A) Survival analysis showing reduced survival in ICB-treated melanoma patients from cohort 1 with and without the HLA-B*15:01 allele. (B) Overview of the three-dimensional structure of the peptide-binding groove of HLA-B*15:01, (light purple), bound peptide (yellow), and bridging residues (light pink). (C) Side view of the bridge-sequestration effect over bound-peptide residue positions P2 (light blue) and P3 (red). (D) MD simulation snapshots of both the isolated HLA B*15:01 molecule and its complex with a 9–amino acid UBCH6 peptide; each trajectory was run over the course of 500 ns of simulation time. (E) Observables from the MD simulations described in (D). The mean bridge distances in the HLA-B*15:01 molecule and in the HLA-B*15:01-peptide complex are comparable. The residue-position root mean square fluctuations (RMSFs) indicate that each of the bridging residues becomes more rigid in the presence of the peptide. (F) On-therapy clonality of TCR CDR3s between HLA heterozygous patients and patients who are HLA-homozygous (in at least one class I locus or at HLA-DP). (G) On-therapy clonality of TCR CDR3s per VJ combination between HLA heterozygous patients and patients with HLA homozygosity (in at least one class I locus or at HLA-DP).
To investigate the clinical relevance of individual HLA-I alleles after ICB therapy, we examined the effects of HLA-I supertypes on overall survival. Individual HLA-I alleles are classified into 12 discrete supertypes (52, 53), on the basis of similar peptide-anchor–binding specificities (26, 52, 53). These supertypes together cover most HLA-A and HLA-B alleles found in distinct populations (52, 53).
To assess the effect of HLA supertype on survival, we focused on melanoma patients because there were a sufficient number of patients in the two patient sets for meaningful analysis. On the basis of the biological definition of supertypes, we classified the 27 HLA-A alleles present in the patients with melanoma into six A super-types and the 50 HLA-B alleles into six B super-types (Fig. 2A and table S1). We found two B supertypes, which were associated with survival outcome in patients with advanced melanoma treated with anti–CTLA-4. Patients with B44 superfamily alleles had significantly better survival (P = 0.01, HR = 0.61, 95% CI 0.42 to 0.89) (Table 1), and patients with B62 alleles had significantly reduced survival (P = 0.0007, HR = 2.29, 95% CI 1.40 to 3.74) (Table 1). The B44 supertype was present at a prevalence of 45%, and the B62 supertype was present at 15% (Fig. 2A). We did not find any supertype significantly associated with overall survival in patients with NSCLC, likely because of the limited sample size.
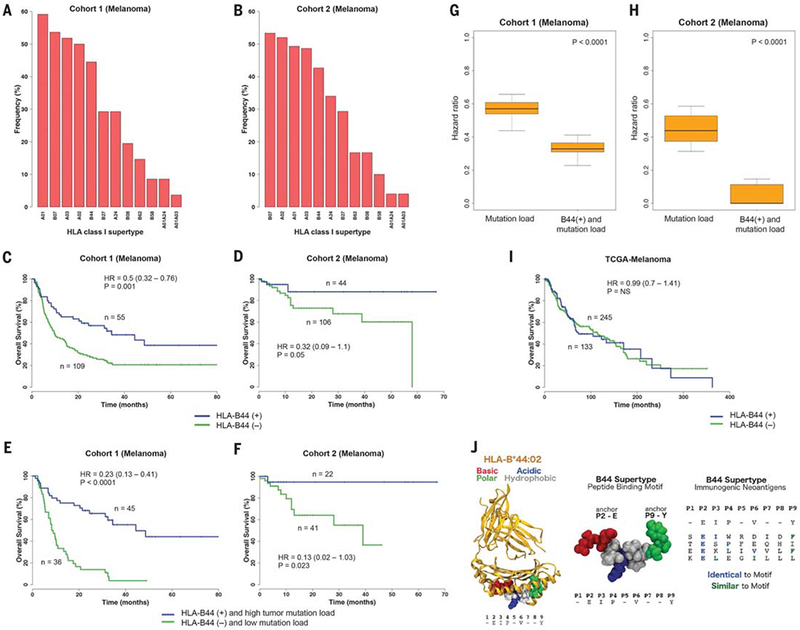
(A) Prevalence of the different HLA supertypes in patients with melanoma from cohort 1. (B) Prevalence of the different HLA supertypes in the patients with melanoma from cohort 2. (C and D) Survival analysis of patients possessing the B44 alleles [B44 (+)] compared with patients without the B44 alleles [B44 (–)] from cohort 1 (C) and cohort 2 (D). (E and F) Survival analysis of patients with the B44 alleles and with high mutation burden versus patients without B44 and with low mutation load, from cohort 1 (E) and cohort 2 (F). (G) Distribution of hazard ratios to stratify cohort 1 patients based on mutation load. The combined effect of B44 and mutation load on increased survival was greater compared with simply considering mutation load alone. (H) Distribution of HRs to stratify cohort 2 patients based on mutation load. A range of cutoffs across the quartiles of mutation load was used. P values were calculated by using the Wilcoxon-rank sum test. (I) Survival analysis of melanoma patients with and without the B44 alleles from the TCGA cohort. (J) (Left) Example of peptide motif common among B44 alleles, docked in complex with HLA-B*44:02 based on an available crystal structure (PDB 1M6O). The five common residues (E2, I3, P4, V6, and Y9) of the motif were reported in (56). Peptide residues are colored according to their properties as basic (red), acidic (blue), polar (green), or hydrophobic (gray). (Center) Close-up view of an example peptide conforming to the B44 motif (54, 56). Residues at positions 2 and 9 are important for anchoring the peptide in the HLA binding groove (54). (Right) Alignment between B44 peptide motif and known immunogenic neoantigens (table S8) restricted to B44 expressed by melanomas. All neoepitopes feature Glu (E) at position 2; neoantigens are also either identical or similar to the motif at one or two additional positions. The neoantigen FAM3C: TESPFEQHI was identified in a melanoma patient with long-term response to anti–CTLA-4 from cohort 1. Sequence similarity was determined by using standard residue classes (GAVLI, FYW, CM, ST, KRH, DENQ, and P).
Table 1.
Data show the influence of specific HLA-I alleles on patient survival.
| HLA-I supertype | Frequency | HR | P value |
|---|---|---|---|
| A24 | 0.29 | 0.67 (0.44 to 1.03) | 0.07 |
| A01 | 0.59 | 0.87 (0.60 to 1.27) | 0.47 |
| A03 | 0.52 | 1.39 (0.96 to 2.03) | 0.08 |
| A02 | 0.5 | 1.13 (0.76 to 1.63) | 0.53 |
| B58 | 0.09 | 0.98 (0.51 to 1.88) | 0.96 |
| B62 | 0.15 | 2.29 (1.40 to 3.74) | 0.0007 |
| B27 | 0.29 | 1.09 (0.73 to 1.63) | 0.67 |
| B44 | 0.45 | 0.61 (0.42 to 0.89) | 0.009 |
| B07 | 0.54 | 1.35 (0.92 to 1.97) | 0.12 |
| B08 | 0.2 | 0.85 (0.52 to 1.39) | 0.51 |
| A01A03 | 0.04 | 1.20 (0.49 to 2.94) | 0.69 |
| A01A24 | 0.09 | 0.89 (0.43 to 1.83) | 0.76 |
| Alleles influencing the significant associations | |||
| B44s, B*18:01, B*44:02, B*44:03, B*44:05, and B*50:01 | 0.34 | 0.49 (0.32 to 0.76) | 0.001 |
| B62s, B*15:01 | 0.13 | 2.21 (1.33 to 3.70) | 0.002 |
We then examined whether these supertype associations were influenced by the presence of specific component HLA-I alleles. The B44 association was influenced by HLA-B*18:01, HLAB*44:02, HLA-B*44:03, HLA-B*44:05, and HLAB*50:01 (P = 0.001, HR = 0.49, 95% CI 0.32 to 0.76) (Fig. 2C and Table 1). And, the B62 association was significantly driven by HLA-B*15:01 (P = 0.002, HR = 2.21, 95% CI 1.33 to 3.70) (Fig. 3A and Table 1). Both of these B44 and B62 allele associations remained statistically significant (P = 0.01 and P = 0.02, respectively) after a Bonferroni correction. The variability in the effect on survival across these B44 alleles might be explained by allele frequency in the cohort or particular differences in the peptide motifs inside or outside primary anchor pockets (54, 55).
In the independent cohort 2, melanoma patients treated with anti–PD-1 or anti–CTLA-4 who had these B44 supertype alleles had signif icantly better overall survival on univariate (P = 0.054, HR = 0.32, 95% CI 0.09 to 1.1) (Fig. 2, B and D) and multivariable analysis (tables S6 and S7). Furthermore, the effect of B44s on extended survival was greater when somatic mutational load was also considered in cohort 1 (P < 0.0001, HR = 0.23, 95% CI 0.13 to 0.41) (Fig. 2E) and in cohort 2 (P = 0.023, HR = 0.13, 95% CI 0.02 to 1.03) (Fig. 2F). The combined effect of the B44 alleles and mutation load was greater than simply considering mutation burden alone (Fig. 2, G and H). In general, outcomes of melanoma patients in cohort 2 tended to be better than in cohort 1 because patients who received ICB and were accrued to our protocol for MSK-IMPACT testing tended to have longer survival. Yet despite this trend, we still observed a significant effect from the B44 alleles. The B44 alleles did not associate with survival in patients with melanoma from The Cancer Genome Atlas (TCGA), suggesting that the presence of B44 was predictive of response to ICB and was not prognostic (Fig. 2I).
Most members of the B44 supertype share a preference for peptides with Glu (E) at anchor position P2 and polar and hydrophobic residues at the C terminus (Fig. 2J) (54, 56). We found that one out of the six enriched amino acid mutations across these tumors was G > E (fig. S3). This observation suggests that there might be an enrichment of presentation of B44-restricted neoantigens. Additionally, a number of previously identified immunogenic antigens expressed by melanomas are HLA-B44–restricted (Fig. 2J and table S8), including the testis antigen MAGEA3, which is restricted to HLA-B*44:03 and HLA-B*18:01 (both members of B44), and a clonal immunogenic neoantigen (FAM3C; TESPFEQHI) that was identified in a melanoma patient with long-term response to CTLA-4 blockade from cohort 1 (table S8) (11, 13).
By contrast, the B62 association with poor survival driven by the HLA-B*15:01 allele was intriguing (Fig. 3A and Table 1). In an exploratory analysis, we sought to determine whether any molecular features in HLA-B*15:01 are associated with its effect on survival. Out of all the HLA-B alleles that were available for three-dimensional structural analysis [n = 119 Protein Data Bank (PDB) crystal structures] (table S9), we identified three alleles at their highest resolutions—HLAB*15:01, HLA-B*07:02, and HLA-B*53:01—as possessing a structural bridge in the peptide-binding groove (Arg62, Ile66, and Leu163) (Fig. 3, B and C).
We postulated that this specific structural feature may modulate the effective T cell recognition of neoepitopes presented on HLA-B*15:01. To evaluate the validity of this hypothesis, we conducted molecular dynamics (MD) simulations following similar protocols used in previous studies (57–59).
In the case of HLA-B*07:02 and HLA-B*53:01, MD simulations demonstrated that the bound peptide expands the respective HLA binding cleft, effectively breaking the bridge (fig. S4, A to D). Conversely, in the HLA-B*15:01 molecule the bridge was largely maintained with the peptide present, and the bridging residues were also made much less flexible (Fig. 3, D and E). Although the mean bridge separation remained nearly constant (~6 Å) in both systems of HLAB*15:01 (Fig. 3E), the fluctuations in this distance were less dramatic in the peptide-bound complex. Altogether, these distinct structural and dynamical elements of HLA-B*15:01 may impair the total strength of the interaction with T cell receptor for effective neoantigen recognition. However, further experimental work will be necessary to test this hypothesis. We found several mutations in genes that have been recently reported to contribute to HLA and cytolytic activity (fig. S5) (4–6, 60). However, we did not find any particular gene mutation associated with decreased overall survival.
Our findings reveal that HLA-I genes influence patient survival to ICB. Both patient-specific HLA-I genotype as well as somatic alterations in tumors affected clinical outcome to ICB, suggesting that these factors could be considered in the design of future clinical trials. The observation that the B44 is associated with extended overall survival may provide an opportunity for the development of therapeutic vaccines that potentially target immunodominant HLA-B44–restricted neoantigens expressed by melanomas. Our findings indicate that HLA-I homozygosity and LOH at HLA-I represent a genetic barrier to effective immunotherapy, and alternative ways to harness the immune system may be necessary to maximize clinical benefit.
Supplementary Material
Supplemental 1
Supplemental Table S1
Table S9
ACKNOWLEDGMENTS
Data reported in this study are tabulated in the main text and supplementary materials. This work was supported by NIH/National Cancer Institute Cancer Center Support Grant P30 CA008748, NIH grant K08 DE024774 (L.G.T.M), Damon Runyon Cancer Research Foundation (L.G.T.M), Pershing Square Sohn Cancer Research Alliance (T.A.C), STARR Cancer Consortium (T.A.C), Stand Up 2 Cancer (T.A.C), and NIH grant 1R01CA205426 (N.A.R and T.A.C). T.A.C. is a cofounder of Gritstone Oncology. We thank the Chan laboratory and members of the Bristol-Myers Squibb (BMS) biomarker team for helpful and stimulating discussions. We thank all the patients for their participation in the clinical trials, which is critical to improving care. We thank members of the melanoma service at MSK, including J. Wolchok, A. Shoushtari, M. Postow, and M. Callahan for their care of clinical trial patients. We thank members of the Columbia University oncology team for their excellent care of trial patients. We thank J. Sims, J. Havel, Y. Shen, and R. Srivastava for very stimulating discussions and helpful suggestions. We thank the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, the Cycle for Survival, and members of the Molecular Pathology and Diagnostics for their help with MSK-IMPACT sequencing. The data are available at the following accession numbers: dbGAP, phs001041.v1.p1; dbGAP, phs000452.v2.p1; SRA, SRP067938 and SRP090294; dbGAP, phs000980.v1.p1; SRA, PRJNA419415, PRJNA419422, and PRJNA419530; cBioPortal for Cancer Genomics, http://cbioportal.org/msk-impact (37). The results published here are in part based on data generated by a TCGA pilot project established by the National Cancer Institute and National Human Genome Research Institute. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov. cancergenome.nih.gov. T.A.C. receives research funding from BMS. N.A.R. is a consultant/advisory board member for AstraZeneca, BMS, Roche, Merck, Novartis, Lilly, and Pfizer. T.A.C. and N.A.R are cofounders of Gritstone Oncology.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/359/6375/582/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S5
Tables S1 to S9
References (61–86)
REFERENCES AND NOTES
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aao4572
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/359/6375/582.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/science.aao4572
Article citations
Cold and hot tumors: from molecular mechanisms to targeted therapy.
Signal Transduct Target Ther, 9(1):274, 18 Oct 2024
Cited by: 0 articles | PMID: 39420203 | PMCID: PMC11491057
Review Free full text in Europe PMC
Unlocking the potential of immunotherapy in platinum-resistant ovarian cancer: rationale, challenges, and novel strategies.
Cancer Drug Resist, 7:39, 15 Oct 2024
Cited by: 0 articles | PMID: 39534871
Review
Perioperative chemoimmunotherapy induces strong immune responses and long-term survival in patients with HLA class I-deficient non-small cell lung cancer.
J Immunother Cancer, 12(10):e009762, 20 Oct 2024
Cited by: 0 articles | PMID: 39428126 | PMCID: PMC11492944
MHC class II genotypes are independent predictors of anti-PD1 immunotherapy response in melanoma.
Commun Med (Lond), 4(1):184, 30 Sep 2024
Cited by: 0 articles | PMID: 39349759 | PMCID: PMC11443121
Integrated multiomics revealed adenosine signaling predict immunotherapy response and regulate tumor ecosystem of melanoma.
Hum Genomics, 18(1):101, 15 Sep 2024
Cited by: 0 articles | PMID: 39278925 | PMCID: PMC11404024
Go to all (580) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject (3)
- (1 citation) BioProject - PRJNA419530
- (1 citation) BioProject - PRJNA419422
- (1 citation) BioProject - PRJNA419415
dbGaP - The database of Genotypes and Phenotypes (3)
- (1 citation) dbGaP - phs001041
- (1 citation) dbGaP - phs000452
- (1 citation) dbGaP - phs000980
Nucleotide Sequences (2)
- (1 citation) ENA - SRP067938
- (1 citation) ENA - SRP090294
Protein structures in PDBe
-
(1 citation)
PDBe - 1M6OView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
CTLA-4 blockade increases antigen-specific CD8(+) T cells in prevaccinated patients with melanoma: three cases.
Cancer Immunol Immunother, 60(8):1137-1146, 05 Apr 2011
Cited by: 59 articles | PMID: 21465316 | PMCID: PMC3654853
Review Free full text in Europe PMC
A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy.
Nature, 551(7681):517-520, 08 Nov 2017
Cited by: 354 articles | PMID: 29132144 | PMCID: PMC6137806
Dendritic cells loaded with mRNA encoding full-length tumor antigens prime CD4+ and CD8+ T cells in melanoma patients.
Mol Ther, 20(5):1063-1074, 28 Feb 2012
Cited by: 53 articles | PMID: 22371843 | PMCID: PMC3345975
Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells.
Science, 348(6236):803-808, 02 Apr 2015
Cited by: 760 articles | PMID: 25837513 | PMCID: PMC4549796
Funding
Funders who supported this work.
Damon Runyon Cancer Research Foundation
NCI NIH HHS (5)
Grant ID: R01 CA205426
Grant ID: P30 CA008748
Grant ID: P30 CA016042
Grant ID: R35 CA232097
Grant ID: R01 CA208403
NIDCR NIH HHS (1)
Grant ID: K08 DE024774
NIH (1)
Grant ID: 1R01CA205426
NIH/NCI Cancer Center Support Grant (1)
Grant ID: P30 CA008748





