Abstract
Free full text

Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments
Abstract
CRISPR (clustered regularly interspaced short palindromic repeats) genome-editing experiments offer enormous potential for the evaluation of genomic loci using arrayed single guide RNARNARNAs (sgRNAs) or pooled sgRNA libraries. Numerous computational tools are available to help design sgRNAs with optimal on-target efficiency and minimal off-target potential. In addition, computational tools have been developed to analyze deep-sequencing data resulting from genome-editing experiments. However, these tools are typically developed in isolation and oftentimes are not readily translatable into laboratory-based experiments. Here, we present a protocol that describes in detail both the computational and benchtop implementation of an arrayed and/or pooled CRISPR genome-editing experiment. This protocol provides instructions for sgRNA design with CRISPOR (computational tool for the design, evaluation, and cloning of sgRNA sequences), experimental implementation, and analysis of the resulting high-throughput sequencing data with CRISPResso (computational tool for analysis of genome-editing outcomes from deep-sequencing data). This protocol allows for design and execution of arrayed and pooled CRISPR experiments in 4–5 weeks by non-experts, as well as computational data analysis that can be performed in 1–2 d by both computational and noncomputational biologists alike using web-based and/or command-line versions.
INTRODUCTION
The CRISPR nuclease system is a facile and robust genome-editing system that was originally identified as the driver of prokaryotic adaptive immunity to allow for resistance to bacteriophages1–3. This system has been subsequently repurposed for eukaryotic genome editing by heterologous expression of the CRISPR components in eukaryotic cells. Site-specific cleavage by Cas9 requires an RNA molecule to guide nucleases to specific genomic loci to initiate double-strand breaks (DSBs)1,2,4. Site-specific cleavage requires Watson–Crick base pairing of the RNA molecule to a corresponding genomic sequence upstream of a protospacer-adjacent motif (PAM)1,2. The required RNA molecule for genome-editing experiments consists of a synthetic fusion of the prokaryotic tracrRNA and CRISPR RNA (crRNA) to create a chimeric sgRNA5. In contrast to Cas9, the Cpf1 nuclease does not require a tracrRNA and engenders DSBs downstream of its PAM sequence. Cpf1 requires a crRNA to create DSBs, which results in 5′ overhangs4.
CRISPR mutagenesis relies on engagement of endogenous DNA repair pathways after nuclease-mediated DSB induction has occurred. The principal repair pathways include nonhomologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ repair is an error-prone pathway, which results in a heterogeneous spectrum of insertions/deletions (indels) primarily in the range of 1–10 bp1,2,6–8. HDR relies on the co-delivery of an extrachromosomal template to be used as a template for DNA repair following DSB, as opposed to an endogenous template such as a sister chromatid. This allows for the insertion of customized sequence into the genome1,2.
Applications of the method and development of the protocol
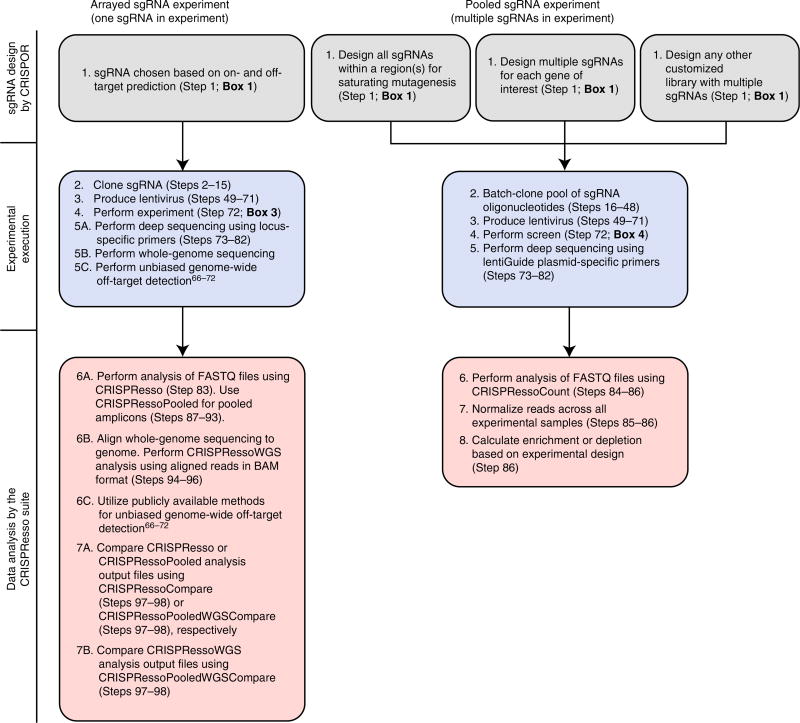
A variety of computational tools have been developed for the design and analysis of CRISPR-based experiments. However, these tools are typically developed in isolation, without features for facile integration with one another, and/or without sufficient consideration to facilitate implementation in a laboratory setting. Here, we offer a protocol to integrate robust, publicly available tools for the design, execution, and analysis of CRISPR genome-editing experiments. Specifically, we have adapted CRISPOR9 and CRISPResso10 to be integrated with one another as well as streamlined for experimental implementation (Figs. 1–3). This protocol has been used in previously published works to functionally interrogate the BCL11A enhancer as well as to evaluate potential functional sequences within all DNase hypersensitive sites (DHSs) in proximity to the MYB gene11,12. CRISPR mutagenesis allows for the study of both coding and noncoding regions of the genome13. This involves usage of one sgRNA for arrayed experiments or multiple sgRNAs for pooled experiments. Arrayed experiments are useful when a target can be mutagenized by one sgRNA. Pooled screening allows for targeting of a handful of genes up to genome-scale gene targeting14–17. It also allows for saturating mutagenesis (tiling of sgRNA) experiments to identify functional sequence within noncoding regions11,12,18. This protocol can be used to design and execute arrayed or pooled genome-editing experiments. Furthermore, it can also be used for the design, implementation, and analysis of pooled screens for gene targeting, saturating mutagenesis of noncoding elements, or any other targeting strategies11,12,19,20. It is important to note that computational skills are required for the command-line and Docker versions of CRISPOR and CRISPResso in this protocol. Specifically, users are required to have a basic understanding of how to execute a command in a terminal and how to navigate a Unix-based file system. However, web-based versions are also available that do not require these skills.
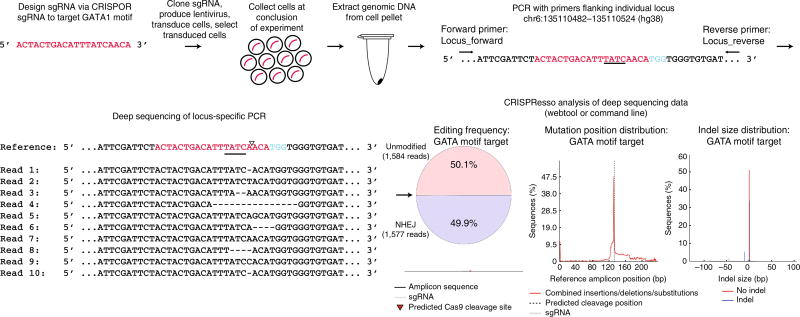
Schematic of an arrayed genome-editing experiment. Arrayed genome-editing experiments are performed by designing one sgRNA using CRISPOR. This schematic demonstrates the design of an sgRNA to mutagenize a GATA motif. After designing the optimal sgRNA, it is cloned into pLentiGuide-puro, lentivirus is produced, cells are transduced, and successful transductants are selected (successful transduction is indicated by red curved lines). After conclusion of the experiment, cells are pelleted, and genomic DNA is extracted. Locus-specific PCR primers are used to amplify regions flanking the double-strand break site. Deep sequencing of the amplicon generated by locus-specific PCR is subsequently performed. Quantification of editing frequency and indel distribution is determined by CRISPResso. The GATA motif is underlined, the triangle indicates the double-strand break position, and the PAM sequence is shown in blue.
CRISPOR: sgRNA and PCR primer design for arrayed and pooled screen experiments
CRISPOR (http://crispor.org) is a computational sgRNA design tool that predicts off-target cleavage sites and offers a variety of on-target efficiency scoring systems to assist sgRNA selection for more than 120 genomes using many different CRISPR nucleases (Supplementary Table 1)9. CRISPOR offers a variety of on- and off-target prediction scores that can aid in optimal sgRNA selection (Box 1)9. Analysis of on-target sgRNA efficiency can be predicted based on available scores and/or investigated experimentally by analysis of editing frequency. In addition to the numerous sgRNA efficiency prediction scores, CRISPOR offers automated primer design to facilitate PCR amplification of regions for deep-sequencing analysis to quantitate editing frequency. This involves PCR amplification of sequences flanking the DSB site for a given sgRNA. Similarly, CRISPOR offers computational prediction of off-target sites as well as PCR primers for deep-sequencing analysis of potential mutagenesis at these predicted off-target sites.
CRISPOR can also be used for the design of gene-targeted pooled screens by including exonic regions for sgRNA design as well as saturating mutagenesis screens. Alternatively, a list of gene names can be input into CRISPOR (‘CRISPOR Batch’) to aid in the design of large scale gene-targeted libraries, which includes non-targeting controls. This analysis includes automated design of the required oligonucleotides for library cloning.
Saturating mutagenesis involves using all PAM-restricted sgRNAs within a given region(s) in a pooled screening format to identify functional sequences11,12,18. Saturating mutagenesis can be used to analyze coding and noncoding elements in the genome or a combination of the two. Screen resolution is a function of PAM frequency and can be enhanced by PAM choice and/or a combination of nucleases with unique PAM sequences12. CRISPOR can be used to design saturating mutagenesis libraries by simply selecting all sgRNAs within the inputted region(s). This analysis also includes automated design of the required oligonucleotides for library cloning. It is particularly important to consider off-target prediction scores offered by CRISPOR for saturating mutagenesis screens, as repetitive sequences can confound screen results12. sgRNAs with high probability of off-target mutagenesis can be excluded at the library design stage or can be appropriately handled at the analysis stage after the experiments have been performed (Box 1).
CRISPResso: analysis of deep sequencing from arrayed or pooled sgRNA experiments
CRISPResso is a computational pipeline that enables accurate quantification and visualization of CRISPR genome-editing outcomes, as well as comprehensive evaluation of effects on coding sequences, noncoding elements, and off-target sites from individual loci, pooled amplicons, and whole-genome deep-sequencing data10. The CRISPResso suite involves multiple tools for analysis, including the CRISPResso webtool and command-line version of CRISPResso. There are also multiple additional command-line-only tools in the CRISPResso suite: CRISPRessoPooled, CRISPRessoWGS, CRISPRessoCount, CRISPRessoCompare, and CRISPRessoPooledWGSCompare. The applications and features of these tools are summarized in Table 1. CRISPResso analysis offers many unique features, such as splice-site or frameshift analysis to quantify the proportion of engendered mutations that result in a frameshift when targeting coding sequence. In addition, features for indel visualization have been added to CRISPResso since its initial publication10. Notably, CRISPResso also provides a variety of features to offer users the opportunity to optimize analysis of sequencing data (Box 2).
Table 1
CRISPResso analysis suite.
| Name | Format | Purpose | Input file formats | Comments |
|---|---|---|---|---|
| CRISPResso | Webtool | Analysis of single amplicon/locus deep sequencing | .fastq or .fastq.gz | http://www.crispresso.rocks |
| CRISPResso | Command-line version | Analysis of single amplicon/locus deep sequencing | .fastq or .fastq.gz | Large file support; batch mode capability |
| CRISPRessoPooled | Command-line version | Analysis of pooled amplicon experiments | fastq or .fastq.gz | — |
| CRISPRessoWGS | Command-line version | Analysis of WGS data or pre-aligned reads | .bam | Useful to interrogate any region of the genome for off-target effects |
| CRISPRessoCompare | Command-line version | Comparison of two CRISPResso analyses | Output for CRISPResso analysis on two different samples | Useful to compare treated and untreated samples or to compare different experimental conditions |
| CRISPRessoPooledWGSCompare | Command-line version | Compare experiments involving several regions analyzed by either CRISPRessoPooled or CRISPRessoWGS | Output from CRISPRessoPooled or CRISPResso WGS analysis on two different samples | — |
| CRISPRessoCount | Command-line version | Enumerate sgRNAs present within a given sample | .fastq or .fastq.gz | Useful to perform enrichment or dropout (‘depletion’) analysis for a pooled screen |
CRISPResso analysis of an individual locus requires PCR amplification of the sequences flanking the genomic position of the DSB for a given sgRNA. The resulting deep-sequencing FASTQ file can be analyzed by CRISPResso to quantitate the indel spectrum as well as visualize individual alleles (see Fig. 4 and ANTICIPATED RESULTS). This analysis can be performed when targeting coding (Fig. 4a–d) or noncoding sequences. When targeting coding (exonic) sequence, CRISPResso can determine the frequency of in-frame and out-of-frame (frameshift) mutations produced (Fig. 4d). This type of analysis may suggest toxicity upon gene knockout by identifying an increased frequency of in-frame mutations12. Two separate CRISPResso analyses with the same amplicon can be directly compared using the CRISPRessoCompare tool (Table 1). CRISPRessoCompare is useful in situations such as comparison of ‘treated’ and ‘untreated’ groups, as well as comparison of different experimental conditions. It can also be used to compare indel distributions created by two different sgRNAs within the same region/amplicon11.

Locus-specific deep-sequencing analysis of coding and noncoding targeting by CRISPResso. (a) Frequency distribution of alleles with indels (shown in blue) and without indels (shown in pink) for an sgRNA targeting BCL11A exon 2. (b) All reads with sequence modifications (insertions, deletions, and substitutions) are mapped to a position within the BCL11A exon 2 reference amplicon. The vertical dashed line indicates the position of predicted Cas9 cleavage. The position of the sgRNA is shown in gray. (c) Distribution of indel sizes when targeting BCL11A exon 2. Percentage of unmodified sequences is shown in red and percentages of modified sequences are shown in blue. (d) Frameshift analysis of BCL11A exon 2 coding sequence targeted reads. Frameshift mutations are shown in red and in-frame mutations are shown in tan. (e) sgRNA enrichment based on analysis of fetal hemoglobin (HbF) levels when performing saturating mutagenesis of BCL11A exon 2 and analysis of the functional core of the BCL11A enhancer using NGG- and NGA-restricted sgRNAs from two previously published studies11,12. Nontargeting sgRNAs are pseudomapped with 5-bp spacing.
CRISPResso analysis can be extended from a single amplicon to multiple amplicons using the CRISPRessoPooled tool (Table 1). This is useful for individual locus experiments that require multiple amplicons for analysis of the full region or when multiple genes are targeted. The CRISPResso suite can also be used to analyze whole-genome sequencing (WGS) data through CRISPRessoWGS. This requires pre-aligned WGS data in BAM format, which can be created using publicly available aligners (e.g., Bowtie2 (ref. 21) or Burrows–Wheeler Aligner22,23). Similar to CRISPRessoCompare, two analyses using either CRISPRessoPooled or CRISPRessoWGS can be directly compared using CRISPRessoPooledWGSCompare (Table 1). This can be particularly useful when using multiple-amplicon sequencing data or WGS data to evaluate off-target cleavages by two different sgRNAs to identify sgRNAs with lower off-target activity.
CRISPResso also offers the ability to analyze deep-sequencing data from pooled CRISPR screens. Sequence/indel-based analysis in a pooled screening format is confounded by non-edited reads from cells containing sgRNAs targeting other regions/loci. Therefore, pooled screens are typically analyzed by enumeration of sgRNAs by PCR-amplifying sequences containing the cloned sgRNA within the lentiviral construct that has been integrated into the cell’s genome (using PCR primers specific to the lentiviral construct). Deep-sequencing data generated using PCR primers specific to the lentiviral construct can be analyzed by CRISPRessoCount for sgRNA enumeration for the purposes of calculating enrichment and/or dropout of sgRNAs under different experimental conditions11,12 (Figs. 2, ,4e).4e). After sgRNA enumeration, it is important to normalize the reads by taking into account the total number of reads when comparing two different deep-sequencing samples. An example strategy for performing normalization involves normalizing all samples to 1 million reads. This can be accomplished by dividing the read count for each sgRNA by the total number of reads in that sample. Then this quotient is multiplied by 1 million, for example: (sgRNAn read count/total read count of sample)×1,000,000. This should be repeated for all samples so that each sample has 1 million total normalized reads. These normalized reads can then be used to calculate enrichment and/or dropout (‘depletion’) ratios, for example: log2(normalized read count of sgRNAn in sample X/normalized read count of sgRNAn in sample Y).
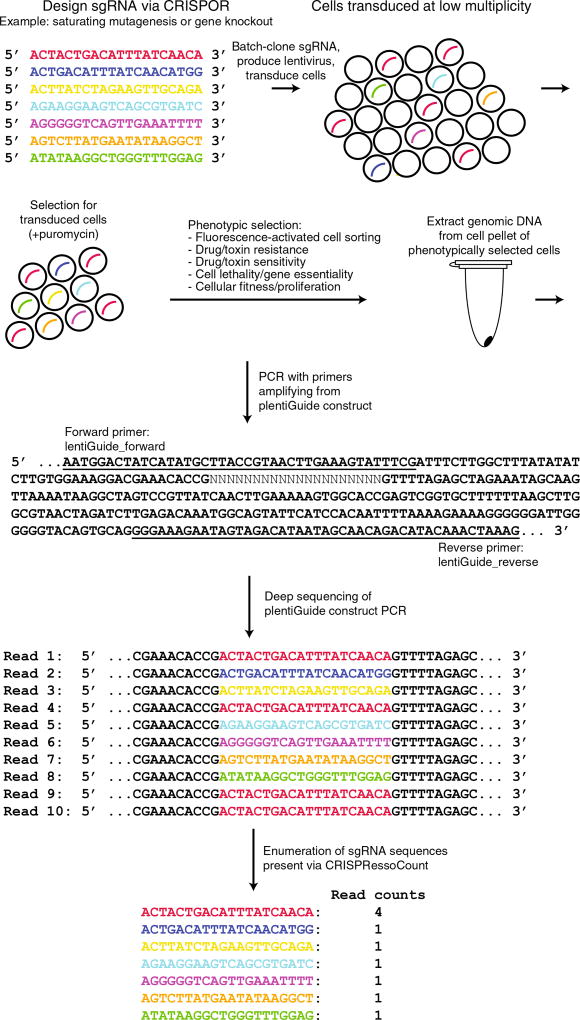
Schematic of a pooled genome-editing experiment. Pooled genome-editing experiments are performed by designing multiple sgRNA using CRISPOR. After designing the sgRNAs, they are batch-cloned into pLentiGuide-puro, lentivirus is produced, cells are transduced at low multiplicity, and successful transductants are selected (successful transduction is indicated by curved lines). Phenotypic selection is performed (e.g., FACS, drug/toxin resistance, drug/toxin sensitivity, cell lethality/gene essentiality, cellular fitness/proliferation). After conclusion of the experiment, cells are pelleted, and genomic DNA is extracted. PCR primers specific (primer sequence is underlined) to the pLentiGuide-Puro construct are used to amplify regions flanking the cloned sgRNA sequence. Deep sequencing of the amplicon generated by construct-specific PCR is subsequently performed. sgRNAs present within the sample are enumerated by CRISPRessoCount.
Comparison with other methods
Numerous computational tools are freely available to aid sgRNA design for a wide spectrum of PAM sequences, as well as for on-target efficiency and off-target cleavage predictions: Broad GPP Portal24, Cas-Database25, Cas-OFFinder26, CasOT27, CCTop28, COSMID29, CHOPCHOP30,31, CRISPRdirect32, CRISPR-DO33, CRISPR-ERA34, CRISPR-P35, CROP-IT36, DNA Striker12, E-CRISP37, flyCRISPR38, GUIDES39, GuideScan40, GT-scan41, MIT CRISPR design tool8, WU-CRISPR42, CRISPRseek43, sgRNAcas9 (ref. 44), and CRISPR multiTargeter45, as well as others offered by companies such as Deskgen46 and Benchling46. CRISPOR offers several unique advantages for designing sgRNAs for genome-editing experiments. First, CRISPOR integrates multiple published on-target sgRNA efficiency scores, including those from Fusi et al.47, Chari et al.48, Xu et al.49, Doench et al.24,50, Wang et al.51, Moreno-Mateos et al.52, Housden et al.53, Prox. GC54, -GG55, and Out-of-Frame56. It also offers previously published off-target prediction (MIT specificity score)8. Second, CRISPOR has been optimized to facilitate experimental implementation by providing automated primer design for both on-target and off-target deep-sequencing analysis. The primers and output files are further designed to be compatible for subsequent analysis by CRISPResso after the experiments have been completed. CRISPOR also offers features to automate oligonucleotide design for cloning pooled sgRNA libraries targeting both genes and noncoding regions. Taken together, CRISPOR provides an sgRNA design platform to facilitate experimental execution and downstream data analysis.
Alternative computational tools to CRISPResso exist to evaluate genome-editing outcomes from deep-sequencing data57–61; however, these tools offer limited analysis functionality for pooled amplicon sequencing or WGS data as compared with the CRISPResso suite. CrispRVariants is another tool that offers functionality to analyze deep-sequencing data by quantifying mosaicism and allele-specific gene editing, as well as multisequence alignment views60.
CRISPR genome-editing reagents have taken many forms, including DNA, RNA, protein, and various combinations of each62,63. Delivery of these reagents has also been attempted using a variety of methods, including electroporation, lipid-based transfection, and viral-mediated delivery62,63. For further discussion of delivery methods for genome-editing reagents, refer to Yin et al.64. Pooled screening relies on the ability to deliver individual reagents to individual cells in batches65. Electroporation and lipid-based transfection methods offer limited ability to control the number of reagents (i.e., sgRNAs) delivered per cell; however, lentiviral transduction at low transduction rates (~30–50%) results in single lentiviral integrants per cell in the majority of cases65. Furthermore, lentivirus offers stable integration of the CRISPR reagents into each cell’s genome. These features of lentivirus allow for pooled CRISPR experiments. Therefore, this protocol describes the use of lentivirus for both arrayed and pooled CRISPR experiments (Boxes 3 and 4).
Limitations of on-target prediction, off-target prediction, and indel analysis
As described above, there are many tools available for both on-target sgRNA efficiency and off-target cleavage prediction. Progress has been made toward enhancing the predictive value of these scores; however, although these predictions are useful to focus sgRNA selection for experimental design, experimental validation provides the definitive analysis of on-target and off-target mutagenesis. Alternatively, experimental approaches have also been developed for unbiased genome-wide detection of off-target cleavages66–72. Continued investigation is necessary to more completely understand the rules governing sgRNA efficiency and off-target mutagenesis (see Box 1 for further discussion).
CRISPResso indel analysis can supplement other common analyses, such as analyses of gene expression or protein-level changes. Although CRISPResso is useful for quantifying editing frequency to demonstrate that editing has successfully occurred and demonstrates the full indel spectrum (substitutions, insertions, and deletions), it is often helpful to perform other techniques to assess for changes beyond genomic DNA (gDNA) for further evaluation (i.e., gene expression or protein-level changes).
Experimental design
Target identification and nuclease choice
CRISPR genome-editing experiments require appropriate target identification to fit experimental objectives, which can include functional analysis of gene- or noncoding sequence (e.g., enhancers, CCCTC-binding factor (CTCF), or other transcription factor binding sites). Gene knockout is usually accomplished by targeting exonic sequences, but it can also be achieved through promoter disruption. Gene knockout is often complicated by alternative splicing and/or expression of multiple isoforms for a given gene of interest. It is difficult to predict the relevant isoform without prior knowledge; however, it is often possible to target exonic sequence common to all isoforms or to design sgRNAs targeting unique isoforms to aid in the identification of a relevant/functional isoform. In the case of gene families, conserved domains can be identified for disruption37. Although exon targeting toward the 5´ end of the gene has been shown to be more effective for functional disruption than targeting the 3´ end50, it is also possible to target functional protein domains. In this case, even in-frame mutations frequently disrupt protein function73. Similar considerations can be applied to CRISPR interference (CRISPRi) or CRISPR activation (CRISPRa) approaches; however, CRISPRi/CRISPRa require sgRNA targeting in close proximity to the transcriptional start site for maximal repression/activation17,74–80; CRISPRi/CRISPRa approaches are not the focus of this protocol. For further discussion and a protocol for CRISPRa approaches, refer to Joung et al.20.
Once target(s) have been identified, it is necessary to determine whether the experiment will be arrayed or pooled. Arrayed experiments are useful when a target can be mutagenized by one sgRNA (e.g., a transcription factor binding motif and gene knockout via exon mutagenesis) (Fig. 1), whereas pooled screen experiments are useful when a target(s) requires > 1 sgRNA for mutagenesis (Fig. 2). Both arrayed and pooled experiments are described in this protocol, as arrayed experiments are typically required to validate the results of pooled experiments. As with arrayed experiments, the first step of pooled experiments is target identification. This can include targeting a single locus with multiple sgRNAs (e.g., saturating mutagenesis (so-called tiling sgRNA) experiments)11,12,18, targeting multiple loci by saturating mutagenesis (e.g., saturating mutagenesis of multiple DHSs)11,12, and gene-targeted pooled screens (e.g., targeting multiple genes with multiple sgRNAs per gene)14–17. In addition, libraries combining gene-based targeting and saturating mutagenesis approaches can also be designed. Experimental designs and workflows in this protocol are summarized in Figure 3.
Finally, each CRISPR nuclease offers a unique PAM sequence with varying frequency of occurrence in the genome depending on the location of targeting such as exons, introns, promoters, DHSs, enhancers, or repressed regions12,65. The optimal nuclease can be chosen based on the density of available PAMs (e.g., for saturating mutagenesis) or proximity of PAMs to a particular genomic position (e.g., for transcription factor binding motifs) (Supplementary Table 1). Finally, high-fidelity nucleases (e.g., HypaCas9, SpCas9-HF1, and eSpCas9) can be used to minimize the probability of off-target mutagenesis81–83.
Positive, negative, and editing controls for arrayed and pooled screen CRISPR experiments
Positive and negative controls are essential for both arrayed and pooled screen experiments. Positive controls are generally experiment-specific; however, some widely used positive controls when performing experiments to identify novel essential genes or when performing dropout (‘depletion’) screens include targeting of known essential genes (e.g., ribosomal genes and housekeeping genes) (see Box 4 for further discussion of dropout screens). When targeting noncoding sequences for the effects on gene expression, one possible positive control is targeting of exonic sequences11,12.
Negative controls typically take the form of nontargeting controls, which are sgRNAs without any perfect matches in a given genome and with low potential for cleavage at genomic loci with imperfect matches (selected by minimization of sites with few mismatches, which is similar to minimizing off-target effects for targeted sgRNA). Nontargeting controls are genome build–, species-, and PAM-specific (unless designed to be compatible with multiple genomes and/or PAMs). Other options for negative controls include targeting a safe harbor locus such as AAVS1 or a gene/region known to have no effect on cellular function or the phenotype of interest. Nontargeting sgRNA can be generated ‘by hand’ via a guess-and-check approach to ensure no genomic matches (and minimal sites with few mismatches) or can be designed using previously published tools12. For both positive and negative controls, one sgRNA is often used for an arrayed experiment and multiple sgRNAs are often used for pooled screen experiments, oftentimes comprising 1–5% of the total number of sgRNAs in the library (see Box 4 for further discussion)11,12.
An editing control can also be included to ensure proper functioning of the CRISPR reagents, particularly nuclease function (see Boxes 3 and 4 for further discussion). One possibility is to include a construct that expresses GFP together with an sgRNA targeting GFP to assess for functional Cas9 expression by flow cytometry12,50. This is particularly informative when using cell lines with stable Cas9 expression and can be helpful for troubleshooting low or absent editing rates. Typically, only one reporter system/sgRNA to serve as an editing control is needed for both arrayed and pooled experiments.
Synthesis of individual or multiple pooled sgRNA libraries
After designing relevant sgRNAs, CRISPOR automates the design of full-length oligonucleotides for pooled screen applications such as saturating mutagenesis (Step 1A and 1C) and gene-targeted libraries generated from a gene list (Step 1B). Barcodes allow for oligonucleotides for multiple unique libraries to be synthesized on the same programmable microarray (Steps 16 and 17), which reduces cost by avoiding purchase of multiple microarrays for multiple libraries. Specifically, the barcodes offer the ability for individual PCR amplification of unique libraries from a single batch of oligonucleotides synthesized on the same microarray. The number of libraries that can be included on a single programmable microarray is limited by the microarray’s oligonucleotide capacity; however, there is theoretically no limit to the number of possible barcodes that can be used (and thus no limit to the number of libraries that can be generated from a single pool of oligonucleotides). Barcodes for ten libraries are provided in Supplementary Table 2. To create additional or new barcodes, generate 10–13 bp of sequence distinct from the sequences to be amplified and the other barcodes used. It is also important to ensure that the barcode results in a primer melting temperature (Tm) compatible with the lsPCR1 reaction (Step 19; see Supplementary Table 3 for examples). Homologous sequence to the lentiGuide-Puro plasmid is required because batch sgRNA library cloning is performed using Gibson assembly (Steps 27 and 28), which relies on homologous flanking sequence for successful cloning.
Sequencing of arrayed experiments and pooled screens
Sanger sequencing is initially required to confirm successful cloning of an individual sgRNA for an arrayed experiment using a U6 sequencing primer (5′-CGTAACTTGAAAGTATTTCGATTTCTTGGC-3′ (ref. 62)). For a pooled screen experiment, deep sequencing is required to confirm successful library cloning.
Deep sequencing can be used to analyze editing outcomes for arrayed experiments. In contrast, deep sequencing is typically required to analyze pooled screen experiments. The choice for appropriate deep-sequencing platform (e.g., MiSeq or HiSeq) should include consideration of the required number of reads for the experiment as well as cost. For an arrayed experiment for indel enumeration/analysis by CRISPResso (Step 83), the number of reads needed to adequately represent a locus may vary widely based on experimental goals; however, > 1,000 reads at a given locus is reasonable. For a pooled sgRNA library experiment, 100–1,000× coverage of the number of sgRNAs in the library is reasonable. The desired read length should be chosen on the basis of the length of the amplicon generated by the primers used in Step 75. Once the deep-sequencing platform has been selected, submit barcoded samples for deep sequencing (Step 82; these represent different barcodes than those described in the ‘Synthesis of individual or multiple pooled sgRNA libraries’ section).
Amplicon length should be compatible with the read length as described above. 100- and 200-bp amplicons are reasonable for paired-end deep sequencing with 75- or 150-bp reads, respectively. Because sequencing quality is lower toward the end of the reads, there must be some overlap of read pairs to allow reliable merging. For example, for a 200-bp amplicon, it is suggested to use 125- to 150-bp reads in order to have sufficient overlap. Some considerations for determining amplicon length include the cost of sequencing for a given read length, the size in base pairs of the region to be sequenced, and the length of reliable sequencing (due to unreliable sequence at read ends). If an amplicon length is too short, it may result in inadequate coverage of the target region, as well as miss larger indels. By contrast, if the amplicon is longer than twice the read length, no overlap will exist between reads, making the merging steps of paired-end reads impossible.
MATERIALS
REAGENTS
Cloning/transformation
E. cloni 10G Elite electrocompetent cells with recovery medium (Lucigen, cat. no. 60052)
Endura electrocompetent cells with recovery medium (Lucigen, cat. no. 60242)
Gibson assembly master mix (New England BioLabs, cat. no. E2611S)
Gene Pulser/MicroPulser electroporation cuvettes, 0.1-cm gap (Bio-Rad, cat. no. 1652089)
Fast Digest Esp3I (Thermo Fisher, cat. no. FD0454)
NEB stable competent E. coli (high efficiency) (New England BioLabs, cat. no. C3040H)
Lysogeny broth (LB) medium base (Thermo Fisher, cat. no. 12780052)
Dehydrated agar (Fisher Scientific, cat. no. DF0140-01-0)
Thermosensitive alkaline phosphatase (TSAP; Promega, cat. no. M9910)
Nuclease-free water (Fisher Scientific, cat. no. AM9937)
T4 polynucleotide kinase (New England BioLabs, cat. no. M0201S)
Ampicillin sodium salt (Sigma-Aldrich, cat. no. A9518)
SOC (Super Optimal broth with Catabolite repression) medium (Thermo Fisher, cat. no. 15544034)
Quick Ligation Kit (New England BioLabs, cat. no. M2200S)
Individual oligonucleotides (e.g., Integrated DNA Technologies, Bio-Rad, custom order)
Oligonucleotide pool (e.g., CustomArray, Twist Bioscience, custom order)
QIAquick PCR Purification Kit (Qiagen, cat. no. 28104)
Gel electrophoresis
SYBR Safe DNA Gel Stain (Thermo Fisher, cat. no. S33102)
Amresco Agarose I (VWR, cat. no. 97062-250)
1-kb Plus DNA Ladder (Thermo Fisher, cat. no. 10787018)
50× Tris-acetate-EDTA (TAE) buffer (Boston Bioproducts, cat. no. BM-250)
PCR
Phusion Hot Start Flex DNA Polymerase (New England BioLabs, cat. no. M0535S)
Q5 High-Fidelity DNA Polymerase (New England BioLabs, cat. no. M0491S)
Herculase II Fusion DNA Polymerase (Agilent Genomics, cat. no. 600675)
DMSO (Sigma-Aldrich, cat. no. D8418)
QIAquick Gel Extraction Kit (Qiagen, cat. no. 28704)
MinElute PCR Purification Kit (Qiagen, cat. no. 28004)
Qubit dsDNA HS Assay Kit (Thermo Fisher, cat. no. Q32854)
DNeasy Blood & Tissue Kit (Qiagen, cat. no. 69504)
Plasmids/plasmid preparation
lentiGuide-Puro (Addgene, plasmid ID 52963; ref. 19)
lentiCas9-Blast (Addgene, plasmid ID 52962; ref. 19)
pXPR_011 (Addgene, plasmid ID 59702; ref. 50)
lenti-Cas9-VQR-Blast (Addgene, plasmid ID 87155; ref. 12)
lenti-Cas9-VQR-GFP_activity_reporter (Addgene, plasmid ID 87156; ref. 12)
Qiagen Plasmid Maxi Kit (Qiagen, cat. no. 12163)
AccuPrep Plasmid Mini Extraction Kit (Bioneer, cat. no. K-3030)
Lentivirus production
pCMV-VSV-G (Addgene, plasmid ID 8454)
psPAX2 (Addgene, plasmid ID 12260)
Polyethylenimine (PEI), branched (Sigma-Aldrich, cat. no. 408727)
Sucrose (Sigma-Aldrich, cat. no. S0389)
Steriflip-HV filter units, 0.45 µm, PVDF, radio-sterilized (Millipore, cat. no. SE1M003M00)
Stericup-GP filter units, 0.22 µm, polyethersulfone, 500 ml, radio-sterilized (Millipore, cat. no. SCGPU05RE)
Phosphate-buffered saline, 1×, without calcium, magnesium (Lonza, cat. no 17-516F)
DMEM (Life Technologies, cat. no. 11995-073)
HEK293FT cells (Thermo Fisher Scientific, cat. no. R70007)
! CAUTION Cell lines should be regularly checked to ensure that they are authentic and are not infected with mycoplasma.
Lenti-X qRT-PCR Titration Kit (Takara Bio, cat. no. 631235)
Lenti-X p24 Rapid Titer Kit (Takara Bio, cat. no. 632200)
EQUIPMENT
Corning untreated 245-mm2 bioassay dishes (Fisher Scientific, cat. no. 431111)
Ultracentrifuge tube, thin-wall, polypropylene, 38.5 ml, 25 × 89 mm (Beckman Coulter, cat. no. 326823)
Falcon 50-ml conical centrifuge tubes (Fisher Scientific, cat. no. 14-432-22)
Corning tissue culture (TC)-treated culture dishes, 15 cm, round (Fisher, cat. no. 08-772-24)
Penicillin–streptomycin (Life Technologies, cat. no. 15140122)
Solid glass beads (Fisher Scientific, cat. no. 11-312-10B)
Electroporation system (e.g., Bio-Rad’s Gene Pulser MXcel, Gene Pulser Xcell, and MicroPulser electroporators)
Corning Falcon bacteriological Petri dishes with lids (Fisher Scientific, cat. no. 08-757-100D)
MiSeq or HiSeq sequencing system (Illumina) or equivalent sequencing platform
Qubit fluorometer (Thermo Fisher Scientific)
UV-visible spectrophotometer (NanoDrop)
Gel visualization system (Alpha Innotech)
Thermocycler (Bio-Rad)
UV-light transilluminator (Fisher Scientific)
UV-filter face mask (Fisher Scientific) ! CAUTION Wear gloves, lab coat, and face shield to avoid harm to eyes and skin caused by UV light.
Microcentrifuge tubes, polypropylene (VWR, cat. no. 87003-294)
8-Strip PCR tubes, 0.2 ml (Fisher Scientific, cat. no. 14-222-250)
Sterile culture tube with attached dual-position cap (VWR, cat. no. 89497-812)
Ultracentrifuge (Beckman Coulter)
SW32Ti rotor with compatible swinging buckets (Beckman Coulter, cat. no. 369694)
Falcon cell scraper with 40-cm handle and 3.0-cm blade (Corning, cat. no. 353087)
COMPUTING EQUIPMENT
Computer with at least 8 GB of RAM, 1 TB of disk space, and a recent central processing unit (CPU) with two or more cores (command-line and Docker version only). Windows (Docker only), OSX, and Linux platforms are supported.
Anaconda Python 2.7 (https://www.continuum.io/downloads (CRISPResso is not compatible with Python 3))
Sequence analysis software, e.g., free software such as ApE (http://biologylabs.utah.edu/jorgensen/wayned/ape/), SerialCloner (http://serialbasics.free.fr/Serial_Cloner.html), or commercial software such as Thermo Scientific Vector NTI (https://www.thermofisher.com/us/en/home/life-science/cloning/vector-nti-software.html), Lasergene DNAstar (https://www.dnastar.com/t-allproducts.aspx), CLC Main Workbench (https://www.qiagenbioinformatics.com/products/clc-main-workbench), or MacVector (http://macvector.com)
CRISPResso command-line software (https://github.com/pinellolab/CRISPResso)
▲ CRITICAL Refer to the ‘Equipment Setup’ section for detailed installation instructions and troubleshooting advice for CRISPResso.
CRISPOR command-line software: http://crispor.org/downloads
▲ CRITICAL Refer to the ‘Equipment Setup’ section for detailed installation instructions and troubleshooting advice for CRISPROR.
FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
REAGENT SETUP
Preparation of sgRNA cloning, sequencing, and primer oligonucleotides
When ordering individual oligonucleotides for sgRNA cloning, add ‘CACC’ onto the sense strand of the sgRNA (5´ CACCNNNNNNNNNNNNNNNNNNNN 3´) and ‘AAAC’ onto the antisense strand of the sgRNA (5´ AAACNNNNNNNNNNNNNNNNNNNN 3´). The two oligonucleotides are designed as reverse complements because they will be annealed together in Step 4. The ‘CACC’ and ‘AAAC’ sequences are required for cloning of the phosphorylated/annealed oligonucleotides into the Esp3l-digested/dephosphorylated lentiGuide-Puro plasmid (Steps 3–10). The addition of a ‘G’ at the 5´ end of the sgRNA sequence can increase transcription from the U6 promoter (5´ CACCGNNNNNNNNNNNNNNNNNNNN 3´ and 5´ AAACNNNNNNNNNNNNNNNNNNNNC 3´); this is not required if the 5´ end of the sgRNA sequence is already a ‘G’. Using CRISPOR, the sequences for sgRNA cloning into lentiGuide-Puro can be downloaded by following the ‘Cloning/PCR primers’ link below the sgRNA sequence. The ready-to-order oligonucleotide sequences are available from the sgRNA results table by selecting the ‘lentiGuide-Puro (Zhang lab)’ plasmid under the ‘U6 expression from an Addgene plasmid’ heading. Dissolve the obtained oligonucleotides in nuclease-free water at a concentration of 100 µM. Store at 20 °C for up to 2 years.
TAE electrophoresis solution
Dilute 50× TAE buffer stock in dH2O to obtain a 1× working solution. Store at room temperature (25 °C) for up to 6 months.
Ampicillin solution
Dissolve ampicillin in a 1:1 mix of 100% ethanol and dH2O to a final concentration of 100 mg/ml. Store at − 20 °C for up to 4–6 months. ▲ CRITICAL Protect from light.
HEK293FT cell culture medium
HEK293FT cell culture medium is DMEM with glucose/sodium pyruvate supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin–streptomycin. Sterilize with a 0.22-µm filter and store it at 4°C for up to 4 weeks. ! CAUTION The HEK293 cells used should be regularly checked to ensure that they are not infected by mycoplasma.
PEI solution
Dissolve PEI in nuclease-free water at 10 µg/µl with pH 7.4. This solution can be stored at 4 °C for years.
20% (wt/vol) sucrose solution
Dissolve sucrose in PBS to create a 20% (wt/vol) solution by mass. Sterilize with a 0.22-µm filter and store at 4 °C for up to 1 year.
EQUIPMENT SETUP
Installation of Docker
Docker is a virtualization technology that allows for packaging software with all dependencies into files called containers to be executed on Windows, Linux, or OSX. This allows for the creation and distribution of a ‘frozen’ version of the software that will always run independent of updates or changes to libraries or the required dependencies on the host machine. In this case, you only need to install Docker and do not need to install any dependencies. Download and install Docker from this link: https://docs.docker.com/engine/installation/.
Sharing disk volumes with Docker
By default, Docker containers cannot share data with the machine on which they run. For this reason, it is necessary to map local folders to folders inside the Docker container. Mapping allows the container to read the input data files to process/write the output files into a folder on the disk of the local machine. First, it is necessary to check whether Docker has permission to access the disk(s) on your machine where analysis data are to be stored and processed. By default, any subfolder within your home directory is automatically shared; however, this behavior may change with future versions of Docker. Check that the drive(s) you want to be available to the container is/are selected in the ‘Settings…/ Shared Drives’ panel (Supplementary Fig. 1a).
To map a local folder (i.e., a folder on the local disk) to a container folder, Docker has a special option with this syntax:
-v local_folder:container_folder
You can specify this option multiple times if it is necessary to map more than one folder. In our examples, the folders to map are all subfolders of the home folder of the user ‘user’, i.e., /home/user/. In addition, some of the Docker commands will be used with the option
–w /DATA
to specify the working directory (i.e., where the command will be executed) to allow for the use of relative paths and to shorten the commands.
Allocation of memory for Docker containers
It is necessary to allocate enough memory to the container. The default assigned by Docker may depend on the version and on the machine on which it is run. To run the CRISPOR/CRISPResso containers, we suggest assigning Docker 6–8 GB of RAM. For analysis involving large sequences or many sites, it may be necessary to increase the amount of allocated memory under the ‘Settings…/ Advanced’ panel (Supplementary Fig. 1b). The execution may halt with an error if enough memory is not allocated to the container (e.g., with ‘ /bin/bash: line 1: 14 Killed/crisporWebsite/bin/Linux/bwa bwas w’). In this instance, the memory allocated to the Docker container is not sufficient to run CRISPOR, and the process is terminated. Therefore, increase the memory as discussed above and run the same command again.
CRISPResso installation
The CRISPResso utility can be used for analysis of deep-sequencing data of a single locus/amplicon on a local machine or proprietary server, using a command-line version or a webtool freely available at http://crispresso.rocks/. The webtool version of CRISPResso does not require installation. For installation of the command-line version of CRISPResso or installation of CRISPResso with Docker, see the instructions below.
Installation of the command-line version of CRISPResso
To install CRISPResso on a local machine, it is necessary to install some dependencies before running the setup script. Download and install Anaconda Python 2.7, following the instructions at this link:
Open a terminal and type
conda config --add channels r conda config --add channels defaults conda config --add channels conda-forge conda config --add channels bioconda conda install CRISPResso fastqc
Close the terminal and open a new one; this will set the PATH variable. Now, you are ready to use the command-line version of CRISPResso.
If you have installed an old version of CRISPResso, please remove it with the following command to avoid conflicts:
rm -Rf /home/user/CRISPResso_dependencies
Installation of CRISPResso with Docker
To install CRISPResso with Docker, it is necessary to verify that Docker is installed before running the setup script (see ‘Installation of Docker’ within the ‘Equipment Setup’ section). After verification of Docker installation, type the following command:
docker pull pinellolab/crispor_crispresso_nat_prot
Check whether the container was downloaded successfully by running the command
docker run pinellolab/crispor_crispresso_nat_prot CRISPResso --help
If CRISPResso was properly installed, you will see the Help for the command-line version of CRISPResso.
PROCEDURE
sgRNA design using CRISPOR ● TIMING 1–4 h
- 1|
Design sgRNAs from a single genomic locus using the CRISPOR webtool by following option A. sgRNAs can be selected manually from the CRISPOR output using on- and off-target predictions (Box 1). Alternatively, follow option B for the ‘CRISPOR Batch’ gene-targeting assistant webtool for gene-targeted pooled library generation from a gene list. If multiple genomic loci are required for sgRNA design or the command-line version is preferred, use option C for the command-line version of CRISPOR.
▲ CRITICAL STEP CRISPOR does not offer the ability to design repair templates for HDR experiments. If the experimental design requires a repair template, other protocols can provide further instruction62.
(A) sgRNA design for arrayed and pooled experiments using the CRISPOR webtool ● TIMING 1–4 h
Open the CRISPOR webtool (http://crispor.tefor.net/).
Input the target DNA sequence. The webtool requires a sequence input (< 2 kb) or genomic coordinates.
▲ CRITICAL STEP The input sequence must be a genomic sequence as opposed to a cDNA sequence, as the latter can include sequences that are not in the genome due to splicing. You can obtain genomic sequences using a website such as the UCSC Genome Browser (https://genome.ucsc.edu, click on ‘View - DNA’ after searching for a gene or genomic region) or Ensembl (http://www.ensembl.org, click on ‘Export data – Text’ after searching for a gene or genomic region). From the UCSC Genome Browser, the current sequence in view can be sent directly to CRISPOR via the menu entry ‘View – In external tools’.
? TROUBLESHOOTING
Select the relevant genome assembly (e.g., hg19, mm9) and PAM sequence for the nuclease (e.g., NGG for Streptococcus pyogenes Cas9).
▲ CRITICAL STEP It is important to pick the appropriate assembly to be consistent with the genomic coordinates of the DNA sequence provided in Step 1A(ii) and for accurate prediction of off-target sites.
Click on the ‘Submit’ button to run the CRISPOR analysis. Select the ‘Cloning/PCR primers’ link underneath each sgRNA sequence for automated primer design for future indel analysis by CRISPResso at on- and off-target loci. If you are conducting a saturating mutagenesis screen, the list of the oligonucleotide pool sequences for Gibson assembly, sequencing primers for validation, sequencing amplicons for CRISPResso, and the full list of sgRNA sequences can be downloaded by following the link ‘Saturating Mutagenesis Assistant’ at the top of the CRISPOR output page. The four files provided by the Saturating Mutagenesis Assistant can be downloaded by selecting from the drop-down menu adjacent to ‘output file’. The outputs are described in Table 2. Alternatively, the full list of sgRNA sequences for saturating mutagenesis can also be found by following the ‘Cloning/PCR primers’ link under each sgRNA sequence and then following the link provided under the ‘Saturating mutagenesis using all guides’ heading.
Table 2
Four output files from CRISPOR sgRNA design analysis.
Output
numberFilename Description File columns 1 REGION_1_sat MutOligos.tsv This file contains the sequences to order from a custom oligonucleotide pool supplier (MATERIALS) GuideId: the identifier of the sgRNA sequence in the input sequence. It consists of the position of the PAM and the strand, e.g., ‘4rev’
targetSeq: the sgRNA sequence, including the PAM
mitSpecScore: the MIT Guide Specificity score (0–100, higher score = lower off-target potential)
off-target count: number of predicted off-target sites (by default at four mismatches)
targetGenomeGeneLocus: gene symbol and sequence location name, e.g., ‘exon:PITX2’
Doench’16EffScore: the Doench 2016 guide efficiency score24
Moreno-MateosEffScore: the Moreno-Mateos 2015 (CRISPRscan) guide efficiency score52
OligoNucleotideAdapterHandle + PrimerFw: the forward oligonucleotide to order from a supplier
OligoNucleotideAdapterHandle + PrimerRev: the reverse oligonucleotide to order from a supplier
2 REGION_1_ontarget Primers.tsv This file contains two primers for each sgRNA that can be ordered from an oligonucleotide supplier. The primers can be used to amplify the DNA fragment around each sgRNA guideId: sgRNA identifier, see above
forwPrimer: forward primer sequence
forwPrimerTm: forward primer Tm
revPrimer: reverse primer sequence
revPrimerTm: reverse primer Tm
ampliconSequence: the genomic sequence between forward and reverse primer
3 REGION_1_ontarget Amplicons.tsv After sequencing using the ontargetPrimers file, this file can be used as input guideId: sgRNA identifier, see above
ampliconSequence: the genomic sequence between forward and reverse primers, see above
For CRISPRessoPooled to determine the cleavage frequency of each sgRNA guideSequence: the sgRNA sequence located within the amplicon
4 REGION_1_tar getSeqs.tsv This file contains a list of all sgRNA sequences (one per line). It can be used to quantify the relative abundance of these sgRNA in a sample of sequenced cells and is one of the input files for CRISPRessoCount List of all sgRNA sequences (one per line) ▲ CRITICAL STEP CRISPOR allows for PCR amplicon lengths of 50–600 bp. The melting temperature (Tm) is calculated for each primer pair.
(B) Gene-targeted library design generated from a gene list using the CRISPOR Batch gene-targeting assistant webtool ● TIMING 1–4 h
To generate a gene-targeted library using the CRISPOR Batch gene-targeting assistant, go to http://crispor.tefor.net/ and click on the ‘CRISPOR Batch’ link.
Select the source genome-wide library for the extraction of sgRNA sequences using the drop-down menu adjacent to ‘Lentiviral screen library’. References are provided for each library to assist with appropriate source library selection.
Select the desired ‘number of guides per gene’ (maximum of six sgRNAs/gene) using the drop-down menu. Note that some libraries have fewer than six sgRNAs per gene. If more sgRNAs are requested than are available for a given gene with the library, the maximum number of sgRNA sequences available within the library will be output instead.
Enter the ‘number of non-targeting control guides’ to include in the library (maximum = 1,000). See the ‘Experimental design’ section for further discussion of controls.
Select a barcode for library preparation as described in Step 16 (see ‘Synthesis of individual or multiple pooled sgRNA libraries’ within the ‘Experimental design’ section for further discussion of barcodes).
Copy and paste the gene list into the entry field. Genes should be entered as official gene symbols, with only one entry per line. Refer to the HUGO Gene Nomenclature Committee (HGNC) (https://www.genenames.org/) and Mouse Genome Informatics (MGI) (http://www.informatics.jax.org/) for official gene symbols for human and mouse, respectively.
▲ CRITICAL STEP This entry is case-insensitive because the libraries are already species-specific. If an entry is not found in the database of genome-wide sgRNAs, sgRNAs for the entered gene will display a warning and will not be included in the output.
? TROUBLESHOOTING
Click on ‘Submit’ for gene-targeted library generation and download the output library.
(C) sgRNA design using command-line CRISPOR ● TIMING 1–4 h
Type the following command into a terminal to download the latest version of the Docker container for this protocol:
docker pull pinellolab/crispor_crispresso_nat_prot
Verify that the container was downloaded successfully by running the following command:
docker run pinellolab/crispor_crispresso_nat_prot crispor.py
Obtain the CRISPOR assembly identifier of the genome (e.g., hg19 or mm9). If you are unsure, the full list of assemblies is available at http://crispor.tefor.net/genomes/genomeInfo.all.tab. This protocol uses ‘hg19’ in its examples; however, this should be switched to the relevant identifier for the assembly of interest as appropriate.
Execute the following commands to download the relevant pre-indexed genome in the folder created in the previous step. Here, we are assuming that the user will store the pre-indexed genomes in the folder /home/user/crispor_genomes.
mkdir -p /home/user/crispor_genomes docker run -v /home/user/crispor_genomes:/crisporWebsite/genomes \ pinellolab/crispor_crispresso_nat_prot downloadGenome hg19 \ /crisporWebsite/genomes
Prepare the input genomic DNA (gDNA) sequences in FASTA format (see a description of FASTA format at https://www.ncbi.nlm.nih.gov/blast/fasta.shtml).
▲ CRITICAL STEP It is important to pick the appropriate genome assembly to be consistent with the genomic coordinates of the input DNA sequences. In particular, it is essential for accurate prediction of off-target sites.
(Optional) Generate a FASTA file with the exonic sequences. If you have a list of genes and you want to target their exons, go to the UCSC Genome Browser page http://genome.ucsc.edu/cgi-bin/hgTables.
Set the following options (refer to the description of each item in the ‘Table Browser’ to assist with setting each parameter). Of note, the ‘Table’ parameter will offer different options depending on the chosen ‘Track’. Step 1C(viii–xii) is based on the example entry from Step 1C(vii):
UCSC Genome Browser table
optionExample entry (adjust as appropriate) Clade Mammal Genome Human, assembly: hg19 Group Genes and gene predictions Track Gencode V19 Table Basic (wgEncodeGencodeBasicV19) Output format Sequence Output file selected_genes_exons.fasta File type returned ‘plain text’ Press the button ‘paste list’ and enter the list of transcript identifiers for each gene.
Press the button ‘get output’. A new page will open.
▲ CRITICAL STEP The ‘Send output to’ Galaxy, GREAT, or GenomeSpace checkboxes are off by default; however, they will remain checked if you have ever checked any of them in the past. Make sure that none of the checkboxes are selected.
Select the option ‘genomic’ and press ‘submit’. A new page will open.
Select ‘CDS: Exons’ and ‘One FASTA record per region (exon, intron, etc.)’.
Press the button: ‘get sequence’ and save the file as selected_genes_exons.fasta.
Run CRISPOR over your single- or multi-fasta input file by entering the following commands. Here, we assume that this file is stored in /home/user/crispor_data/crispor_input.fasta and contains a sequence with id > REGION_1.
docker run \ -v /home/user/crispor_data:/DATA \ -v /home/user/crispor_genomes:/crisporWebsite/genomes \ -w /DATA \pinellolab/crispor_crispresso_nat_prot \ crispor.py hg19 crispor_input.fasta crispor_output.tsv --satMutDir=./
The sgRNA sequences will be written to the file crispor_output.tsv and four files for each sequence ID in the FASTA file will be created as in the web version in the folder /home/user/crispor_data/ (see Table 2 for output files).
To select a subset of sgRNAs from the file REGION_1_satMutOligos.tsv as generated in Step 1A(iv) or 1C(xiii) (Table 2), select the lines corresponding to sgRNAs with the desired scores/attributes (Box 1) and save them to a new file called REGION_1_satMutOligos_filtered.tsv. Filter the sgRNAs by running the following commands:
docker run -v /home/user/crispor_data/:/DATA -w /DATA pinellolab/ crispor_crispresso_nat_prot bash -c "join -t $'\t' −1 1 −2 1 REGION_1_satMutOligos_filtered.tsv REGION_1_ontargetAmplicons.tsv -o 2.1,2.2,2.3 > CRISPRessoPooled_amplicons.tsv" docker run -v /home/user/crispor_data/:/DATA -w /DATA pinellolab/ crispor_crispresso_nat_prot bash -c "join -t $'\t' −1 1 −2 1 REGION_1_satMutOligos_filtered.tsv REGION_1_ontargetAmplicons.tsv -o 2.3 | sed 1d > CRISPRessoCounts_sgRNA.tsv" docker run -v /home/user/crispor_data/:/DATA -w /DATA pinellolab/ crispor_crispresso_nat_prot bash -c "join -t $'\t' −1 1 −2 1 REGION_1_satMutOligos_filtered.tsv REGION_1_ontargetPrimers.tsv -o 2.1,2.2,2.3,2.4,2.5,2.6,2.7 > REGION_1_ontargetPrimers_filtered.tsv"
This will create a filtered version of the files described in Table 2 to use for designing amplicons and for performing CRISPResso analysis.
▲ CRITICAL STEP If the input file to CRISPOR was a multi-fasta file (Step 1C(xiii)), repeat this step for the other regions as appropriate.
Synthesis and cloning of individual sgRNA into a lentiviral vector ● TIMING 3 d to obtain sequence-confirmed, cloned sgRNA lentiviral plasmid
- 2|
Individually resuspend lyophilized primer oligonucleotides at 100 µM in nuclease-free water.
■ PAUSE POINT Resuspended oligonucleotides can be stored at −20 °C for years.
- 3|
Set up the phosphorylation reaction:
Components Amount (µl) Final concentration Oligonucleotide 1 (at 100 µM) 1 – Oligonucleotide 2 (at 100 µM) 1 10× Ligation buffer 1 1× T4 polynucleotide kinase (10 units/µl) 1 10 U Nuclease-free water To 10 - 4|
Perform phosphorylation and annealing of the oligonucleotides in a thermocycler:
Step Temperature (°C) Time Comment 1 37 30 min 2 95 5 min 3 95–25 30 s Ramp down 5 °C/min ■ PAUSE POINT Resuspended phosphorylated/annealed oligonucleotides can be stored at − 20 °C for years.
- 5|
Prepare the digestion reaction by combining the following components in a 0.2-ml or 1.5-ml tube.
Components Amount Final concentration lentiGuide-Puro plasmid 1 µg – 10× FastDigest buffer 2 µl 1× DTT (20 mM) 1 µl 1 mM Esp3l restriction enzyme 1 µl Nuclease-free water To 20 µl - 6|
Digest the sgRNA plasmid (lentiGuide-Puro) with Esp3l (an isoschizomer of BsmBI) restriction enzyme for 15 min at 37 °C.
- 7|
Add 1 µl of TSAP to dephosphorylate the reaction for 15 min at 37 °C, followed by heat inactivation for 15 min at 74 °C.
- 8|
Perform gel purification of the Esp3l-digested/dephosphorylated lentiGuide-Puro plasmid. See MATERIALS for gel purification kit and follow the manufacturer’s instructions. For further information on gel purification, refer to this previously published protocol84.
- 9|
Determine the concentration of the Esp3l-digested/dephosphorylated lentiGuide-Puro plasmid after gel purification using a UV-visible spectrophotometer (e.g., NanoDrop) or other equivalent method, following the manufacturer’s instructions.
- 10|
Perform a ligation reaction for 5–15 min at room temperature using 30–70 ng of Esp3l-digested/dephosphorylated lentiGuide-Puro plasmid (generated in Steps 5–9) with the phosphorylated–annealed oligonucleotides produced in Steps 2–4 diluted 1:500 with nuclease-free water.
Components Amount Final concentration 2× Quick ligase reaction buffer 5 µl 1× Diluted oligonucleotides (from Step 4) 1 µl LentiGuide-Puro plasmid (from Steps 5 to 9) 30–70 ng 1× Quick ligase 1 µl 10 U Nuclease-free water To 10 µl - 11|
Heat-shock-transform the ligation reaction into stable competent E. coli by incubating samples at 42 °C for 45 s, followed by a 10-min incubation on ice. Add 250 µl of SOC medium and incubate at 37 °C with 250-r.p.m. shaking for 1 h. Use of recombination-deficient E. coli is important to minimize recombination events of repetitive elements in lentiviral plasmid (e.g., long terminal repeats). For more information about performing heat-shock transformation of competent E. coli, refer to previously published protocols85,86.
- 12|
Plate the transformation product from Step 11 on a 10-cm ampicillin-resistant (0.1 mg/ml ampicillin) agar plate. Incubate at 32 °C for 14–16 h. It is recommended that transformation products be incubated at 32 °C to reduce recombination between lentiviral long terminal repeats, but incubation at 37 °C can also be performed.
- 13|
Picking three to five colonies is reasonable for individual incubation of each colony in 2 ml of ampicillin-resistant (0.1 mg/ml ampicillin) LB medium at 32 °C for 14–16 h with shaking at 250 r.p.m. It is recommended that transformation products be incubated at 32 °C to reduce recombination between lentiviral long terminal repeats, but incubation at 37 °C can also be performed.
? TROUBLESHOOTING
- 14|
Perform mini-scale plasmid preparation of each 2-ml culture from Step 13 (see MATERIALS for a mini-scale plasmid preparation kit; follow the manufacturer’s instructions).
■ PAUSE POINT Successfully cloned sgRNA plasmids can be stored long term (years) at − 20 °C before creating lentivirus (Steps 49–71).
- 15|
Sanger-sequence each colony-derived plasmid with the U6 sequencing primer to identify correctly cloned plasmids (see ‘Experimental design’ section for U6 primer sequence)62. Subsequent midi- or maxi-scale plasmid preparation may be required for lentivirus production (Steps 49–71).
Synthesis and cloning of pooled sgRNA libraries into a lentiviral vector ● TIMING 2–4 d to obtain cloned sgRNA lentiviral plasmid library
- 16|
Use the sgRNAs from Step 1 to design full-length oligonucleotides (96–99 bp) flanked by barcodes and homologous sequence to the lentiGuide-Puro plasmid (Supplementary Table 2).
- 17|
Order the designed DNA oligonucleotides on a programmable microarray (MATERIALS).
- 18|
Obtain the resuspended oligonucleotides for use as the template for the PCR reaction in Step 19. The concentration will be determined by the manufacturer, so refer to the manufacturer for identifying the concentration.
■ PAUSE POINT Oligonucleotide pools can be stored at − 20 °C for short-term storage (months to years) and at − 80 °C for long-term storage (years).
- 19|
Set up the barcode-specific PCR to amplify using the barcode-specific primers (Supplementary Table 3). This PCR will herein be referred to as ‘Library Synthesis PCR1’ (lsPCR1).
▲ CRITICAL STEP It is important to use a proofreading DNA polymerase to minimize introduction of PCR errors.
Components Amount (µl) Final concentration Synthesized oligonucleotide template from Step 18 1 – 5× Phusion HF buffer or 5× Q5 reaction buffer 10 1× Deoxynucleoside triphosphates (dNTPs; at 10 mM) 1 200 µM Barcode-specific forward primer (at 10 µM) 2.5 0.5 µM Barcode-specific reverse primer (at 10 µM) 2.5 0.5 µM Phusion Hot Start Flex DNA Polymerase or Q5 High-Fidelity DNA Polymerase 0.5 1.0 U/50-µl PCR Nuclease-free water To 50 ? TROUBLESHOOTING
- 20|
Perform multiple cycles (e.g., 10, 15, and 20 cycles) of lsPCR1 in a thermocycler as follows:
Cycle number Denature Anneal Extend 1 98 °C, 30 s 2-x (e.g., 10, 15, 20) 98 °C, 10 s 63 °C, 30 s 72 °C, 30 s x + 1 72 °C, 5 min ▲ CRITICAL STEP The number of PCR cycles should be limited to reduce PCR bias. It is difficult to predict the required number of cycles for a given library. Therefore, it is important to perform multiple PCRs with different numbers of cycles to empirically determine the optimal number of cycles.
■ PAUSE POINT The PCR product can be stored for multiple days at 4 °C in the thermocycler at the end of the PCR program. Long-term storage (months to years) should be at − 20 °C.
- 21|
Run a fraction (1–5 µl of the 50-µl PCR reaction from Step 20) of each lsPCR1 product with a different number of PCR cycles on a 2% (wt/vol) agarose gel. Analyze the gel to determine if the products occur at the expected size of ~100 bp (Supplementary Fig. 2a). If the bands run at the expected size, determine the minimal number of PCR cycles required to produce a visible band (Supplementary Fig. 2a). For example, 15 cycles would be used based on the gel presented in Supplementary Figure 2a; however, it may be possible to reduce the cycle number even further (i.e., another experiment with 5 and 10 PCR cycles). Use the remaining reaction volume for the cycle number chosen for Step 22 based on the gel from this step.
- 22|
Using nuclease-free water, dilute the lsPCR1 reaction chosen in Step 21 1:10.
- 23|
Set up ‘Library Synthesis PCR2’ (herein referred to as lsPCR2) with the universal lsPCR2 primers listed in Supplementary Table 4. lsPCR2 amplification removes the library-specific barcodes and adds a sequence homologous to the lentiGuide-Puro plasmid for Gibson assembly-based cloning of the library.
▲ CRITICAL STEP It is important to use a proofreading DNA polymerase to minimize introduction of PCR errors.
Components Amount (µl) Final concentration Diluted lsPCR1 product from Step 22 1 – 5× Phusion HF buffer or 5× Q5 reaction buffer 10 1× dNTPs (at 10 mM) 1 200 µM lsPCR2_forward primer (at 10 µM) 2.5 0.5 µM lsPCR2_reverse primer (at 10 µM) 2.5 0.5 µM Phusion Hot Start Flex DNA Polymerase or Q5 High-Fidelity DNA Polymerase 0.5 1.0 U/50-µl PCR Nuclease-free water To 50 - 24|
Perform lsPCR2 using the same cycling conditions and multiple cycle numbers as Step 20.
■ PAUSE POINT The PCR product can be stored for multiple days at 4 °C in the thermocycler at the end of the PCR program. Long-term storage (months to years) should be at − 20 °C.
- 25|
Run lsPCR2 products on a 2% (wt/vol) agarose gel to determine whether the products occur at the expected size of ~140 bp (Supplementary Fig. 2b). Choose the sample with the minimal number of PCR cycles that produces a visible band in order to minimize PCR bias (Supplementary Fig. 2b). For example, 15 cycles would be used based on the gel presented in Supplementary Figure 2b; however, it may be possible reduce cycle number even further (i.e., another experiment with 5 and 10 PCR cycles).
- 26|
Gel-purify the ~140-bp band. See MATERIALS for gel purification kit and follow the manufacturer’s instructions. For further information on gel purification, refer to this previously published protocol84.
▲ CRITICAL STEP If synthesizing multiple libraries simultaneously, leave empty lanes between different samples during gel electrophoresis to minimize the risk of sample contamination during gel purification.
- 27|
Set up a Gibson assembly reaction using gel-purified Esp3l-digested/dephosphorylated lentiGuide-Puro plasmid from Steps 5 to 9:
Components Amount Final concentration Gel-purified lsPCR2 from Step 26 10 ng Esp3l-digested/dephosphorylated lentiGuide-Puro plasmid from Steps 6 to 9 25 ng 2× Gibson assembly master mix 10 µl 1× Nuclease-free water To 20 µl - 28|
Incubate the Gibson assembly reaction mix in a thermocycler at 50 °C for 60 min.
- 29|
Thaw one 25-µl aliquot of electrocompetent bacterial cells on ice until they are completely thawed (~10–20 min).
- 30|
Mix thawed electrocompetent bacterial cells by gently tapping on the side of the tube.
- 31|
Add 25 µl of thawed electrocompetent bacterial cells to a prechilled 1.5-ml microcentrifuge tube on ice.
- 32|
Add 1 µl of the Gibson assembly reaction mixture from Step 28 to 25 µl of thawed electrocompetent bacterial cells (from Step 31).
▲ CRITICAL STEP It is reasonable to aim for > 50× representation of the sgRNA library. Depending on the achieved transformation efficiency as calculated in Step 41 and the number of sgRNAs in the library, it may be necessary to set up more than 1 transformation reaction from Step 32. It is important to set up multiple reactions using 1 µl of Gibson assembly reaction mixture and 25 µl of thawed electrocompetent bacterial cells. Increasing the amount of Gibson assembly reaction mixture beyond 1 µl for a given electroporation can alter the chemistry of the electroporation and reduce transformation efficiency.
? TROUBLESHOOTING
- 33|
Carefully pipette 25 µl of the electrocompetent bacterial cell/Gibson assembly reaction mixture from Step 32 into a chilled 0.1-cm-gap electroporation cuvette without introducing bubbles. Quickly flick the cuvette downward to deposit the cells across the bottom of the cuvette’s well.
▲ CRITICAL STEP Minimize introduction of air bubbles, as they can interfere with plasmid electroporation efficiency.
- 34|
Using an electroporator, electroporate the sample at 25 µF, 200 Ω, and 1,500 V. The expected time constant during electroporation is ~4.7 (typical range of 4.2–4.8).
? TROUBLESHOOTING
- 35|
Add 975 µl of recovery or SOC medium to the cuvette as soon as possible after electroporation (to a final volume of 1 ml in the cuvette). Pipette up and down enough times to resuspend the cells within the cuvette (probably a single pipetting up and down will be sufficient). Recovery medium is preferred here; however, SOC medium can be used in its place.
▲ CRITICAL STEP Electroporation is toxic to bacterial cells. Addition of recovery or SOC medium as quickly as possible enhances bacterial cell survival after electroporation.
- 36|
Transfer the cell mixture from Step 35 (~1 ml) to a culture tube already containing 1 ml of SOC medium.
- 37|
Shake each culture tube (containing a total of 2 ml of recovery/SOC medium) at 250 r.p.m. for 1 h at 37 °C.
▲ CRITICAL STEP If multiple transformations were set up in Step 32 using the same Gibson assembly reaction mixture, the 2-ml culture mixes generated in Step 37 can be pooled/mixed together so that one set of dilution plates is sufficient for calculation of transformation efficiency (Step 38).
- 38|
Remove 5 µl from the 2-ml mixture (or 5 µl from a larger volume, if samples were pooled/mixed as described in Step 37) from Step 37 and add it to 1 ml of SOC medium. Mix well and plate 20 µl of the mixture (20,000× dilution) onto a prewarmed 10-cm ampicillin-resistant (0.1 mg/ml ampicillin) agar plate and then plate the remaining 200 µl (2,000× dilution) onto a separate 10-cm ampicillin-resistant (0.1 mg/ml ampicillin) agar plate. These two dilution plates can be used to estimate transformation efficiency, which will help ensure full library representation of the sgRNA plasmid library. It is reasonable to aim for > 50× representation of the sgRNA library.
▲ CRITICAL STEP Adjust the dilutions as necessary to obtain plates with a colony density that allows for accurate counting of the number of colonies on the 10-cm plates.
- 39|
Plate the remaining 2-ml transformation mixture (or 2 ml from a larger volume, if samples were pooled/mixed) from Step 37 onto a prewarmed square 24.5-cm ampicillin-resistant (0.1 mg/ml ampicillin) agar plate using beads to ensure even spreading. Spread the liquid culture until it is largely absorbed into the agar and will not drip when inverted.
▲ CRITICAL STEP If multiple samples were mixed/pooled together in Step 37, it is still important to only plate 2 ml onto each square 24.5-cm ampicillin-resistant agar plate, as volumes > 2 ml may not be fully absorbed into the agar.
- 40|
Invert all three (or more) plates (2,000× dilution plate, 20,000× dilution plate, and square 24.5-cm nondilution plate(s)) from Steps 38 and 39, and grow for 14–16 h at 32 °C. It is recommended to incubate transformations at 32 °C to reduce recombination between lentiviral long terminal repeats, but incubation at 37 °C can also be performed.
- 41|
Count the number of colonies on the two dilution plates from Step 38. Multiply this number of colonies by the dilution factor (2,000× or 20,000×) and by the increased area of the square 24.5-cm plate (~7.6-fold increase in area) for estimation of the total number of colonies on the square 24.5-cm plate. It is reasonable to aim for > 50× representation of the sgRNA library.
? TROUBLESHOOTING
- 42|
Select ~10–20 colonies from the dilution plates (from Step 41) for screening by mini-scale plasmid preparation and subsequent Sanger sequencing (as in Steps 14 and 15) to determine whether the sgRNA library has been PCR-amplified. Given that the oligonucleotides for multiple libraries can be synthesized on the same programmable microarray (Steps 16 and 17), it is important to confirm that the correct library intended for amplification has been obtained. This is also useful for preliminary evaluation of library representation. The expectation is to identify unique sgRNA sequences among all sequenced mini-scale plasmid preparations, given that the probability of obtaining the same sgRNA from a full library of sgRNAs should be low.
▲ CRITICAL STEP This step is intended to offer qualitative re-assurance that library synthesis is proceeding in the expected manner by determining that the correct library has been PCR-amplified and offering an initial assessment of library representation. Ultimately, the deep-sequencing step (Step 48) provides a more comprehensive assessment of library composition.
? TROUBLESHOOTING
- 43|
Pipette 10 ml of LB medium onto each square 24.5-cm plate from Steps 39 and 40 and scrape the colonies off with a cell scraper. The LB medium aids in removal of the colonies from the agar.
- 44|
Pipette the LB medium/scraped bacterial colony mixture into a preweighed 50-ml tube and repeat the procedure a second time on the same plate with an additional 5–10 ml of LB to maximize removal of bacterial colonies.
- 45|
Centrifuge the LB medium/scraped bacterial colony mixture at 400g for 5 min at room temperature to pellet the bacteria and then discard the supernatant.
- 46|
Weigh the bacterial pellet in the tube (and subtract the preweighed tube to determine the weight of the bacterial pellet) to determine the proper number of columns for maxi-scale plasmid preparation of the library. Each column can support ~0.45 g of bacterial pellet.
? TROUBLESHOOTING
- 47|
Perform a sufficient number of maxi preps, following the manufacturer’s instructions and combine the eluted library plasmid DNA in a 1.5-ml tube.
- 48|
(Optional) To confirm successful library representation, it is recommended to deep-sequence the batch-cloned plasmid library produced in Steps 16–47 by following the deep-sequencing procedure found in Steps 73–82 and using library plasmid DNA as the template DNA for laPCR1 in Step 75.
Lentivirus production from individual sgRNA plasmid or pooled sgRNA plasmid library ● TIMING 4 d
! CAUTION Take all necessary precautions for the handling of lentivirus and ensure proper disposal of lentiviral waste. Lentivirus is capable of integrating into the genome of human cells. This caution should be exercised during Steps 49–71.
- 49|
Passage and maintain HEK293 cells in a 15-cm round plate with 16 ml of HEK293 medium as previously described62.
- 50|
Perform transfection when HEK293 cells reach ~80% confluency in a 15-cm round plate using polyethylenimine (PEI) as a transfection reagent. The plasmid/PEI ratio should be 1 µg of the total transfected plasmid to 3 µg of PEI. Total plasmid consists of the sum of micrograms of VSV-G + micrograms of psPAX2 + library plasmid.
- 51|
Mix PEI, VSV-G, psPAX2, and library plasmid in 1 ml of filtered DMEM without supplements in a sterile microcentrifuge tube:
Reagent Amount (µg) VSV-G plasmid 8.75 psPAX2 plasmid 16.25 Individual sgRNA plasmid or sgRNA library plasmid from Step 15 or 47 25 Branched PEI (at 10 µg/µl) 150 ? TROUBLESHOOTING
- 52|
Invert the microcentrifuge tube several times to mix. Allow the tube to incubate at room temperature for 20–30 min.
▲ CRITICAL STEP The DMEM/plasmid DNA/PEI mixture should change from translucent to opaque during this incubation period. If the color change does not occur, it is likely that one (or more) of the components is missing, and Steps 51 and 52 should be repeated.
- 53|
Add the full volume (~1 ml) of DMEM/plasmid DNA/PEI mixture dropwise to the ~80% confluent HEK293 cells from Steps 49 and 50 and incubate the mixture at 37 °C for 16–24 h.
- 54|
Replace the medium with 16 ml of fresh HEK293 medium 16–24 h after transfection in Step 53.
- 55|
Lentiviral supernatant harvest no. 1.24 h after replacing the medium in Step 54, collect the 16 ml of medium that now contains lentiviral particles (herein referred to as ‘viral supernatant’) in a 50-ml tube. Replace the medium with 16 ml of fresh HEK293 medium. Store the viral supernatant at 4 °C until Step 56 is performed. The viral supernatant can be stored at 4 °C for up to 7 d before ultracentrifugation; however, storage at 4 °C may result in reduction of viral titer.
- 56|
Lentiviral supernatant harvest no. 2.24 h after lentiviral supernatant harvest no. 1, collect the 16 ml of viral supernatant and put it into the same 50-ml tube from Step 55 (should now contain ~32 ml of viral supernatant). Appropriately discard the plate of HEK293 cells.
- 57|
Centrifuge the collected viral supernatant from Step 56 at 500–700g for 5 min at 4 °C to pellet the HEK293 cells and other debris.
- 58|
Filter the viral supernatant through a 0.45-µm 50-ml filter.
? TROUBLESHOOTING
- 59|
Physical or functional titering can be performed. Physical titering involves detection of viral DNA (e.g., qPCR-based amplification of the viral genome, see MATERIALS) or viral proteins (e.g., ELISA for p24, see MATERIALS). Functional titering involves determination of the transduction rate using cells. Either physical or functional titering can be performed based on experimental needs. Refer to Kutner et al. for various titering protocols87.
- 60|
If sufficient viral titer is obtained with the viral supernatant from Step 59, the filtered viral supernatant can be stored at − 80 °C. It is recommended to freeze aliquots with volumes appropriate for future experimental needs to minimize freeze–thaw cycles, as they result in a reduction of viral titer. Aliquots also help to ensure consistent titer for each use of the same batch of virus. If insufficient viral titer is achieved based on the desired multiplicity of infection, it can be improved via ultracentrifugation (Steps 61–71).
■ PAUSE POINT The filtered viral supernatant can be stored at 4 °C for up to 7 d before ultracentrifugation; however, storage at 4 °C may result in reduction of viral titer. If ultracentrifugation is required (Steps 61–71), it is recommended to perform ultracentrifugation as soon as possible (minimize storage time at 4 °C). Lentiviruses can be stored at −80 °C for months. Longer storage will result in a decrease of viral titer.
(Optional) Ultracentrifugation of viral supernatant ● TIMING 2.5 h
- 61|
Transfer the filtered viral supernatant to ultracentrifugation tubes.
- 62|
Add 4–6 ml of 20% (wt/vol) sucrose solution to the bottom of the ultracentrifugation tube to create a sucrose layer (also known as a ‘sucrose cushion’) below the viral supernatant. This sucrose layer helps to remove any remaining debris from the viral supernatant, as only the high-density viral particles can pass through the sucrose layer to pellet, whereas the low-density debris remains in the supernatant.
? TROUBLESHOOTING
- 63|
Add sterile PBS to the top of the ultracentrifuge tube until the final liquid volume within the ultracentrifuge tube is ~2–3 mm from the top.
▲ CRITICAL STEP The volume must be within 2–3 mm of the top of the tube to prevent the tube from collapsing because of the force of ultracentrifugation.
- 64|
Place ultracentrifuge tubes into ultracentrifuge buckets.
- 65|
Weigh all ultracentrifuge buckets containing ultracentrifuge tubes before ultracentrifugation to ensure appropriate weight-based balancing. Use sterile PBS to correct weight discrepancies.
- 66|
Centrifuge at 100,000g for 2 h at 4 °C in an ultracentrifuge.
- 67|
Remove ultracentrifuge tubes from ultracentrifuge buckets. The sucrose gradient should still be intact.
? TROUBLESHOOTING
- 68|
Discard the supernatant by inverting the ultracentrifugation tube. Allow the inverted ultracentrifuge tube to stand on sterile paper towels to dry for 1–3 min.
? TROUBLESHOOTING
- 69|
Revert the ultracentrifugation tube to an upright orientation and add 100 µl of sterile DMEM supplemented with 1% (vol/vol) FBS.
- 70|
Incubate for 3–4 h (or overnight) on a rotator/rocker at ~10–15 rotations per minute at 4 °C to resuspend the lentiviral pellet.
■ PAUSE POINT Resuspension at 4 °C can be left overnight on a rotator/rocker.
- 71|
Store concentrated lentivirus in a microcentrifuge tube at − 80 °C. Owing to loss of lentiviral titer with repeated freeze–thaw cycles, consider aliquoting at this point with volumes appropriate for experimental needs (i.e., aliquot size with enough lentivirus for one experiment). Aliquots frozen at the same time can be assumed to have the same titer, which can be advantageous for multiple experiments using the same lentivirus.
■ PAUSE POINT Lentiviruses can be stored at − 80 °C for months. Longer storage will result in a decrease of viral titer.
Execution of arrayed or pooled screen experiments ● TIMING 1–2 weeks
- 72|
Perform arrayed or pooled screen experiments. Experiments/screens using lentivirus should be performed as appropriate for the experimental design/objectives and the cells being used for study. Refer to Boxes 3 and 4 for discussions about and considerations for performing an arrayed or pooled screen experiment.
■ PAUSE POINT After experiments/screens have been completed, cell pellets can be stored at − 20 or − 80 °C for weeks to months before processing for deep-sequencing analysis of the samples (Steps 73–82).
Deep sequencing of arrayed or pooled screen experiments ● TIMING 1 d
- 73|
Extract gDNA from fresh cell pellets after completion of genome-editing screens/experiments or from frozen pellets from previous experiments. See MATERIALS for genomic DNA extraction kit and follow the manufacturer’s instructions.
▲ CRITICAL STEP For arrayed experiments intended for indel analysis by CRISPResso (Step 83), it is recommended to include a non-edited sample from the same cell type. This sample will be a useful negative control when performing CRISPResso indel analysis/enumeration. Specifically, a non-edited sample is useful in regard to optimization of the CRISPResso analysis parameters summarized in Box 2. Furthermore, a non-edited sample can be useful in identifying single-nucleotide polymorphisms (SNPs) or other variants present in the cells under study that can confound CRISPResso analysis. If a variant/indel is identified in the non-edited sample, the CRISPResso settings can be altered to account for the variant/indel during CRISPResso indel enumeration (Box 2).
- 74|
Determine the concentration of each gDNA sample using a UV-visible spectrophotometer (e.g., NanoDrop) or other equivalent method using the manufacturer’s instructions.
▲ CRITICAL STEP The amount of gDNA can vary based on experimental needs for an arrayed experiment and based on the number of sgRNAs in the library for a pooled experiment. On average, a genome from a single cell is ~6.6 pg88. Use adequate gDNA to represent the desired number of cells. For an arrayed experiment for indel analysis of a single locus using CRISPResso, > 1,000 cells at a given locus is a reasonable number. For a pooled sgRNA library experiment, 100–1,000× coverage of the sgRNAs in the library is reasonable. For example, 6.6 µg of gDNA is estimated to represent 1 million cells. For a library with 1,000 sgRNAs, 6.6 µg of gDNA would provide 1,000× coverage.
- 75|
Set up the ‘Library Analysis PCR1’ reaction (herein referred to as laPCR1). Use lentiGuide-Puro-specific primers (Supplementary Table 5) to enumerate the sgRNAs present for enrichment/dropout analysis or locus-specific primers to quantitate indels (Supplementary Table 6). lentiGuide-Puro-specific primers are suggested in Supplementary Table 5; however, other primer pairs to PCR-amplify from the lentiGuide-Puro construct are possible.
▲ CRITICAL STEP It is important to use a proofreading DNA polymerase to minimize introduction of PCR errors.
▲ CRITICAL STEP Optimization of DMSO concentration may be required (typically 1–10% (vol/vol)). 8% (vol/vol) is used in the reaction below.
▲ CRITICAL STEP It is important to sequence the cloned plasmid library as generated in Steps 16–47. Sequencing of the plasmid library allows for confirmation of successful cloning/representation of the library and can be used as an initial time point for a dropout (‘depletion’) screen (see Box 4 for details of this type of screen). Given that plasmids contain many fewer base pairs than a full genome, ≥50 pg of plasmid DNA will be sufficient to represent a pooled plasmid library.
? TROUBLESHOOTING
Components Amount (µl) Final concentration gDNA or plasmid DNA X – 5× Herculase II reaction buffer 10 1× dNTPs (at 100 mM) 1 2 mM lentiGuide_forward or Locus_forward primer (at 10 µM) 2.5 0.5 µM lentiGuide_forward or Locus_reverse primer (at 10 µM) 2.5 0.5 µM DMSO 4 8% Herculase II Fusion DNA Polymerase 0.5 Nuclease-free water To 50 - 76|
Perform multiple cycles (e.g., 10, 15, and 20 cycles) of laPCR1 in a thermocycler as follows. Gradient PCR may be required to determine the optimal annealing temperature (Supplementary Fig. 2c).
Cycle number Denature Anneal Extend 1 95 °C, 30 s 2-x (e.g., 10, 15, and 20) 95 °C, 10 s 60 °C, 30 s 72 °C, 30 s x + 1 72 °C, 5 min ? TROUBLESHOOTING
■ PAUSE POINT The PCR product can be stored at 4 °C in a thermocycler at the end of the PCR program for multiple days. Longer-term storage (months to years) should be at − 20 °C.
- 77|
Run a fraction (1–5 µl of the 50-µl PCR reaction from Steps 75 and 76) of the laPCR1 product with a different number of PCR cycles on a 2% (wt/vol) agarose gel.
▲ CRITICAL STEP The number of PCR cycles should be limited to reduce PCR bias. It is difficult to predict the required number of cycles for a given library. Therefore, it is important to perform multiple PCRs with different cycle numbers to empirically determine the optimal number of cycles.
? TROUBLESHOOTING
- 78|
Set up ‘Library Analysis PCR2’ (herein referred to as laPCR2) using the barcode-specific primers (Supplementary Tables 7 and 8). Each sample will need a unique Illumina Nextera index to allow demultiplexing of multiple samples sequenced together. Set up two PCRs for each sample.
▲ CRITICAL STEP Two separate 10-µl reactions are performed, as opposed to a single 20-µl reaction, to minimize PCR bias.
Components Amount (µl) Final concentration laPCR1 product (diluted 1:5 in nuclease-free water) from Steps 75–77 1 - 5× Herculase II reaction buffer 2 1× dNTPs (at 100 mM) 0.1 1 mM Forward primer (at 2 µM) 1 0.2 µM Reverse primer (at 2 µM) 1 0.2 µM Herculase II Fusion DNA Polymerase 0.1 Nuclease-free water To 10 - 79|
Perform laPCR2 using the same cycling conditions and multiple cycle numbers as in Step 76.
▲ CRITICAL STEP The number of PCR cycles should be limited to reduce PCR bias. It is difficult to predict the required number of cycles for a given library. Therefore, it is important to perform multiple PCRs with different cycle numbers to empirically determine the optimal number of cycles.
■ PAUSE POINT The PCR product can be stored at 4 °C in a thermocycler at the end of the PCR program for multiple days. Longer-term storage (months to years) should be at − 20 °C.
- 80|
Combine both 10-µl reactions from the same sample for a total volume of 20 µl. Run laPCR2 products on a 2% (wt/vol) agarose gel to determine whether the products occur at the expected size (expected size varies based on primers used in laPCR1). Choose the sample with the minimal number of PCR cycles that produces a visible band in order to minimize PCR bias (Supplementary Fig. 2d). For example, 20 cycles would be used based on the gel presented in Supplementary Fig. 2d; however, it may be possible to reduce the cycle number even further (i.e., another experiment with 5, 10, and 15 cycles). Then gel-purify the relevant band of the expected size.
▲ CRITICAL STEP Leave empty lanes between different samples to minimize the risk of sample contamination during gel purification.
- 81|
Quantitate DNA concentration by Qubit or another equivalent method, following the manufacturer’s instructions. Refer to the manufacturer’s instructions for the sequencing platform to be used (Step 82) for the minimum amount of DNA required. If an inadequate amount of DNA is obtained, repeat Steps 75–81.
- 82|
Sequence the libraries on a MiSeq, HiSeq, or equivalent sequencing platform (see ‘Experimental design’ for details).
▲ CRITICAL STEP It can be useful to discuss experimental goals with the sequencing facility or individual performing the sequencing prior to sequencing samples to help ensure that the desired number of reads are obtained to adequately represent the sequenced locus/loci or sgRNA library.
CRISPResso analysis of deep-sequencing data ● TIMING 10–120 min
- 83|
The CRISPResso utility can be used for analysis of deep-sequencing data of a single locus/amplicon on a local machine or proprietary server, using a command-line version or a webtool freely available at http://crispresso.rocks/. Installation instructions are provided in the ‘Equipment Setup’ section for command-line and Docker versions. No installation is required for the webtool. The following step will present detailed instructions for workflows using the webtool (option A), command-line (option B), or Docker (option C) versions:
(A) Analysis of deep-sequencing data using the CRISPR esso webtool ● TIMING 10–120 min
▲ CRITICAL An illustrative video of the entire process is provided here: http://www.youtube.com/embed/dXblIIiAe00?autoplay=1.
Open a web browser and go to the CRISPResso page http://crispresso.rocks/.
Select the option for single-end (one FASTQ file) or paired-end reads (two FASTQ files) based on the type of deep sequencing performed. Analysis of paired-end reads requires overlapping sequence, as the paired reads will be merged. Upload the relevant FASTQ file(s) (as .fastq or .fastq.gz) for analysis. An example data set to test the tool with previously generated FASTQ files is provided at http://crispresso.rocks/help (see the ‘Try it now!’ section).
? TROUBLESHOOTING
Input the amplicon sequence (required), sgRNA sequence (optional), and coding sequence(s) (optional input for when targeting exonic sequence). The reads uploaded in the previous step will be aligned to the provided amplicon sequence. If an sgRNA sequence is provided, its position will be indicated in all output analyses. If a coding sequence is provided, it will allow CRISPResso to perform frameshift analysis. The provided exonic sequence must be a subsequence of the amplicon sequence and not the sequence of the entire exon.
? TROUBLESHOOTING
Adjust the parameters for window size (bp around each side of cleavage site) to quantify NHEJ edits (if sgRNA sequence is provided), minimum average read quality (Phred33 scale), minimum single-bp quality (Phred33 scale), exclude bp from the left side of the amplicon sequence for the quantification of the mutations, exclude bp from the right side of the amplicon sequence for the quantification of the mutations, and trimming adapter (see Box 2 for a discussion of these parameters).
Submit samples for analysis and download analysis reports.
(B) Analysis of deep-sequencing data using command-line CRISPR esso ● TIMING 10–120 min
Gather the required information and files (see Step 1A(iv) or 1C(xiii); the CRISPOR utility will output all the required information to run CRISPResso).
Input Example entry (adjust as appropriate) or description FASTQ files for analysis (.fastq or .fastq.gz) reads1.fastq.gz reads2.fastq.gz Reference amplicon sequence AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCACAGGTGA GGAGGTCATGATCCCCTTCTGGAGCTCCCAACGGGCCGTGGTCT GGTTCATCATCTGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT sgRNA(s) used TGAACCAGACCACGGCCCGT Expected repaired amplicon sequence Only provide this if it is necessary to quantify HDR efficiency Desired single-base-pair or average read quality in Phred33 score We suggest using 20 for single-base-pair quality and 30 for the average read quality Exonic sequence (for frameshift analysis) Only provide this if the reference amplicon sequence contains exonic sequence Experiment name BCL11A_exon2 Open a terminal and run the CRISPResso utility by running the following command. Here, we are assuming that the FASTQ files reads1.fastq.gz and reads2.fastq.gz are stored in the folder /home/user/amplicons_data/.
CRISPResso \ -r1 /home/user/amplicons_data/reads1.fastq.gz \ -r2 /home/user/amplicons_data/reads2.fastq.gz \ -a AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCACAGGTGAGG\ AGGTCATGATCCCCTTCTGGAGCTCCCAACGGGCCGTGGTCTGGTTCAT\ CATCTGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT \ -g TGAACCAGACCACGGCCCGT \ -s 20 \ -q 30 \ -n BCL11A_exon2
▲ CRITICAL STEP Be sure to check whether or not the sequencing files were trimmed for adapters (refer to Box 2 for further discussion of the trimming process and options).
▲ CRITICAL STEP The command is based on the example data from Step 83B(i) and must be amended before use. After the execution of the command, a new folder with all the results will be created; in this example, the folder will be in /home/user/amplicons_data/CRISPResso_on_BCL11A_exon2. A summary of the different events discovered in the sequencing data is presented in the Alleles_frequency_table.txt file, and in several illustrative plots (.pdf files). Refer to the online documentation for the full description of the output: https://github.com/pinellolab/CRISPResso.
? TROUBLESHOOTING
(C) Analysis of deep-sequencing data using CRISPR esso with Docker ● TIMING 10–120 min
Gather the required information and files as in Step 83B(i).
Assuming that the FASTQ files reads1.fastq.gz and reads2.fastq.gz are stored in the folder /home/user/amplicons_data/, run the command
docker run \ -v /home/user/amplicons_data:/DATA \ -w /DATA pinellolab/crispor_crispresso_nat_prot \ CRISPResso \ -r1 reads1.fastq.gz \ -r2 reads2.fastq.gz \ -a AATGTCCCCCAATGGGAAGTTCATCTGGCACTGCCCACAGGTGAGG\ AGGTCATGATCCCCTTCTGGAGCTCCCAACGGGCCGTGGTCTGGTTCAT\ CATCTGTAAGAATGGCTTCAAGAGGCTCGGCTGTGGTT \ -g TGAACCAGACCACGGCCCGT \ -s 20 \ -q 30 \ -n BCL11A_exon2
After the execution of the command, a new folder with all the results will be created; in this example, the folder will be in /home/user/amplicons_data/CRISPResso_on_BCL11A_exon2. A summary of the different events discovered in the sequencing data is presented in the Alleles_frequency_table.txt file, and in several illustrative plots (.pdf files). Refer to the online documentation for the full description of the output: https://github.com/pinellolab/CRISPResso.
? TROUBLESHOOTING
Analysis of a pooled sgRNA experiment using CRISPRessoCount ● TIMING 20 min–4 h
▲ CRITICAL CRISPRessoCount is a utility for the enumeration of sgRNA (Fig. 2; Box 4). It is necessary to use the command-line or Docker version of CRISPResso to run CRISPRessoCount. CRISPRessoCount can enumerate sgRNAs from a user-generated list or can empirically identify all sgRNAs present in the FASTQ file.
- 84|
Gather the required data and information (adjust the entries as appropriate for the experiment):
Input Example entry (adjust as appropriate) or description FASTQ files for sgRNA enumeration analysis (.fastq or .fastq.gz) sample_screen.fastq.gz File containing only the sgRNA sequences to enumerate, one per line. This is an optional input. If not provided, all unique sgRNA sequences will be enumerated library_NGG.txt (or the file created by CRISPOR, see Step 1A(iv) or 1C(xiii)) The sgRNA scaffold sequence immediately downstream of the cloned sgRNA sequence GTTTTAGAGCTAGAAATAGC sgRNA length 20 Desired single-base-pair or average read quality in Phred33 score We suggest 20 for single-base-pair quality and 30 for the average read quality Experiment name SAMPLE_SCREEN - 85|
Run the CRISPRessoCount analysis. The following step will present detailed instructions for running CRISPRessoCount from the command line (option A) or Docker (option B). Here, we are assuming that the FASTQ file and the optional sgRNA file are stored in the folder home/user/sgRNA _data/.
(A) Using the command-line version for CRISPR essoCount analysis ● TIMING 10–120 min
Run the CRISPRessoCount analysis with the following commands:
CRISPRessoCount -r /home/user/sgRNA_data/sample_screen.fastq.gz \ -s 20 -q 30 -f library_NGG.txt \ -t GTTTTAGAGCTAGAAATAGC -l 20 --name SAMPLE_SCREEN
(B) Using the Docker container for CRISPR essoCount analysis ● TIMING 10–120 min
Run the CRISPRessoCount analysis with the following commands:
docker run \ -v /home/user/sgRNA_data:/DATA \ -w /DATA \ pinellolab/crispor_crispresso_nat_prot \ CRISPRessoCount -r sample_screen.fastq.gz \ -s 20 -q 30 -f library_NGG.txt \ -t GTTTTAGAGCTAGAAATAGC -l 20 --name SAMPLE_SCREEN
After the execution of the command, a new folder with all the results will be created; in this example, the folder will be in /home/user/amplicons_data/CRISPRessoCount_on_SAMPLE_SCREEN. This folder contains two files, the execution log (CRISPRessoCount_RUNNING_LOG.txt) and a file (CRISPRessoCount_only_ref_guides_on_sample_screen.fastq.gz.txt) containing a table with the raw and normalized counts for each sgRNA in the input FASTQ file:
| Guide sequence | Read counts | Read (%) | Reads per million (RP M) |
|---|---|---|---|
| CTCTGCCCTTCTGACATTGT | 2,592 | 0.312315496488 | 3123.15496488 |
| ATGTGAGCATATGTATTCAT | 2,553 | 0.30761630499 | 3076.1630499 |
| CCTGCTATGTGTTCCTGTTT | 2,455 | 0.2958080802 | 2958.080802 |
| TTCTCGTGCCTCAGCCTCCT | 2,430 | 0.292795777957 | 2927.95777957 |
| ACCCTGTGTATTTCACACAT | 2,349 | 0.283035918692 | 2830.35918692 |
| TTCCATTTAATACACAATGT | 2,235 | 0.269299820467 | 2692.99820467 |
- 86|
Calculate the enrichment and/or dropout (‘depletion’) between two conditions, and calculate the log2 ratio using the normalized sgRNA counts (this takes into account the total number of reads within each sequenced sample) using the ‘Reads_Per_Millions_(RPM)’ column for the two conditions. Refer to the ‘CRISPResso: Analysis of deep sequencing from arrayed or pooled sgRNA experiments’ section for further discussion of read normalization.
▲ CRITICAL STEP The log2 transformation is not necessary, but is often used when displaying the data, as it will center the data at ~0 for an equal ratio; positive values will represent enrichment and negative values will represent depletion/dropout.
? TROUBLESHOOTING
CRISPR esso analysis using CRISPRessoPooled ● TIMING 20 min–4 h
▲ CRITICAL CRISPRessoPooled is a utility for analyzing and quantifying targeted sequencing CRISPR experiments involving sequencing with pooled amplicons. To use CRISPRessoPooled, it is necessary to use the command-line version of CRISPResso (see Step 83B). Although CRISPRessoPooled can be run in different modes (amplicon only, genome only, and mixed), in this protocol, we use the most reliable mode for quantification, called ‘mixed mode’. In this mode, the amplicon reads are aligned to both reference amplicons and the genome to discard ambiguous or spurious reads. For more details regarding the different running modes, consult the online help (help is located below the list of files at the following link): https://github.com/pinellolab/CRISPResso.
- 87|
Gather the required data and information, including FASTQ files that contain reads for a pooled experiment with multiple sgRNAs, for example:
Input Example entry (adjust as appropriate) or description FASTQ files for analysis (.fastq or .fastq.gz) Reads_pooled_1.fastq.gz Reads_pooled_2.fastq.gz Reference amplicon sequences used in the pooled experiment sgRNA sequences used, one for each amplicon Expected repaired amplicon sequence Only provide this if it is necessary to quantify HDR efficiency Desired single-base-pair or average read quality in Phred33 score We suggest 20 for single-base-pair quality and 30 for the average read quality Exonic sequence (for frameshift analysis) Only provide this if the reference amplicon sequence contains exonic sequence Amplicon names All the names should be unique A global name for the pooled analysis Pooled_amplicons ▲ CRITICAL STEP Be sure to check whether or not the sequencing files were trimmed for adapters (refer to Box 2 for further discussion of the trimming options).
- 88|
Create a folder for the reference genome and genomic annotations (this step is necessary only one time):
mkdir -p /home/user/crispresso_genomes/hg19
- 89|
Download the desired genome sequence data and precomputed index from https://support.illumina.com/sequencing/sequencing_software/igenome.html. In this example (Steps 90 and 91), we download the hg19 assembly of the human genome.
- 90|
Move to the folder containing the genome:
cd /home/user/crispresso_genomes/hg19 wget ftp://igenome:[email protected]\ /Homo_sapiens/UCSC/hg1938/Homo_sapiens_UCSC_hg19.tar.gz
- 91|
Extract the data with the following command:
tar -xvzf Homo_sapiens_UCSC_hg19.tar.gz \ Homo_sapiens/UCSC/hg19/Sequence/Bowtie2Index \ --strip-components 5
- 92|
Create an amplicon description file or use the amplicon file created by CRISPOR in Step 1A(iv) or 1C(xiii). This file is a tab-delimited text file with up to five columns (the first two columns are required) (Supplementary Fig. 3a):
Column no. Description 1 An identifier for the amplicon (must be unique) 2 Amplicon sequence used in the design of the experiment 3 (optional) sgRNA sequence used for this amplicon, without the PAM sequence. If not available, enter ‘NA’ 4 (optional) Expected amplicon sequence in the case of HDR. If more than one, separate by commas and not spaces. If not available, enter ‘NA’ 5 (optional) Subsequence(s) of the amplicon corresponding to coding sequences. If more than one, separate by commas and not spaces. If not available, enter ‘NA’ Here, we are assuming that the file is saved in the folder /home/user/pooled_amplicons_data/ and is called AMPLICONS_FILE.txt
- 93|
Open a terminal and run the CRISPRessoPooled utility from the command line (option A) or Docker (option B). Here, we are assuming that the FASTQ files Reads_pooled_1.fastq.gz and Reads_pooled_1.fastq.gz are stored in the folder /home/user/pooled_amplicons_data/
(A) Using the command-line version for CRISPR essoPooled analysis ● TIMING 10–120 min
Run CRISPRessoPooled by executing the following commands:
CRISPRessoPooled \ -r1 /home/user/pooled_amplicons_data/Reads_pooled_1.fastq.gz \ -r2 /home/user/pooled_amplicons_data/Reads_pooled_2.fastq.gz \ -f /home/user/amplicons_data/AMPLICONS_FILE.txt \ -x /home/user/crispresso_genomes/hg19/genome \ -s 20 -q 30 \ --name Pooled_amplicons
? TROUBLESHOOTING
(B) Using the Docker container for CRISPR essoPooled analysis ● TIMING 10–120 min
Run CRISPRessoPooled by executing the following commands:
docker run \ -v /home/user/amplicons_data/:/DATA -w /DATA \ -v /home/user/crispresso_genomes/:/GENOMES \ pinellolab/crispor_crispresso_nat_prot \ CRISPRessoPooled \ -r1 Reads_pooled_1.fastq.gz \ -r2 Reads_pooled_2.fastq.gz \ -f AMPLICONS_FILE.txt \ -x /GENOMES/hg19/genome \ -s 20 -q 30 \ --name Pooled_amplicons
After the execution of the command in the previous step, a new folder with all the results will be created. In this example, the folder will be in /home/user/amplicons_data/CRISPRessoPooled_on_Pooled_amplicons. In this folder, the user can find the following files:
Output Description REPORT_READS_ALIGNED_TO_GENOME_AND_AMPLICONS.txt This file contains the same information provided in the input description file, plus some additional columns: Amplicon_Specific_fastq.gz_filename: name of the file containing the raw reads recovered for the amplicon
n_reads: number of reads recovered for the amplicon
chr_id: chromosome of the amplicon in the reference genome
bpstart: start coordinate of the amplicon in the reference genome
bpend: end coordinate of the amplicon in the reference genome
Reference_Sequence: sequence in the reference genome for the region mapped for the amplicon
MAPPED_REGIONS (folder) This folder contains all the .fastq.gz files for the discovered regions Set of folders with CRISPResso reports CRISPResso analysis for the recovered amplicons with enough reads SAMPLES_QUANTIFICATION_SUMMARY.txt This file contains a summary of the quantification and the alignment statistics for each region analyzed (read counts and percentages for the various classes: unmodified, NHEJ, and HDR) CRISPRessoPooled_RUNNING_LOG.txt Execution log and messages for the external utilities called ? TROUBLESHOOTING
CRISPResso analysis using CRISPRessoWGS ● TIMING 20 min–4 h
- 94|
Gather the required data and information:
Input Example entry (adjust as appropriate) or description Genome-aligned WGS data in BAM format sample_WGS.bam The sequence of the reference genome used for the alignment, in FASTA format (Steps 88–91) Human hg19 genome assembly in FASTA format (Optional) gene annotation file (Step 1C(vi–xii)) gencode_v19.gz Coordinates of the regions to analyze REGIONS_FILE.txt Expected repaired amplicon sequence Only provide this if it is necessary to quantify HDR efficiency Desired single-base-pair or average read quality in Phred33 score We suggest 20 for single-base-pair quality and 30 for the average read quality Exonic sequences (for frameshift analysis) Only provide this if the reference amplicon sequence contains exonic sequence Region names All the names should be unique A global name for the WGS analysis WGS_regions - 95|
Create a regions description file. This file is a tab-delimited text file with up to seven columns (four are required) and contains the coordinates of the regions to analyze and some additional information (Supplementary Fig. 3b):
Column no. Description 1 Chromosome of the region in the reference genome 2 Start coordinate of the region in the reference genome 3 End coordinate of the region in the reference genome 4 An identifier for the region (must be unique) 5 (optional) sgRNA sequence used for this genomic segment without the PAM sequence. If not available, enter ‘NA’ 6 (optional) Expected genomic segment sequence in case of HDR. If more than one, separate by commas and not spaces. If not available, enter ‘NA’ 7 (optional) Subsequence(s) of the genomic segment corresponding to coding sequences. If more than one, separate by commas and not spaces. If not available, enter ‘NA’ Here, we are assuming that the file is saved in the folder /home/user/wgs_data/ and it is called REGIONS_FILE.txt.
- 96|
Open a terminal and run the CRISPRessoWGS utility. The following step will present detailed instructions for the command-line version (option A) and Docker (option B). Here, we are assuming that the BAM file is stored in the folder /home/user/wgs_data/ and was aligned using the human hg19 reference genome.
(A) Using the command-line version for CRISPR essoWGS analysis ● TIMING 10–120 min
Run CRISPRessoWGS by executing the following commands:
CRISPRessoWGS\ -b /home/user/wgs_data/sample_WGS.bam \ -f /home/user/wgs_data/REGIONS_FILE.txt \ -r /home/user / crispresso_genomes/hg19/hg19.fa \ -s 20 -q 30 \ --name WGS_regions
? TROUBLESHOOTING
(B) Using the Docker container version for CRISPR essoWGS analysis ● TIMING 10–120 min
Run CRISPRessoWGS by executing the following commands:
docker run \ -v /home/user / wgs_data/:/DATA -w /DATA \ -v /home/user / crispresso_genomes/:/GENOMES \ pinellolab/crispor_crispresso_nat_prot \ CRISPRessoWGS \ -b sample_WGS.bam \ -f REGIONS_FILE.txt \ -r /GENOMES/hg19.fa \ -s 20 -q 30 \ --name WGS_regions
After the execution of the command, a new folder with all the results will be created; in this example, the folder will be in /home/user/wgs_data/CRISPRessoWGS_on_WGS_regions. In this folder, the user can find the following files:
Output filename Description REPORT_READS_ALIGNED_TO_SELECTED_REGIONS_WGS.txt This file contains the same information provided in the input description file, plus some additional columns: sequence: sequence in the reference genome for the region specified
n_reads: number of reads recovered for the region
bam_file_with_reads_in_region: file containing only the subset of the reads that overlap, also partially, with the region. This file is indexed and can be easily loaded, for example, onto the Integrative Genomics Viewer (IGV; http://software.broadinstitute.org/software/igv/) for visualization of single reads or for the comparison of two conditions
fastq.gz_file_trimmed_reads_in_region: file containing only the subset of reads fully covering the specified regions and trimmed to match the sequence in that region. These reads are used for the subsequent analysis with CRISPResso
ANALYZED_REGIONS (folder) This folder contains all the BAM and FASTQ files, one for each region analyzed A set of folders with the CRISPResso report on the regions provided in the input Refer to description of the output of CRISPResso (http://software.broadinstitute.org/software/igv/) CRISPRessoWGS_RUNNING_LOG.txt Execution log and messages for the external utilities called ? TROUBLESHOOTING
Direct comparison of CRISPResso analyses using CRISPRessoCompare and CRISPRessoPooledWGSCSCompare ● TIMING 20 min–4 h
- 97|
Gather the required data and information: two completed CRISPResso, CRISPRessoPooled or CRISPRessoWGS analyses:
Input Example entry (adjust as appropriate) or description Path to two completed CRISPResso, CRISPRessoPooled, or CRISPRessoWGS analyses /home/user/results/CRISPResso_on_sample1 /home/user/results/CRISPResso_on_sample2 or home/user/results/CRISPRessoPooled_on_sample1 home/user/results/CRISPRessoPooled_on_sample2 or /home/user/results/CRISPRessoWGS_on_sample1 /home/user/results/CRISPRessoWGS_on_sample2 A name to use for the report comparison_sample1_sample2 - 98|
Open a terminal and run the CRISPRessoCompare or CRISPRessoPooledWGSCompare utility from the command line (option A) or Docker (option B). Here, we are assuming that output folders from those utilities were saved in /home/user/results/.
(A) Using the command-line version for CRISPRessoCompare or CRISPRessoPooledWGSCSCompare analysis ● TIMING 10–120 min
Execute the following command for CRISPRessoCompare:
CRISPRessoCompare \ /home/user/results/CRISPResso_on_sample1 \ /home/user/results/CRISPResso_on_sample2 \ -n comparison_sample1_sample2
Or execute the following command for CRISPRessoPooledWGSCompare:
CRISPRessoPooledWGSCompare \ /home/user/results/CRISPRessoPooled_on_sample1 \ /home/user/results/CRISPRessoPooled_on_sample2 \ -n comparison_sample1_sample2
(B) Using the Docker container version for CRISPRessoCompare or CRISPRessoPooledWGSCSCompare analysis ● TIMING 10–120 min
Execute the following command for CRISPRessoCompare:
docker run \ -v /home/user/results:/DATA -w /DATA \ pinellolab/crispor_crispresso_nat_prot \ CRISPRessoCompare \ CRISPResso_on_sample1 \ CRISPResso_on_sample2 \ -n comparison_sample1_sample2
Or execute the following command for CRISPRessoPooledWGSCompare:
docker run \ -v /home/user/results:/DATA -w /DATA \ pinellolab/crispor_crispresso_nat_prot \ CRISPRessoPooledWGSCompare \ CRISPRessoPooled_on_sample1 \ CRISPRessoPooled_on_sample2 \ -n comparison_sample1_sample2
The syntax for the utility CRISPRessoPooledWGSCompare is exactly the same for results obtained with CRISPRessoWGS.
After the execution of the command CRISPRessoPooledWGSCompare, a new folder with subfolders (one for each region) will be created. In this example, the folder will be in /home/user/results/CRISPRessoPooledWGSCompare _on_comparison_sample1_sample 2.
In the folder created by CRISPRessoPooledWGSCompare, the user will see the following files:
| Output filename | Description |
|---|---|
| COMPARISON_SAMPLES_QUANTIFICATION_SUMMARIES.txt | This file contains a summary of the quantification for each of the two conditions for each region and their differences (read counts and percentages for the various classes: unmodified, NHEJ, MIXED NHEJ–HDR and HDR) |
| A set of folders with CRISPRessoCompare reports on the common regions with enough reads in both conditions | Please refer to the table below for the description of the files contained in this folder |
| CRISPRessoPooledWGSCompare_RUNNING_LOG.txt | Detailed execution log |
A single folder will be created if CRISPRessoCompare was used. In the folder created by CRISPRessoCompare, the user will see the following files:
| Output filename | Description |
|---|---|
| Comparison_Efficiency.pdf | A figure containing a comparison of the editing frequencies for each category (NHEJ, MIXED NHEJ–HDR and HDR), as well as the net effect of subtracting the second sample (second folder in the command line) provided in the analysis from the first sample (first folder in the command line) |
| Comparison_Combined_Insertion_Deletion_Substitution_Locations.pdf | A figure showing the average profile for the mutations for the two samples in the same scale and their differences with the same convention used in the previous figure (first sample–second sample) |
| CRISPRessoCompare_RUNNING_LOG.txt | Detailed execution log |
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
Table 3
Troubleshooting table.
| step | problem | possible reason | solution |
|---|---|---|---|
| 1A(ii) | Regions >2 kb or multiple regions | Large region or multilocus design | For batch mode or genomic regions >2 kb, the command-line version can be used (Step 1C) |
| 1A(ii) | Genome of interest not available | – | If the genome of interest is not available at crispor.org, ten.rofet@ropsirc. It is recommended to provide a link to download the relevant genome and the gene transcript models or the National Center for Biotechnology Information (NCBI) Assembly accession |
| 1B(vi) | One or several genes from the gene list are not included in the gene-targeted library output from the CRISPOR Batch gene-targeting assistant | This probably results from incorrect entry of the gene symbol, Entrez Gene ID, or Refseq ID | Many genes have multiple names. Be sure to use the official gene symbol. This entry is case-insensitive |
| 13 | Low cloning efficiency | Inadequate lentiGuide-Puro plasmid dephosphorylation and/or Esp3l restriction enzyme digestion | Repeat lentiGuide-Puro plasmid dephosphorylation and/or Esp3l restriction enzyme digestion (Steps 5–9). Alternatively, increase the duration of dephosphorylation and/or restriction enzyme digestion |
| 19 | PCR failure | Too much oligonucleotide template | Dilute a subset of the oligonucleotide pool from Steps 17 and 18 based on the manufacturer’s recommendations or try different dilutions (e.g., 1:10 dilution with nuclease-free water) |
| 19 | PCR bias | Suboptimal DNA polymerase | Q5 High-Fidelity DNA Polymerase may result in less PCR bias as compared with Phusion Hot Start Flex DNA Polymerase, so this reagent can be considered as an alternative |
| 32, 34, 41 | Low transformation efficiency/abnormal electroporation time constant | Abnormal reaction chemistry from use of too much Gibson assembly reaction mixture | Minimize Gibson assembly reaction mixture volume added (< 2 gl, but 0.5–1 µl is recommended) |
| 41 | Low cloning/transformation efficiency | Recombination of lentiviral long terminal repeats | Incubation at 32 °C instead of 37 °C can minimize recombination events and result in increased transformation efficiency |
| Low efficiency of Gibson assembly reaction or no identifiable cause | Perform multiple Gibson assembly reactions. Combine Gibson assembly reaction mixtures and concentrate using minimum elution kit to elute in 10 µl of nuclease-free water (MATERIALS) | ||
| Suboptimal electrocompetent cells | Endura electrocompetent cells have demonstrated a higher efficiency; however, consider performing direct head-to-head comparison experiment with E. Cloni electrocompetent cells | ||
| 42 | Overrepresentation of single or a few sgRNA sequences | This probably results from PCR biases | Repeat library synthesis, beginning with lsPCR1 (Step 19), and ensure usage of the correct primers for the lsPCR1 and lsPCR2 reactions. It is possible that the full-length oligonucleotides have been incorrectly synthesized; however, this possibility is a less likely alternative |
| sgRNA from different libraries identified | This probably results from using the incorrect barcode-specific primers for the lsPCR1 reaction (Step 19) or contamination from other libraries during gel purification (Step 25 and 26) | Repeat the lsPCR1 reaction with the correct barcode-specific primers. Separate lsPCR2 samples with multiple empty lanes and use separate gel purification supplies for each sample | |
| Identified sgRNA sequence(s) not present in the library being synthesized or any other barcoded libraries | This probably represents error in the oligonucleotide synthesis pipeline | Proceed with the protocol, as long as the majority (50–75%) of sgRNAs identified match sequences within the library. Contact the oligonucleotide manufacturer to inquire about its synthesis error rate | |
| 46 | Low yield from maxi-scale plasmid preparation | This probably results from overloading the maxi-scale plasmid preparation column | Ensure that < 0.45 g of bacterial pellet is loaded onto each column |
| 51 | Low lentiviral titer | Mixing of plasmid DNA and PEI in DMEM with supplements | Charged proteins (e.g., from FBS) can interfere with the charge-based complexation of PEI with plasmid DNA. Repeat with filtered (0.22-µM filter) DMEM without supplements |
| 58 | Viral supernatant becomes stuck in the filter | This is probably due to a large amount of HEK293 cells and other debris in the viral supernatant clogging the filters | After Step 57, transfer the viral supernatant to a fresh 50-ml tube and repeat centrifugation from Step 57 to further remove HEK293 cells and debris before proceeding to the filtration step |
| 62 | Disrupted or mixed sucrose/viral supernatant layers in the ultracentrifugation tube | This is probably due to the release of air bubbles while creating the layers. Alternatively, the layers are unlikely to form correctly if the pipette tip is not at the bottom of the tube while the sucrose is being dispensed | Minimize the release of air bubbles by pipetting carefully while layering sucrose at the bottom of the ultracentrifugation tube. Ensure that the pipette tip is at the bottom of the ultracentrifugation tube when layering sucrose |
| 67 | Debris at the bottom of lentivirus-containing ultracentrifuge tubes after ultracentrifugation | Absent, disrupted, or inappropriate sucrose gradient | See troubleshooting advice for Step 62. Handle lentivirus-containing ultracentrifuge tubes and buckets carefully to avoid disrupting the layers. Ensure that the sucrose solution is 20% (wt/vol) by mass |
| 68 | Low lentiviral titer | Loss of lentiviral pellet | Invert the tube as quickly as possible in a single motion/movement. Once the tube is inverted, do not revert to the upright orientation to avoid resuspension of viral pellet and subsequent loss of resuspended viral particles |
| 75–77 | PCR failure | Suboptimal DMSO concentration and/or annealing temperature | Separately perform DMSO concentration gradient and annealing temperature gradient for each primer set used (supplementary Fig. 2c) |
| 83 | Multiple sequencing files | Examination of multiple unique amplicons | Use command-line CRISPResso, as opposed to the webtool, for batch mode to expedite the analysis of all files |
| 83 | Low number of reads analyzed by CRISPResso despite high number of reads in the input file | If using paired-end reads, verify that the reads have sufficient overlapping sequence. In addition, verify that the correct reference amplicon has been provided | See considerations for amplicon design presented in the ‘Sequencing of arrayed experiments and pooled screens’ section |
| 83 | Low number of reads analyzed by CRISPResso despite high number of reads in the input file | This probably results from low-sequence-quality reads (low Phred33 scores) being removed from the analysis | Phred33-based read filtering is too stringent based on sequencing data quality. Adjust the following two parameters: ‘Minimum average read quality (phred33 scale)’ and ‘Minimum single-bp quality (phred33 scale)’. Refer to box 2 for further discussion of these parameters |
| 83 | Many indels created far from the predicted DSB position | This probably results from low sequencing quality, whereby sequencing errors are interpreted as indels. If a particular indel(s) is consistently present, it may represent a variant/mutation in the cells | Use the ‘Window size (bp around each side of cleavage site) to quantify NHEJ edits (if sgRNA sequence provided)’ feature to restrict the analysis of indels to a set interval centered around the predicted DSB position. Refer to box 2 for further discussion of this parameter |
| 83 | CRISPResso cannot align any reads to the reference amplicon | The amplicon sequence provided is not correct, the wrong FASTQ file was uploaded, or the sample was inappropriately demultiplexed (if demultiplexing was performed) | Inspect the first few lines of your FASTQ file. The start of the reference amplicon sequence should match the start of each read (or the reverse complement of the read) |
| 83 | CRISPResso reports spurious indels localized at the end(s) of the amplicon | The reads may not be trimmed for adapters/barcodes | If reads are not already trimmed, select the adapters used for trimming under the ‘Trimming Adapter’ heading under the ‘Optional Parameters’. Failure to trim adapters may result in false positives. For the command line version, it is necessary to add the option ‘--trim sequences’. Refer to box 2 for further discussion of adapter trimming |
| 83 | CRISPResso cannot align any sequence to the reference amplicon for paired-end reads if provided as a single FASTQ file | The paired-end reads are provided in a single file instead of the two that are required (a format usually referred to as interleaved) | In this case, it is necessary to use the command-line version of CRISPResso and add the option ‘--split_paired_end’ |
| 83 | CRISPResso is not performing frameshift analysis | The subsequence of the amplicon corresponding to the exon(s) is not provided | It is required to add the option ‘-e’ to the command line to enable frameshift analysis. In addition, the sequence of the amplicon corresponding to the exon must be a subsequence of the reference amplicon. Refer to box 2 for further discussion of this parameter |
| 83A | Web-version analysis is very slow | It is likely that the web version is experiencing high traffic, which is causing the prolonged analysis times | The analysis speed of CRISPResso is dependent on the number of users running the analysis at the same time on the site. If the webpage is very unresponsive, we suggest using the command-line version, attempting the analysis at a later time, or providing an email address when submitting samples for analysis so that CRISPResso can email you when your analysis is complete |
| 83A | Web version fails to complete the analysis because of the size of the uploaded FASTQ file(s) | The combined maximum size for the FASTQ file(s) must be <100 MB for the web version of CRISPResso | To analyze large file(s) (>100 MB), it is necessary to use the command-line version of CRISPResso (skip to Step 83B) |
| 83B | The command-line version of CRISPResso is not running correctly | Be sure to install Anaconda Python 2.7, not Python 3.x, as CRISPResso is not compatible with Python 3 | Install Anaconda Python 2.7, and then run the setup again |
| 83B,C | CRISPResso analysis is taking too much time | The high number of reads analyzed is slowing down the computation | Consider speeding up the computation by adding the option ‘-p <number_or_processes >’ to enable the parallel option feature. For example, in a machine with four cores, you can add the following flag to the command line to use all four cores (default is to use one core): ‘-p 4’ |
| 86 | The enrichment score is not defined (contains not a number (NAN) or infinitive (INF)) | If one of the two conditions has a value of 0, the ratio may be not defined | Add a pseudocount to both conditions before taking the ratio. For example, you can add 0.1 to all the scores. This can be accomplished with Excel or similar software |
| 93 | Some regions are not analyzed and reported using CRISPRessoPooled | By default, a region is analyzed and reported if it has at least 1,000 reads | If an amplicon(s) is not reported in the analysis, check whether the sequence used in the amplicon description file is correct. In addition, check the ‘--min_reads_to_use_region’ parameter. This parameter allows for control over which amplicons have sufficient reads to be analyzed (default = 1,000). |
| 96 | Some regions are not analyzed and reported using CRISPRessoWGS | By default, a region is analyzed and reported if it has at least 10 reads | Adjust the ‘--min_reads_to_use_region’ parameter. This parameter allows for control over which regions have sufficient reads to be analyzed (default 10) |
● TIMING
Step 1, sgRNA design using CRISPOR: 1–4 h
Steps 2–15, synthesis and cloning of individual sgRNA into a lentiviral vector: 3 d to obtain sequence-confirmed, cloned sgRNA lentiviral plasmid
Steps 16–48, synthesis and cloning of pooled sgRNA libraries into a lentiviral vector: 2–4 d to obtain cloned sgRNA lentiviral plasmid library
Steps 49–60, lentivirus production from individual sgRNA plasmid or pooled sgRNA plasmid library: 4 d
Steps 61–71, (optional) ultracentrifugation of viral supernatant: 2.5 h
Step 72, execution of arrayed or pooled screen experiments: 1–2 weeks
Steps 73–82, deep sequencing of arrayed or pooled screen experiments: 1 d
Step 83, CRISPResso analysis of deep-sequencing data: 10–120 min
Steps 84–86, analysis of a pooled sgRNA experiment using CRISPRessoCount: 20 min–4 h
Steps 87–93, CRISPResso analysis using CRISPRessoPooled: 20 min–4 h
Steps 94–96, CRISPResso analysis using CRISPRessoWGS: 20 min–4 h
Steps 97 and 98, direct comparison of CRISPResso analyses using CRISPRessoCompare and CRISPRessoPooledWGSCompare: 20 min–4 h
ANTICIPATED RESULTS
Indel enumeration/analysis of a deep-sequencing amplicon generated by locus-specific PCR primers can be performed by CRISPResso for an arrayed experiment (Fig. 4a–d). As a reference for assessment of locus-specific edits, we provide data demonstrating the quantification of editing (Fig. 4a) and distribution of combined insertions/deletions/substitutions, to show successful editing of BCL11A exon 2 with indels flanking the DSB site (Fig. 4b–c). In this example, 36.1% of reads had a mutation present, whereas 63.9% of reads were unmodified (Fig. 4a). Mutation position analysis demonstrated that the majority of indels clustered at the predicted sgRNA DSB site (Fig. 4b), and indel size analysis demonstrated that most of the identified indels were small (< 10 bp) (Fig. 4c). Frameshift analysis was performed because coding sequence was targeted, which revealed a predominance of frameshift mutations (79.3% of reads) as compared with in-frame mutations (20.7% of reads) (Fig. 4d). The distribution of all identified alleles can be directly visualized as well (Supplementary Fig. 4). Using CRISPRessoCompare for direct comparison of the sample treated with CRISPR/Cas9 with a non-edited control can offer reassurance that the identified mutations are likely to have resulted from CRISPR-mediated mutagenesis. Specifically, this analysis was performed by sequencing the mutagenized sample and a wild-type control, followed by analysis via CRISPRessoCompare (Supplementary Fig. 5).
As a reference for assessment of a pooled CRISPR screen experiment, data are provided from two separate saturating mutagenesis experiments for identification of functional sequence within the BCL11A enhancer11,12. All sgRNAs were designed within the core of the BCL11A enhancer. All sgRNAs were designed within BCL11A exon 2 to serve as positive controls and nontargeting controls were included as negative controls. sgRNAs were batch-cloned into pLentiGuide-puro, and lentivirus was produced. HUDEP-2 erythroid cells with stable Cas9 expression were transduced at low multiplicity. After puromycin selection and allowing 1–2 weeks for editing to occur, phenotypic selection occurred by FACS. Specifically, cells with high and low fetal hemoglobin (HbF) were sorted by FACS. gDNA was extracted from the high- and low-HbF sorted cell pellets. PCR was performed with primers specific to the pLentiGuide-Puro construct. After deep sequencing, FASTQ files were analyzed by CRISPRessoCount to determine read counts for each sgRNA in the library. Read counts were subsequently normalized for each sample. Normalized read counts from the high-HbF sample were divided by the normalized read counts from the low-HbF sample for each sgRNA, followed by a log2 transformation to provide an enrichment score. The enrichment score for each sgRNA was plotted at the genomic position for the predicted DSB (Fig. 4e). Notably, this analysis shows pooled experiments using NGG- and NGA-restricted sgRNA (Supplementary Table 1), and demonstrates reproducibility of the experimental results using this procedure across multiple studies. On the basis of the analysis presented in Figure 4e, enrichment of sgRNA based on HbF levels can be observed for sgRNAs targeting BCL11A coding sequence as compared with the nontargeting controls. Notably, the nontargeting sgRNAs cluster around an enrichment score of zero, suggesting no effect on HbF. The sgRNAs targeting the BCL11A enhancer demonstrate comparable enrichment to targeting BCL11A coding sequence and demonstrate substantial enrichment as compared with the nontargeting controls. Taken together, these saturating mutagenesis pooled screen experiments suggest that functional sequence exists within the BCL11A enhancer.
Acknowledgments
M.C.C. was supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Award (F30DK103359). M.H. was funded by National Institutes of Health (NIH)/National Human Genome Research Institute (NHGRI) grant 5U41HG002371-15 and NIH/National Cancer Institute (NCI) grant 5U54HG007990-02 and by a grant from the California Institute of Regenerative Medicine, CIRM GC1R-06673C. D.E.B. was supported by the National Heart, Lung, and Blood Institute (NHLBI) (DP2OD022716, P01HL032262), the Burroughs Wellcome Fund, and a Doris Duke Charitable Foundation Innovations in Clinical Research Award. S.H.O. was supported by an award from the NHLBI (P01HL032262) and an award from the NIDDK (P30DK049216, Center of Excellence in Molecular Hematology). N.E.S. was supported by the NIH through the NHGRI (R00-HG008171). L.P. was supported by an NHGRI Career Development Award (R00HG008399) and the Defense Advanced Research Projects Agency (HR0011-17-2-0042).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS M.C.C., M.H., and L.P. conceived this project. M.H. and J.-P.C. created CRISPOR. L.P., M.C.C., D.E.B., and G.-C.Y. created CRISPResso. M.C.C. and D.E.B. performed the experiments.
M.C.C., D.E.B., S.H.O., N.E.S., O.S., G.-C.Y., F.Z., and L.P. analyzed the experimental data. M.C.C., M.H., and L.P. wrote the manuscript with input from all authors.COMPETING INTERESTS
The authors declare no competing interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nprot.2018.005
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc6182299?pdf=render
Citations & impact
Impact metrics
Article citations
CRISPR-Cas and CRISPR-based screening system for precise gene editing and targeted cancer therapy.
J Transl Med, 22(1):516, 30 May 2024
Cited by: 1 article | PMID: 38816739 | PMCID: PMC11138051
Review Free full text in Europe PMC
Aggresome formation promotes ASK1/JNK signaling activation and stemness maintenance in ovarian cancer.
Nat Commun, 15(1):1321, 13 Feb 2024
Cited by: 0 articles | PMID: 38351029 | PMCID: PMC10864366
Next-generation forward genetic screens: uniting high-throughput perturbations with single-cell analysis.
Trends Genet, 40(2):118-133, 20 Nov 2023
Cited by: 4 articles | PMID: 37989654
Review
CRISPR/SaCas9-based gene editing rescues photoreceptor degeneration throughout a rhodopsin-associated autosomal dominant retinitis pigmentosa mouse model.
Exp Biol Med (Maywood), 248(20):1818-1828, 14 Oct 2023
Cited by: 1 article | PMID: 37837380 | PMCID: PMC10792415
Stem cell-like reprogramming is required for leukemia-initiating activity in B-ALL.
J Exp Med, 221(1):e20230279, 06 Nov 2023
Cited by: 1 article | PMID: 37930337 | PMCID: PMC10626194
Go to all (41) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
OffScan: a universal and fast CRISPR off-target sites detection tool.
BMC Genomics, 21(suppl 1):872, 05 Mar 2020
Cited by: 9 articles | PMID: 32138651 | PMCID: PMC7057453
[CRISPR/CAS9, the King of Genome Editing Tools].
Mol Biol (Mosk), 51(4):582-594, 01 Jul 2017
Cited by: 4 articles | PMID: 28900076
Review
In Silico Meets In Vivo: Towards Computational CRISPR-Based sgRNA Design.
Trends Biotechnol, 35(1):12-21, 11 Jul 2016
Cited by: 47 articles | PMID: 27418421
Review
New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae.
Yeast, 32(12):711-720, 21 Sep 2015
Cited by: 160 articles | PMID: 26305040 | PMCID: PMC4715497
Funding
Funders who supported this work.
NHGRI NIH HHS (4)
Grant ID: R00 HG008171
Grant ID: U54 HG007990
Grant ID: R00 HG008399
Grant ID: U41 HG002371
NHLBI NIH HHS (1)
Grant ID: P01 HL032262
NIDDK NIH HHS (2)
Grant ID: P30 DK049216
Grant ID: F30 DK103359