Abstract
Free full text

IG-MYC+ neoplasms with precursor B-cell phenotype are molecularly distinct from Burkitt lymphomas
Key Points
preBLL shows molecular features of pB-ALL/LBL.
preBLL shows recurrent NRAS/KRAS mutations, lack of functional B-cell receptor, and IG-MYC translocations due to aberrant VDJ recombination.
Abstract
The WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue notes instances of Burkitt lymphoma/leukemia (BL) with IG-MYC rearrangement displaying a B-cell precursor immunophenotype (termed herein “preBLL”). To characterize the molecular pathogenesis of preBLL, we investigated 13 preBLL cases (including 1 cell line), of which 12 were analyzable using genome, exome, and targeted sequencing, imbalance mapping, and DNA methylation profiling. In 5 patients with reads across the IG-MYC breakpoint junctions, we found evidence that the translocation derived from an aberrant VDJ recombination, as is typical for IG translocations arising in B-cell precursors. Genomic changes like biallelic IGH translocations or VDJ rearrangements combined with translocation into the VDJ region on the second allele, potentially preventing expression of a productive immunoglobulin, were detected in 6 of 13 cases. We did not detect mutations in genes frequently altered in BL, but instead found activating NRAS and/or KRAS mutations in 7 of 12 preBLLs. Gains on 1q, recurrent in BL and preB lymphoblastic leukemia/lymphoma (pB-ALL/LBL), were detected in 7 of 12 preBLLs. DNA methylation profiling showed preBLL to cluster with precursor B cells and pB-ALL/LBL, but apart from BL. We conclude that preBLL genetically and epigenetically resembles pB-ALL/LBL rather than BL. Therefore, we propose that preBLL be considered as a pB-ALL/LBL with recurrent genetic abnormalities.
Introduction
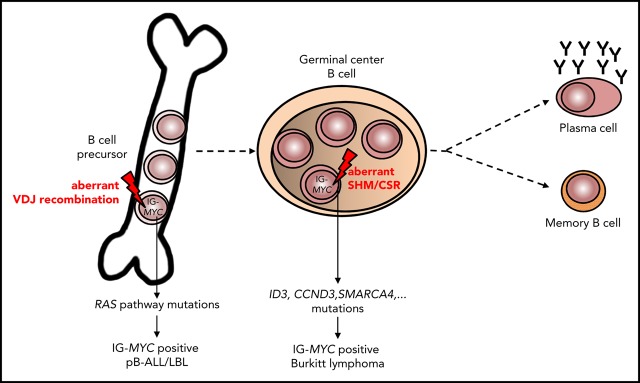
The molecular hallmark of Burkitt lymphoma (BL) is a MYC rearrangement involving as partner the IGH locus, in the form of t(8;14)(q24;q32), or less commonly the IGK or IGL locus, as t(2;8) or t(8;22), respectively.1,2 The tumor cells of BL, including its leukemic counterpart, typically display a germinal center B-cell phenotype.3 Nevertheless, the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue notes infrequent cases of Burkitt leukemia that “have a phenotype of precursor B-cells, with expression of terminal deoxynucleotidyltransferase, and sometimes CD34, and absence of CD20 and surface immunoglobulin expression. The reason for this aberrant phenotype remains unclear.”3(p333) Notably, the precursor B-cell immunophenotype and the clinical presentation of this possible BL variant (termed herein “preBLL”) would also be compatible with classification as precursor lymphoid neoplasms, particularly preB lymphoblastic leukemia/lymphoma (pB-ALL/LBL). Nevertheless, IGH translocations are rare (~5%) chromosomal aberrations in pB-ALL,4,5 whereas IG-MYC rearrangements seem to be underrepresented in pB-ALL series.
A subset of the preBLL cases described to date carries a t(14;18)/IGH-BCL2 translocation in addition to the IG-MYC translocation.6,7 Therefore, initial presentation as primary preBLL has to be differentiated from secondary evolution of a mature B-cell lymphoma to a precursor B-cell lymphoblastic lymphoma, which has been described as a rare but recurrent mode of progression, particularly in follicular lymphoma (FL).8,9
To better characterize the molecular pathogenesis of primary preBLL, as currently defined, we analyzed 12 primary cases and 1 cell line showing typical features, including MYC rearrangement and immature immunophenotype, using a range of genomic and epigenomic approaches.
Study design
In this retrospective study, we included 16 samples from 12 patients (for 3 patients, initial diagnosis [ID] and corresponding relapse samples; for 1 patient, 2 samples at ID) with primary preBLL defined as immunophenotype compatible with pB-ALL/LBL, including expression of terminal deoxynucleotidyltransferase,10 detection of IG-MYC translocation by conventional and/or molecular cytogenetics, and absence of a preceding or simultaneous mature B-cell lymphoma. Clinical and immunophenotypical data have been published for 4 of these cases.6,11,12 DNA was obtained from 15 samples (11 patients). Moreover, a representative cell line (380) was analyzed,13 adding to a total of 13 patients. DNA was subjected to whole-exome (6 samples from 5 patients) or whole-genome sequencing (n = 1; cell line 380). Selected findings (breakpoint sequences and mutations) were validated by Sanger sequencing or pyrosequencing. IGHV sequence analysis was performed as described.14 For copy number analyses, we applied the OncoScan CNV FFPE assay (n = 13), and for DNA methylation analysis, the HumanMethylationEPIC BeadChip array (n = 2; supplemental Table 1; details provided in supplemental methods).
Results and discussion
We collected tumor materials from 13 patients, including 1 cell line (380), fulfilling the outlined criteria for primary preBLL; 11 samples derived from leukemic and 2 from nodal presentation. In 6 cases, an L3/BL-like morphology was reported (Table 1). Median age at diagnosis was 21 years (range, 3-75 years), and the male/female ratio was 11:2. Three patients (cases 1, 4, and 11) developed a relapse from which samples were also analyzed. IGH-MYC, IGK-MYC, and IGL-MYC translocations were present in 10, 1, and 2 patients, respectively. Four of 13 preBLLs (cases 10-12 and cell line 38013,15) had (molecular) cytogenetic evidence for a concurrent t(14;18)/IGH-BCL2 translocation. Remarkably, median age at diagnosis of those 4 preBLL cases was 18 years (range, 13-43 years), which argues against a transformation of an occult FL. In line with this, the cell line 380 does not harbor mutations in genes recurrently mutated in FL.16
Table 1.
Overview of clinical, immunophenotypical, and cytogenetic (conventional and/or molecular cytogenetic) characteristics of preBLL samples studied
| Case | Age, y/sex | Site* | Diagnosis | Morphology | Time to relapse | Conventional and/or molecular cytogenetic characteristics | Immunophenotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IG-MYC status | BCL2 | BCL6 | Karyotype | TdT | CD34 | CD10 | BCL6 | CD19 | CD20 | sIg | Ki-67 | ||||||
| 1 ID† | 6/M | BM | B-ALL | BL-like | 8 mo | IGH-MYC | − | − | 47,XY,+idic(1)(p12),t(8;14)(q24;q32)[1]/ 47,idem,del(15)(q24)[22]/46,XY[2] | + | − | + | NA | + | +/−, weak | − | NA |
| 1 R† | BM | B-ALL | BL-like | IGH-MYC | − | − | 53,XY,+X,+Y,+idic(1)(p12),+6,+7,+8, t(8;14)(q24;q32)x2,+14,del(15)(q24)[25] | NA | − | + | NA | NA | + | − | NA | ||
| 2 ID | 75/M | Skin | B-LBL | NA | — | IGH-MYC | − | − | NA | + | − | + | NA | + | − | − | 80% |
| 3 ID | 69/M | PB | B-ALL | NA | — | IGH-MYC | − | − | 46,XY,+1,+1,der(1;13)(q10;q10), der(1;15)(q10;q10),t(8;14)(q24;q32)[20] | + | NA | NA | NA | − | +/− | NA | 100% |
| 4 ID | 13/M | BM | B-ALL | L2/3‡ | 14 mo | IGK-MYC | − | − | 47,XY,t(2;8)(p12;q24),+21[11] | + | NA | + | NA | + | − | − | NA |
| 4 R | BM | NA | NA | NA | NA | NA | + | NA | − | − | + | +/− | − | NA | |||
| 5 ID | 3/M | BM | B-ALL | L3‡ | — | IGH-MYC | − | − | 47,XY,trp(1)(q21q43),t(8;14)(q24;q32), +mar[10] | + | − | + | − | + | + | − | NA |
| 6 ID | 11/F | BM | B-ALL | L3‡ | — | IGL-MYC | − | − | 46,XX,t(8;22)(q24;q11)[20] | + | − | + | − | + | − | NA | NA |
| 7 ID§ | 70/M | BM | B-ALL | BL-like | — | IGH-MYC | − | − | NA | + | − | + | − | + | − | + | High |
| 8 ID§ | 56/M | BM | B-ALL | BL-like | — | IGH-MYC | − | − | NA | +/− | − | + | − | + | −/+ | NA | 95% |
| 9 ID‖ | 44/M | LN | B-LBL | NA | — | IGH-MYC | − | − | NA | + | − | + | − | NA | + | NA | ~95% |
| 10.1 ID | 43/M | BM | B-LBL | NA | — | IGH-MYC | + | − | 47,XY,t(8;14)(q24;q32),t(14;18)(q32;q21), der(16)t(1;16)(q21;p13.3),+21[16]/ 46,XY[4] | + | NA | + | NA | NA | NA | NA | NA |
| 10.2 ID | BM | B-LBL | NA | — | IGH-MYC | + | − | NA | NA | NA | + | − | NA | + | NA | NA | |
| 11 ID | 13/F | BM | B-ALL | NA | 142 d | IGL-MYC | + | − | 48,XX,i(1)(q10),+7,t(8;22)(q24;q11), t(14;18)(q32;q21)[10] | + | − | + | NA | + | NA | NA | NA |
| 11 R | BM | B-ALL | NA | IGL-MYC | + | − | 48,XX,i(1)(q10),+7,t(8;22)(q24;q11), t(14;18)(q32;q21)[20] | NA | NA | NA | NA | NA | NA | NA | NA | ||
| 12 ID¶ | 21/M | BM | B-ALL | L2‡ | — | IGH-MYC | + | − | 46,XY,t(8;14)(q23;q32),t(14;18)(q32;q21)[3]/46,XY,idem,t(13;17)(q12;p11),-17[7]/ 46,XY,idem,+t(13;13)(p11;q13), t(13;17)(q12;p11),-17[2]/46,XY[2] | + | NA | NA | NA | NA | NA | NA | NA |
| 380 cell line# | 15/M | PB | B-ALL | L2‡ | — | IGH-MYC | + | − | 46(43-47)<2n>XY,-14,t(8;14;18)(q24; q32;q21), +der(14)t(8;14;18)(q24;q32; q21) 1.8% polyploidy | + | − | + | NA | + | + | − | NA |
+, positive; −, negative; −/+, negative with positive parts; BM, bone marrow; LN, lymph node; NA, not available; PB, peripheral blood; sIg, surface immunoglobulin; TdT, terminal deoxynucleotidyltransferase.
The breakpoint junctions of the IG-MYC translocations could be retrieved from 4 of 5 patients (5 of 6 samples) submitted to exome sequencing (cases 1-4) and from the whole-genome data of cell line 380 (supplemental Table 2). The IG-MYC breakpoint junctions were identical in ID and relapse samples of cases 1 and 4. Junctions were verified by polymerase chain reaction–based Sanger sequencing in 4 of 6 samples, and a junction could be detected in the relapse sample of case 4 (Figure 1A; supplemental Figures 1-3). In all cases, the junctional sequences indicated aberrant VDJ recombination at the IGH locus as mechanism generating the IG-MYC translocation (details provided in supplemental data). The same held true for the IGH-BCL2 translocation in cell line 380, as previously published17 (supplemental Figures 2 and 3; supplemental Table 3). Thus, the translocation architecture suggests that the IG-MYC translocation in preBLL occurs in a premature B cell undergoing VDJ recombination and not in a germinal center B cell undergoing class switch or somatic hypermutation as in BL.18
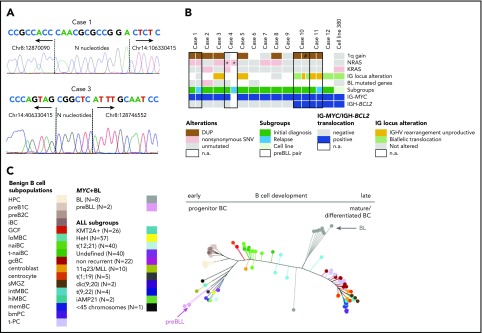
Genetic and epigenetic characterization of the preBLL samples. (A) Verification of IG-MYC breakpoint junctions retrieved from exome sequencing data by Sanger sequencing in cases 1 and 3. Both breakpoints showed features of aberrant VDJ rearrangement, including localization within the VDJ region and occurrence of N nucleotides. Details provided in supplemental Figure 1. (B) Summary of recurrent copy number aberrations (CNAs) and single-nucleotide variants (SNVs) identified by whole-exome sequencing/whole-genome sequencing and OncoScan analysis. Of note is that only 1 preBLL case harbored a mutation (SMARCA4) in those genes recurrently mutated in BL (overview provided in supplemental Figure 2). (C) Phylogenetic tree based on the DNA methylation pattern of 1404 CpG loci described to be differentially methylated during B-cell development.26 In the analysis, 17 B-cell subpopulations from various differentiation states were included, which arranged themselves along the main trunk of the phylogenetic tree according to their differentiation stadia. The 2 preBLL cases clustered with the pB-ALL/LBL samples24,25,26 near the precursor B-cell subpopulations, whereas the BL samples were closer to the mature B-cell subpopulations. *Indicates that the NRAS mutation in case 4 differed between the ID and relapse samples. #Indicates that the 1q gain in sample 2 of case 10 occurred in comparison with sample 1 of case 10 in a subset of cells. DUP, duplication; n.a., not available.
The clonal IGH rearrangements from 7 of 14 samples (6 of 12 patients) were successfully amplified. A total of 5 of 7 samples (5 of 6 patients) showed unmutated IGHV rearrangements (<2% mutations), and none showed signs of intraclonal diversity, pointing to ongoing somatic hypermutation (supplemental Table 4). In line with this, the IGH rearrangement sequences derived from ID and relapse samples of case 11 were identical and lacked evidence of clonal evolution (supplemental Table 4). Interestingly, in 2 of 6 informative patients, only an out-of-frame rearrangement was obtained, and in 2 additional cases, an originally productive rearrangement was rendered nonfunctional by mutations within the IGHV genes. Remarkably, in 3 of 13 cases (cases 10 and 12, cell line 380), we found evidence of involvement of both IGH alleles in chromosomal translocations resulting from the cooccurring IG-MYC and IG-BCL2 translocations (Figure 1B; supplemental Table 5). These data indicate that clonal genomic changes (out-of-frame V[D]J rearrangements and translocations targeted into the V[D]J region of the IGH locus) target both alleles of the IGH locus and hence prevent expression of a functional B-cell receptor (BCR) in a substantial subset of preBLLs (supplemental Table 5). This is in contrast to the situation in BL in which expression of a functional BCR and its signaling are important for survival.19
Mining the whole-exome sequencing data, as detailed in “Study design,” in the 5 primary samples from ID, we detected a median of 77 (range, 64-108) potentially protein-changing variants. The relapse of case 1 harbored 90 nonsynonymous exonic variants, in contrast to 77 in the corresponding ID sample, with 70 variants occurring in both (supplemental Figure 4; supplemental Table 6). Strikingly, none of the genes recurrently mutated in BL (≥15% of BL patients; C.L., K.K., S.M.A., M. Rohde, S. H. Bernhart, R.W., D.H., U.H.T., F. Raimondi, M. Kreuz, S. M. Waszak, Z. Huang, L. Sieverling, N. Paramasivam, J.S., S. Sungalee, R. B. Russell, J. Bausinger, H. Kretzmer, O. Ammerpohl, A. K. Bergmann, H. Binder, A. Borkhardt, B. Brors, A. Claviez, G. Doose, L. Feuerbach, A. Haake, M.-L. Hansmann, J. Hoell, M. Hummel, J. O. Korbel, C. Lawerenz, D. Lenze, B. Radlwimmer, J. Richter, P. Rosenstiel, A. Rosenwald, M. B. Schilhabel, H. Stein, S.S., P. F. Stadler, M. Szczepanowski, M. A. Weniger, M. Zapatka, R. Eils, P. Lichter, M. Loeffler, P. Möller, L. Trümper, W.K., S. Hoffmann, R.K., B.B., M.S., R.S., manuscript submitted August 2018),20-22 including ID3, CCND3, or MYC, were recurrently mutated in preBLL (supplemental Figures 5 and 6). In contrast, 5 of 6 cases contained either NRAS and/or KRAS mutations, which affected well-known mutational hotspots (supplemental Figure 7). To further investigate, we performed targeted analysis of NRAS codons 12 and 13 and KRAS codons 12 and 13 in 6 preBLLs not subjected to exome sequencing. Taken together, 7 of 12 analyzed cases (9 of 16 samples) carried NRAS and/or KRAS mutations (Figure 1B; supplemental Table 7). Whereas mutations of KRAS and NRAS are recurrent in pB-ALL/LBL, only 1 (0.9%) of 112 BLs harbored a RAS mutation in a recent study.19 Interestingly, it has been shown that in MYC-driven B-cell lymphomas, RAS mutations are a mechanism to substitute for the lack of BCR signaling,19 which might explain the independence of preBLL from BCR signaling. In line with this, the ID sample of case 3, as well as cell line 380, carried RAS mutations and harbored IG alterations, likely inactivating the BCR function (Figure 1B). Remarkably, ID and relapse samples of case 4, which shared the identical IGK-MYC translocation, harbored different NRAS mutations, indicating these occurred later than the MYC translocation in clonal evolution (supplemental Figure 8).
Next, we analyzed the copy number profiles of 11 preBLL cases (13 samples). Median number of CNAs in the 9 ID samples was 4 (range, 1-11) (supplemental Table 8), demonstrating a low genomic complexity in agreement with the conventional cytogenetics. The ID and relapse samples of case 11 showed exactly the same CNA profile, whereas in case 1, an increase of genomic complexity was reported in the relapse sample.12 The only highly recurrent imbalance (>2 cases) was a gain in 1q21.1-q44, present in 7 of 11 preBLLs (minimal region chr1:144,936,742-249,212,878 bp), which constitutes a common alteration in BL and pB-ALL/LBL11,12 (supplemental Figure 9). Moreover, in cell line 380, we detected focal CNAs in 5q33.3 and 13q14.2 affecting EBF1 and RB1, respectively (supplemental Figure 10). This might point to a role of alterations in B-cell developmental genes in the pathogenesis of preBLL similar to pB-ALL/LBL.23 We might have missed such focal deletions in the primary preBLL samples because of the resolution of the imbalance mapping.
Finally, we compared the DNA methylation profile of 2 preBLLs with that of benign B-cell subsets as well as with BL and pB-ALL/LBL samples of various subtypes (supplemental Tables 9 and 10).24,25 Focusing on loci becoming differentially methylated during B-cell development,26 we showed that preBLLs clustered with pB-ALL/LBL samples close to the precursor B-cell populations. In contrast, BL samples clustered close to the germinal center B cells but apart from preBLLs (Figure 1C).
In summary, by combined genomic and epigenomic profiling, we provide evidence that primary preBLL resembles pB-ALL/LBL rather than BL, not only with regard to immunophenotype but also with regard to the B-cell maturation stage at which the IG-MYC translocation occurred; the frequent lack of a productive IGH rearrangement; the mutational landscape, with frequent activation of the RAS pathway, mutations in B-cell developmental genes, and absence of mutations in genes recurrently altered in BL; and the DNA methylation pattern. Therefore, we propose that although morphology and male predominance are similar to those of BL-like disease, IG-MYC–translocated precursor cell leukemias/lymphomas should be classified as a subtype separate from the group of preB lymphoblastic leukemias/lymphomas with recurrent genetic abnormalities. The clinical implications of these findings warrant additional studies.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the teams of the tumor genetic laboratories at the Institutes of Human Genetics in Kiel and Ulm for expert technical assistance and R. Marienfeld for valuable information regarding design of mutational assays.
This study was supported by the German Ministry of Science and Education in the framework of the MMML-MYC-SYS project (036166B) and the ICGC MMML-Seq and ICGC DE Mining projects (01KU1002A-J and 01KU1505G, respectively). Part of the work was performed in association with SFB1074 (particularly subproject B1), funded by DFG. KinderKrebsInitiative Buchholz Holm-Seppensen provided infrastructural support, and the Deutsche Kinderkrebsstiftung provided support of the NHL-BFM Registry (DKS 2014.11 A/B).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.A., D.O., R.H., E.S.J., E.K., I.O., M.K., U.K., L.d.L., W.K., B.B., and W.W. provided tumor samples and clinical data; H.G.D. provided the cell line; S.M.A. and I.N. contributed to the collection of samples; C.L., I.N., B.S., and E.M.M.P. performed cytogenetic analysis; J.A., H.T., and P.N. performed whole-exome sequencing (WES); K.K., J.P., J.S., U.H.T., D.H., and M.S. analyzed the whole-genome sequencing and WES data; C.S. and S.S. performed IGHV sequence analysis; J.K. performed pyrosequencing; C.L. analyzed IG-MYC breakpoints; R.K. interpreted the IG-MYC breakpoint and IGHV sequence data; R.W. analyzed the OncoScan data and performed the DNA methylation analysis; J.B. and R.W. interpreted the WES data and performed validation by Sanger sequencing; R.S. designed the study and coordinated the project; R.W., C.L., and R.S. interpreted the data and wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliations for D.H. are Division of Stem Cells and Cancer, German Cancer Research Center, Heidelberg, Germany, and Heidelberg Institute for Stem Cell Technology and Experimental Medicine, Heidelberg, Germany.
Correspondence: Reiner Siebert, Institute of Human Genetics, Ulm University and Ulm University Medical Center, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: [email protected].
REFERENCES
Articles from Blood are provided here courtesy of The American Society of Hematology
Full text links
Read article at publisher's site: https://doi.org/10.1182/blood-2018-03-842088
Read article for free, from open access legal sources, via Unpaywall:
https://ashpublications.org/blood/article-pdf/132/21/2280/1747314/blood842088.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Latent Epstein-Barr virus infection collaborates with Myc over-expression in normal human B cells to induce Burkitt-like Lymphomas in mice.
PLoS Pathog, 20(4):e1012132, 15 Apr 2024
Cited by: 0 articles | PMID: 38620028 | PMCID: PMC11045125
Cytogenetic and pathologic characterization of MYC-rearranged B-cell lymphomas in pediatric and young adult patients.
J Hematop, 17(2):51-61, 02 Apr 2024
Cited by: 0 articles | PMID: 38561469 | PMCID: PMC11127862
Burkitt Lymphoma With Aberrant Expression of Cytoplasmic Terminal Deoxynucleotidyl Transferase: A Case Report.
Cureus, 16(2):e53972, 10 Feb 2024
Cited by: 0 articles | PMID: 38348200 | PMCID: PMC10859684
Diagnostic and prognostic molecular pathology of lymphoid malignancies.
Virchows Arch, 484(2):195-214, 25 Sep 2023
Cited by: 2 articles | PMID: 37747559 | PMCID: PMC10948535
Review Free full text in Europe PMC
The Gene Expression Classifier ALLCatchR Identifies B-cell Precursor ALL Subtypes and Underlying Developmental Trajectories Across Age.
Hemasphere, 7(9):e939, 25 Aug 2023
Cited by: 11 articles | PMID: 37645423 | PMCID: PMC10461941
Go to all (29) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genetic features of precursor B-cell phenotype Burkitt leukemia with IGH-MYC rearrangement.
Cancer Rep (Hoboken), 5(7):e1545, 02 Sep 2021
Cited by: 1 article | PMID: 34472720 | PMCID: PMC9327653
Molecular characteristics of terminal deoxynucleotidyl transferase negative precursor B-cell phenotype Burkitt leukemia with IGH-MYC rearrangement.
Genes Chromosomes Cancer, 59(4):255-260, 26 Nov 2019
Cited by: 3 articles | PMID: 31705772
B Lymphoblastic Leukemia/Lymphoma With Burkitt-like Morphology and IGH/MYC Rearrangement: Report of 3 Cases in Adult Patients.
Am J Surg Pathol, 42(2):269-276, 01 Feb 2018
Cited by: 10 articles | PMID: 29112016 | PMCID: PMC5762415
Burkitt Leukemia With Precursor B-Cell Immunophenotype and Dual Translocation of t(14;18) and t(8;14) in a Child: Case Report and Review of the Literature.
J Pediatr Hematol Oncol, 42(1):e61-e63, 01 Jan 2020
Cited by: 0 articles | PMID: 30499905
Review
 1,2
1,2