Abstract
Free full text

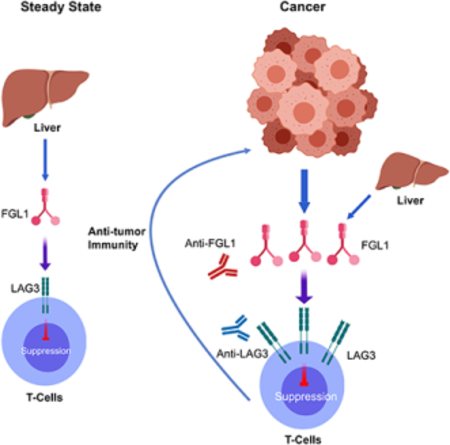
Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG3
SUMMARY
Lymphocyte-activation gene 3 (LAG3) is an immune inhibitory receptor, with major histocompatibility complex class II (MHC-II) as a canonical ligand. However, it remains controversial whether MHC-II is solely responsible for the inhibitory function of LAG3. Here, we demonstrate that fibrinogen-like protein 1 (FGL1), a liver-secreted protein, is a major LAG3 functional ligand independent from MHC-II. FGL1 inhibits antigen-specific T-cell activation and ablation of FGL1 in mice promotes T-cell immunity. Blockade of the FGL1/LAG3 interaction by monoclonal antibodies stimulates tumor immunity and is therapeutic against established mouse tumors in a receptor-ligand inter-dependent manner. FGL1 is highly produced by human cancer cells and elevated FGL1 in the plasma of cancer patients is associated with a poor prognosis and resistance to anti-PD-1/B7-H1 therapy. Our findings reveal an immune evasion mechanism and have implications for the design of cancer immunotherapy.
Graphical Abstract
In Brief
FGL1 is identified as a major ligand for the inhibitory receptor LAG3 and its blockade can potentiate anti-tumor T cell responses
INTRODUCTION
Lymphocyte-activation gene 3 (LAG3, CD223) is a transmembrane protein primarily found on activated T cells (Anderson et al., 2016; Andrews et al., 2017; Triebel et al., 1990). LAG3 protein consists of four extracellular immunoglobulin (Ig)-like domains (D1-D4) with high homology to CD4 (Triebel et al., 1990). LAG3 expression can be upregulated by IL-2 and IL-12 on activated T cells (Annunziato et al., 1997; Annunziato et al., 1996; Bruniquel et al., 1998), where it mainly functions as a receptor that delivers inhibitory signals (Huard et al., 1996; Huard et al., 1994; Workman et al., 2002a). LAG3 negatively regulates the proliferation, activation, effector function, and homeostasis of both CD8+ and CD4+ T cells, as shown in LAG3 knockout mice and antibody studies (Huard et al., 1994; Workman et al., 2004; Workman et al., 2002a; Workman et al., 2002b; Workman and Vignali, 2003, 2005). LAG3 may represent an “exhaustion” marker for CD8+ T cells similar to PD-1 in response to repetitive antigen stimulation in chronic viral infections or cancers (Blackburn et al., 2009; Chihara et al., 2018; Grosso et al., 2009; Grosso et al., 2007; Matsuzaki et al., 2010; Williams et al., 2017). Additionally, LAG3 is also constitutively expressed on a subset of regulatory T-cells and contributes to their suppressive function (Camisaschi et al., 2010; Gagliani et al., 2013; Huang et al., 2004). Currently, monoclonal antibodies (mAbs) that block the interaction of LAG3 with its canonical ligand, MHC-II, are being evaluated for their antitumor activity in clinical trials (Anderson et al., 2016; Ascierto et al., 2017; Rotte et al., 2018).
The major ligand that mediates the immune suppressive functions of LAG3, however, remains controversial. Initial studies by Baixeras and colleagues showed an interaction between MHC-II and LAG3 via a cell-cell adhesion assay (Baixeras et al., 1992), which was further extended by studies indicating LAG3 fusion protein binding to MHC-II+ B cell lines (Huard et al., 1996; Huard et al., 1995). However, there is a lack of direct evidence for the protein-protein interaction between LAG3 and MHCII. MHC-II was proposed to interact with LAG3 through the residues on the membrane-distal, top face of the LAG3 D1 domain (Huard et al., 1997). Functionally, the MHC-II/CD4 interaction supported helper T cell activation, while overexpression of LAG3 downregulated antigen-dependent CD4+ T cell responses in vitro (Workman and Vignali, 2003). However, several mAbs that do not block the binding of LAG3 to MHC-II nonetheless promoted T cell functions. For example, C9B7W, a specific mAb against the murine LAG3 D2 domain, enhanced the proliferation and effector functions of T cells in vitro and in vivo (Workman et al., 2004; Workman et al., 2002b; Workman and Vignali, 2005). This antibody also increased the accumulation and effector function of tumor-specific CD8+ T cells in several tumor models (Grosso et al., 2007; Woo et al., 2012). The effects of C9B7W mAb on T cells are largely similar, if not identical, to those produced by LAG3 genetic deficiency (Woo et al., 2012; Workman and Vignali, 2005). A recent study also showed that anti-LAG3 mAb that do not block MHCII binding could still stimulate T cell activation and anti-tumor activity (Cemerski, 2015). Given that LAG3 also suppresses the function of CD8+ T cells and NK cells, which do not interact with MHC-II (Anderson et al., 2016), these studies raise the possibility that the immunological functions of LAG3 might be mediated via an unknown ligand.
Here we report that fibrinogen-like protein 1 (FGL1) is a major functional ligand of LAG3. FGL1 belongs to the fibrinogen family with high amino acid homology to the carboxyl terminus of the fibrinogen beta- and gamma-subunits, but it does not have the characteristic platelet-binding site, crosslinking region, and thrombin-sensitive site necessary for fibrin clot formation (Yamamoto et al., 1993). Under normal physiological conditions, FGL1 protein is primarily secreted from hepatocytes and contributes to its mitogenic and metabolic functions (Demchev et al., 2013; Hara et al., 2001; Li et al., 2010; Liu and Ukomadu, 2008; Yamamoto et al., 1993; Yan et al., 2002). The immunological function of FGL1, however, remains unknown. Our results demonstrate that FGL1 is a major inhibitory ligand for LAG3, revealing a new mechanism of immune evasion.
RESULTS
FGL1 is an MHC class II-independent high affinity ligand of LAG3
We employed our Genome-Scale Receptor Array (GSRA) technology to search for LAG3 binding protein(s) using an immunoglobulin (Ig) Fc-tagged LAG3 extracellular domain fusion protein (LAG3-Ig) (Figure 1a). The GSRA is a semi-automatic gene expression and detection system for rapidly identifying protein-protein interactions, which has been modified from our previous report (Yao et al., 2011). In this updated system, individual human cDNA encoding transmembrane and secreted proteins (upon addition of a transmembrane domain) were overexpressed on the surface of 293T cells. Several adaptor genes were also expressed in 293T (293T.2A cells) to facilitate protein expression on the cell surface (Figure 1a). An Ig-tagged protein of interest can then be screened for interaction(s) using the GSRA system in a high throughput fashion by the mix-and-read laser scanning macro-confocal fluorescent plate reader. The current version of the GSRA contains over 90% of annotated genes encoding human transmembrane (~5,600) and secreted (~1,000) proteins (Table S1). FGL1 was identified as a major binding protein for LAG3-Ig in the GSRA system (Figure S1a and Figure 1b). The FGL1/LAG3 interaction is conserved across species in both human and mouse (Figure 1b). This interaction was further validated by flow cytometry, as indicated by a linear association between FGL1Ig and anti-LAG3 staining on LAG3+ cells (Figure 1c). The FGL1/LAG3 interaction was shown to have a Kd value of ~1.5 nM by Octet bio-layer interferometry analysis (Figure 1d). Using an SEC650 size exclusion column, the purified recombinant FLAG-tagged FGL1 (FLAG-FGL1) showed an oligomeric state (peak1–2) and a dimeric peak (peak 3) (Figure S1b), which was validated via size exclusion chromatography with Multi-Angle Light Scattering analysis (SEC-MALS, data not shown). We observed stronger binding of the oligomeric forms of FLAG-FGL1 (peak 1–2) than the dimeric form (peak 3) to immobilized LAG3-Ig in the Octet analysis (Figure S1c). In addition, the slow disassociation rate hints at a stable interaction between FGL1 and LAG3 in both human and mouse (Figure 1d and Figure S1d). FGL2, a homolog of FGL1 previously implicated in Treg functions (Shevach, 2009), as well as other fibrinogen domain-containing family members such as angiopoietin-related proteins, did not bind LAG3 (Figure 1b and data not shown), indicating that the FGL1/LAG3 interaction is highly specific.
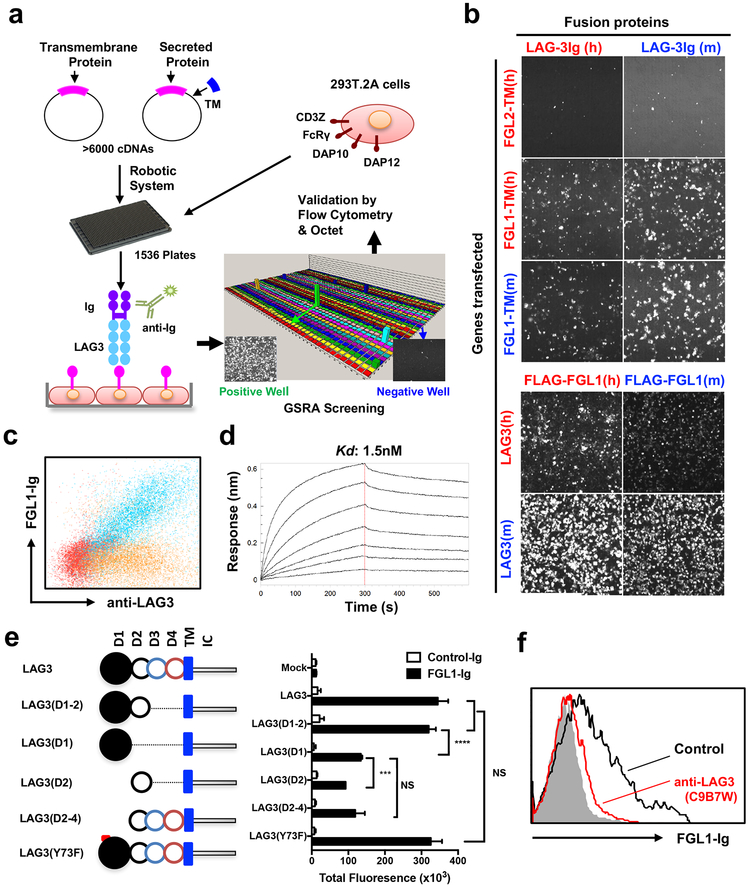
(a) Schematic representation of the GSRA system. Individual plasmids of genes encoding both transmembrane and secreted proteins were transfected into 293T.2A cells (see methods and Table S1) in 1,536-well plates. LAG3-Ig as well as fluorescence labeled anti-Fc mAb were added into each well for rapid detection of LAG3-Ig binding. Human Fc receptors served as internal positive controls within each plate. Positive hits were confirmed by flow cytometry or Octet bio-layer interferometry. TM, transmembrane domain.
(b) Image of the FGL1/LAG3 interaction in GSRA system. 293T.2A cells were transfected with human (h) or mouse (m) FGL1-TM or full-length LAG3 as indicated on the Y-axis. Human FGL2-TM was included as a negative control. The indicated fusion proteins shown on the X-axis were added to the culture to evaluate binding to the transfectants by the cellular detection system (CDS).
(c) Representative flow cytometry dot plot of FGL1-Ig binding to mouse LAG3+ 293T.2A (blue) or mock cells (red). Control Ig binding to mouse LAG3+ 293T.2A is also shown (brown).
(d) Representative Octet sensorgrams showing various amounts of FLAG-tagged mouse FGL1 (starting from 10ug/ml, two-fold serial dilutions) binding to immobilized mouse LAG3-Ig.
(e) Schematic representation of constructs coding full-length mouse LAG3, LAG3 Y73F mutant, or LAG3 with different extracellular domain deletions (left). LAG3 full-length protein consists of four extracellular Ig domains (D1 to D4), the transmembrane domain (TM), and intracellular domain (IC) (left). Quantification of FGL1-Ig binding to 293T.2A cells transfected to express LAG3 with domain deletion/mutation (right). Data were analyzed by CDS software and presented as the mean ± SEM. *** p<0.001, **** p<0.0001, NS, not significant by Student’s t-test.
(f) FGL1-Ig binding to mouse LAG3+ 293T.2A cells in the presence of control mAb (black line) or antiLAG3 (red line) by flow cytometry. Cells stained with control Ig (shadow) served as a negative control. All data are representative of at least two independent experiments.
FGL1 is composed of a coil-coil domain (CCD) and a fibrinogen-like domain (FD) (Yamamoto et al., 1993). Through domain deletion studies, we demonstrated that the FD, but not CCD, is responsible for LAG3 binding (Figure S1e). The LAG3 protein consists of four Ig-like extracellular domains, D1-D4 (Huard et al., 1997; Triebel et al., 1990) (Figure 1e, left panel). The deletion of the D3-D4 domain in LAG3 did not affect FGL1 binding, while either D1 or D2 alone partially decreased the binding (Figure 1e, right panel), suggesting that both D1 and D2 contribute to the FGL1/LAG3 interaction. A single point mutation (Y73F) in the C’ strand of LAG3 D1 domain was previously shown to disrupt MHC-II binding (Huard et al., 1997; Workman et al., 2002a). However, this mutation did not affect FGL1-Ig binding (Figure 1e, right panel), indicating that the FGL1/LAG3 interaction is nonredundant with MHC-II/LAG3 binding. Furthermore, pre-incubation of LAG3+ 293T cells with C9B7W, an anti-LAG3 mAb that binds the LAG3 D2 domain without blocking the FGL1/MHC-II interaction (Andrews et al., 2017; Cemerski, 2015; Workman et al., 2002b), led to complete abrogation of FGL1/LAG3 binding (Figure 1f). Finally, LAG3+ cells stained with MHC-II (I-Ab) fusion protein did not show a significant decrease in binding even in the presence of a 100-fold excess of FGL1-Ig (Figure S1f). Taken together, our results indicate that FGL1 interacts with LAG3 in an MHC-II independent manner, and that this interaction involves the FGL1 fibrinogen-like domain and the LAG3 D1-D2 domain.
FGL1 inhibits antigen-mediated T cell responses via LAG3 in vitro and in vivo
LAG3 is not found on resting T cells other than a subset of Tregs but can be upregulated under various antigen stimulation conditions (Baixeras et al., 1992; Triebel et al., 1990; Workman et al., 2002b). FGL1-Ig fusion protein did not bind resting T cells which express minimal LAG3 levels (data not shown), though it did bind activated T cells from wild type (WT) but not from LAG3-KO mice, as determined via flow cytometry analysis (Figure 2a). Inclusion of FGL1-Ig partially suppressed WT splenic T cell proliferation under suboptimal anti-CD3 stimulation, but this suppression was diminished using LAG3-KO splenocytes (Figure 2b), indicating that the suppressive effect of FGL1-Ig is dependent on LAG3. Similarly, FGL1 better suppressed the antigen-specific induction of IL-2 from a murine LAG3 overexpressing 3A9 T cell line (3A9-LAG3) in a dose dependent fashion compared to the parental 3A9 cell line with low endogenous LAG3 expression (Figure 2c). We generated a mAb specific for mouse FGL1 (clone 177R4) that blocks FGL1-Ig binding to LAG3+ 293T cells in a similar manner to anti-LAG3 mAb C9B7W (Figure S2a–b). Both mAbs abrogated the suppression of FGL1 on IL-2 production from 3A9-LAG3 cells (Figure 2d). Upon CD8+ OT-1 transgenic T cell transfer into syngeneic mice and subsequent immunization with chicken ovalbumin peptide antigen, administration of anti-FGL1 mAb 177R4 significantly promoted antigen-specific OT-1 T cell activation in a manner similar to anti-LAG3 mAb, as determined by increased plasma levels of TNF-α and IFN-γ (Figure S2c–d). Thus, our results support that FGL1 is an inhibitory ligand of LAG3.
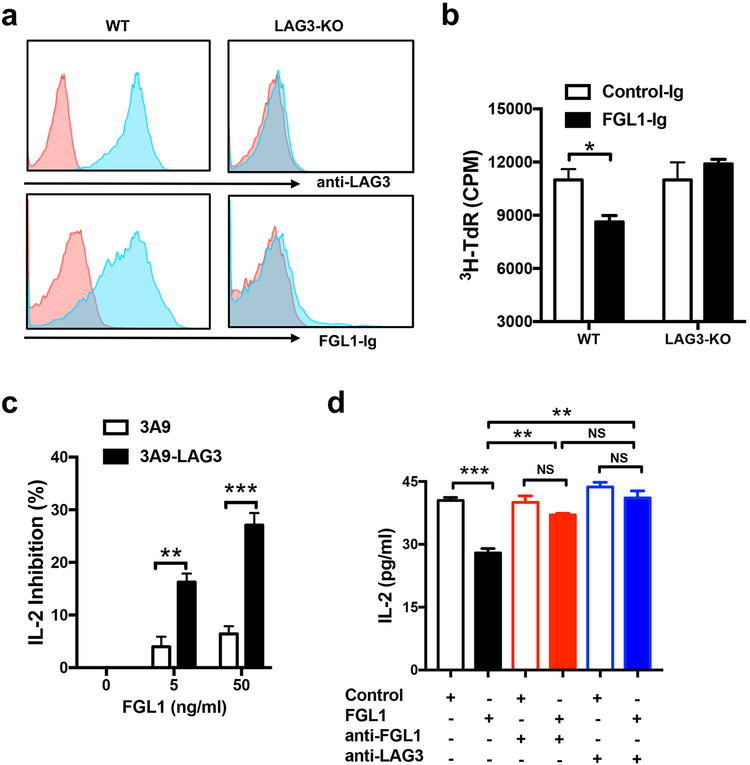
(a) Splenic T cells from WT or LAG3-KO mice were activated by immobilized anti-CD3 mAb for 24 hours, stained with anti-LAG3 mAb or FGL1-Ig fusion protein (blue) or control antibody/Ig (red), and analyzed by flow cytometry.
(b) Splenic T cells from WT or LAG3-KO mice were activated by immobilized anti-CD3 mAb at suboptimal concentration in the presence of soluble FGL1-Ig or control-Ig (5ug/ml) for 3 days before the addition of 3H-dTR. Thymidine incorporation of proliferated T cells was analyzed 16 hours later.
(c) The 3A9-LAG3 or parental 3A9 mouse T cell hybridoma cells were co-cultured with LK35.2 B cell line in CellGro serum free medium in the presence of HEL peptide and the indicated concentrations of FLAG-tagged FGL1. Shown is the normalized % of inhibition on the IL-2 levels in the supernatant at 24 hours normalized to levels with 0 ng/ml FGL1.
(d) The 3A9-LAG3 mouse T cell hybridoma cells were co-cultured with LK35.2 B cell line in the presence of HEL peptide, FLAG tagged FGL1 (50ng/ml), anti-FGL1, or anti-LAG3 mAb (1ug/ml). Shown are the IL-2 levels in the supernatant at 24 hours.
Data are representative of at least two independent experiments and are presented as the mean ± SEM. *p < 0.05, ** p < 0.01; *** p<0.001, NS, not significant by Student’s t-test.
Fgl1 deficient mice slowly develop spontaneous autoimmune symptoms
We generated a Fgl1 gene knockout mouse strain (FGL1-KO) on the C57BL/6 background using an agouti color gene modified mouse embryonic stem cell line (JM8) (Pettitt et al., 2009). In WT mice, Fgl1 mRNA was detected in the liver but not in other organs or hematopoietic cells (Figure S3a). Soluble FGL1 was also detected in mouse blood (Figure S3b) as previously reported (Liu and Ukomadu, 2008). In contrast, FGL1 was not detected in the plasma or liver of FGL1-KO mice via specific sandwich ELISA and western blot analysis, respectively (Figure S3b–c).
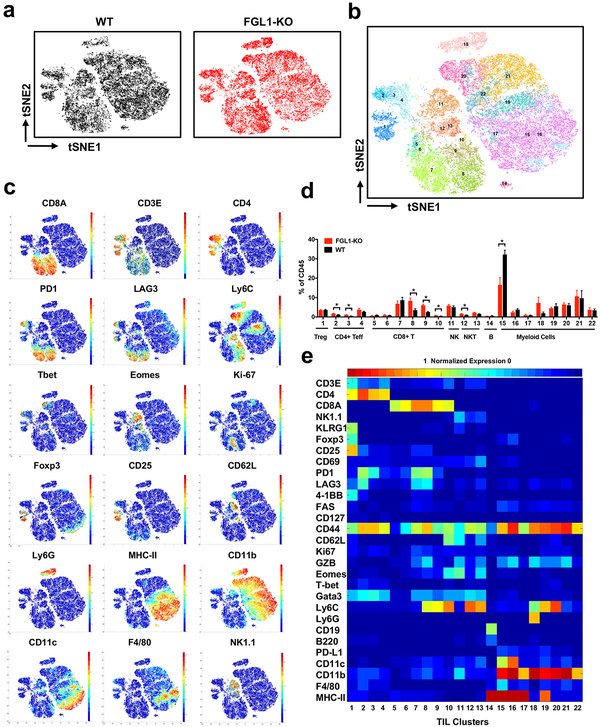
FGL1-KO mice have an overall normal appearance, organ size, and litters, indicating that FGL1 does not globally affect the development and growth of mice. However, up to 40% (8/20) of FGL1-KO mice developed spontaneous dermatitis at the age of 8 months or older, showing lymphocyte infiltration in the dermis (Figure S3d–e). At 14–16 months of age, 5/8 female, but not male mice, had elevated levels of anti-double-stranded DNA autoantibodies in their plasma compared to WT mice (Figure S3f). These findings are consistent with the role of FGL1 as an immune suppressive molecule. To evaluate overall changes in the immune system of this KO strain, we profiled mouse peripheral blood cells by mass cytometry (CyTOF), a single cell high dimensional analysis tool using 32 metal-conjugated mAbs to determine immune cell lineages as well as functional molecules. A recently described unsupervised clustering method named x-shift was also employed (Samusik et al., 2016). Analysis of the total CD45+ hematopoietic cells revealed 22 distinct cell type/subsets (clusters), with small but significant increases in central memory-like CD8+ T cells subsets (cluster 14–15, CD44+ CD62L+ Ly6C+ CD127med Tbet+ Eomes+) and decreases in two B cell subsets (cluster 2 and 4) (Figure 3a–e). All other clusters were similar in FGL1-KO compared to WT mice (Figure 3a–e). There were no major differences in T cells or myeloid cell subsets in peripheral lymphoid tissues including the spleen or liver (data not shown).
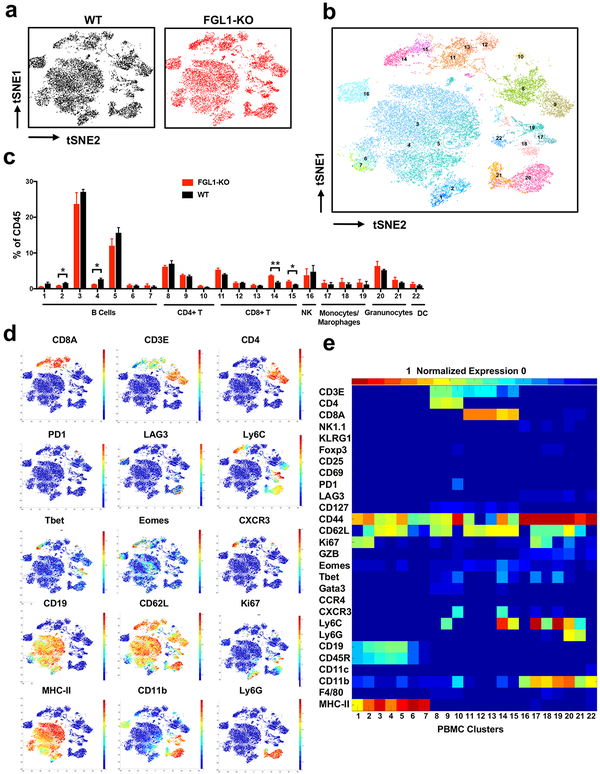
(a) Density t-SNE plots of an equal number of CD45+ compartment in the peripheral blood from WT and FGL1-KO mice (n=3).
(b) t-SNE plot of CD45+ compartment overlaid with color-coded clusters.
(c) Frequency of clusters grouped by indicated immune cell subsets. Data were shown as the mean ± SEM. *, p<0.05; ** p<0.01 by unpaired t-test.
(d) t-SNE plot of CD45+ compartment overlaid with the expression of selected markers.
(e) Heatmap displaying normalized marker expression of each immune cluster.
These findings indicate that endogenous FGL1 does not affect mouse development and growth, although it may participate in regulating autoimmunity and immune homeostasis in aged mice.
Fgl1 silencing promotes T cell immunity against tumor growth in mouse models
FGL1-KO and LAG3-KO mice were inoculated subcutaneously (s.c.) with syngeneic murine MC38 colon cancer cells. Similar to LAG3-KO mice, FGL1-KO mice showed significantly slower tumor growth in comparison to WT mice (Figure 4a). Whereas all of the WT mice reached an endpoint (average mean tumor diameter of 15 mm) within 60 days, ~50% of FGL1-KO or LAG3-KO mice were tumor free beyond 200 days upon MC38 inoculation (Figure 4b). Similarly, both anti-FGL1 and antiLAG3 mAbs significantly controlled tumor growth of established MC38 murine colon (Figure 4c) and Hepa1–6 murine liver cell lines inoculated s.c. in syngeneic C57BL/6 mice (Figure S4a). In contrast, the anti-FGL1 and anti-LAG3 mAb antitumor effect was abrogated in Rag1-KO C57BL/6 mice, which are devoid of T and B cells (Figure 4d). Consistent with these findings, depletion of either CD8+ or CD4+ T cells by specific mAbs completely eliminated the anti-tumor effect of both anti-FGL1 and anti-LAG3 mAb in the MC38 tumor model, indicating that the anti-tumor effect of these mAbs is dependent on both CD8+ and CD4+ T cells (Figure S4b).
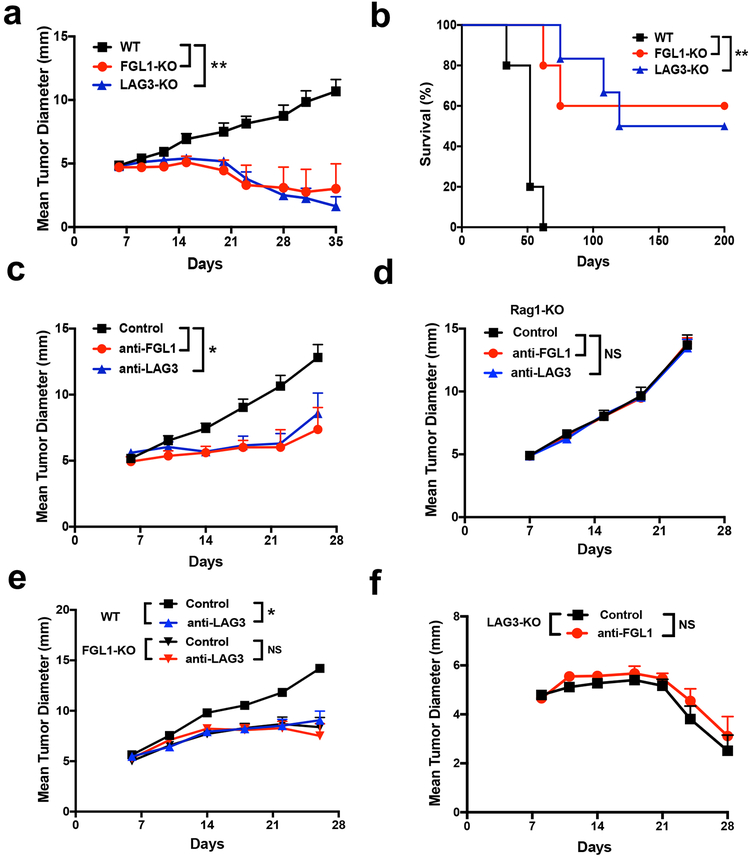
(a-b) FGL1-KO, LAG3-KO, or WT littermates were inoculated with MC38 cells (0.5×106/mouse). The mean tumor diameters (a) and survival (b) of mice in each group (n=6) are shown.
(c-d) B6 (c) or Rag1-KO (d) mice were inoculated with MC38 cells (0.5×106/mouse) at day 0 and treated with anti-FGL1, anti-LAG3, or control mAbs every four days from day 6 to day 18. The mean tumor diameters in each group (n=6) are shown.
(e) WT or FGL1-KO mice were inoculated with MC38 cells and were treated with anti-LAG3 or control mAb as in (c). The mean tumor diameters in each group (n=6–8) are shown.
(f) LAG3-KO mice were inoculated with MC38 cells and were treated with anti-FGL1 (n=8) or control mAb (n=7) as in (c). The mean tumor diameters in each group are shown.
Data were representative of at least two independent experiments and are shown as the mean ± SEM. *, p<0.05; **, p<0.01; NS, not significant. a, c-f by two-way ANOVA; b by Log-rank test.
To exclude the possibility that additional ligands for LAG3 are functionally redundant to FGL1 and contribute to the anti-tumor effect of the anti-LAG3 mAb, we tested the effect of anti-LAG3 mAb in FGL1-KO mice. While the anti-LAG3 mAb suppressed MC38 tumor growth in WT mice, this antitumor effect was completely eliminated in FGL1-KO mice (Figure 4e). The effect of anti-FGL1 was also dependent on LAG3, as this mAb did not have additive effects on tumor growth in LAG3-KO mice (Figure 4f). Therefore, the anti-tumor effect of anti-FGL1 mAb is dependent on LAG3, whereas the effect of anti-LAG3 relies on FGL1 but not MHC-II or other LAG3 ligands. Altogether, our findings support FGL1 as a major ligand for LAG3 to induce T cell suppressive function and immune evasion.
The deficiency of FGL1 significantly reduced MC38 tumor growth while spleen size and the number of lymphocytes in either tumor-draining or non-tumor-draining lymph nodes remained similar (Figure S4c). Analysis of tumor-infiltrating leukocytes (TIL) in tumors excised on day 17 from FGL1KO and WT mice by mass cytometry revealed a significant increase of CD45+ leukocytes in FGL1-KO tumors (Figure S4d). In 22 clusters across CD45+ cells (Figure 5a–b), we found a significant expansion of CD44+ CD62L- PD-1+ Gata3+ effector memory-like CD4+ TIL (cluster 2 and 3), as well as CD44+ Ly6C+ memory-like CD8+ (DeLong et al., 2018; Pihlgren et al., 1996; Walunas et al., 1995) TIL populations (clusters 8–10) in FGL1-KO tumors (Figure 5b–e). In contrast, Treg (cluster 1), NK (cluster 11) or B cells (cluster 14) did not change significantly (Figure 5b–e). Interestingly, a natural killer T (NKT) population (cluster 12) was highly expanded in the FGL1-KO tumors, in comparison with a significant decrease of F4/80+ CD11b+ MHC-II+ CD11cmed tumor-associated macrophages (Clusters 15) (Figure 5c–e). Confirming this data, we also observed a significant increase in the absolute number of leukocytes (CD45+ cells), CD8+, and CD4+ TIL per mg of tumor tissues in mice treated with antiFGL1 or anti-LAG-3 mAbs compared to control treated mice (Figure S4e). Furthermore, there was a significant increase in activation/functional markers, such as CD69, Ly6C, granzyme B (GZB), CD4 and FAS, in CD4+ or CD8+ TIL from anti-FGL1 or anti-LAG3 treated mice (Figure S4f). Our results indicate that silencing the FGL1/LAG3 interaction by either genetic knockout or antibody blockade promotes tumor immunity by stimulating T-cell expansion and activation preferentially in the tumor microenvironment.
(a) Density t-SNE plots of an equal number of CD45+ MC38 tumor-infiltrating leukocytes in WT (n=4) and FGL1-KO (n=5) mice. Size of unsupervised clusters denotes the relative number of cells in that grouping.
(b) t-SNE plot of tumor infiltrating leukocytes overlaid with color-coded clusters.
(c) t-SNE plot of tumor infiltrating leukocytes overlaid with the expression of selected markers.
(d) Frequency of clusters grouped by indicated immune cell subsets. Data were shown as the mean ± SEM. *, p<0.05 by unpaired t-test.
(e) Heatmap displaying normalized marker expression of each immune cluster.
FGL1 is upregulated in human cancers
FGL1 mRNA and protein expression is largely limited to the liver and pancreas of human normal tissues according to the BioGPS tissue microarray database and proteome analysis (Kim et al., 2014) (Figure S5a). Meta-analysis of the Oncomine databases revealed the upregulation of FGL1 mRNA in human solid tumors including lung cancer, prostate cancer, melanoma and colorectal cancer compared to normal tissues, with the highest percentage of upregulation (8/23, or 35%) in lung cancer datasets, while its expression appeared to be downregulated in pancreas, liver, and head and neck cancers (Figure S5b). Furthermore, FGL1 is one of the most upregulated genes in The Cancer Genome Atlas (TCGA) database for lung adenocarcinoma (Figure S5c and Table S2). FGL1 is also significantly upregulated in prostate or breast cancer but is downregulated in liver cancer within this database (Figure S5c).
We established a multiplex quantitative immunofluorescence (QIF) assay to detect FGL1 protein expression on cells and tissues. In addition, a quantitative sandwich ELISA was also established to detect secreted FGL1 in human plasma (see Methods). A 293T line constitutively expressing FGL1 and human tissue samples were utilized to standardize the assay in FFPE preparations and establish the signal detection threshold (Figure S6a). As expected, levels of FGL1 protein were highest in a cell line transfected to express the FGL1 gene (Figure S6a) and human liver (data not shown), but low/undetectable in mock-transfected cells (Figure S6a) or samples from human testis and skeletal muscle (data not shown). We then evaluated the localized expression of FGL1 in 275 non-small cell lung carcinomas (NSCLC) presented in tissue microarray format (cohort #1, from Yale University, also see Table S3) by simultaneous staining of FGL1 and pan-cytokeratin using multiplex quantitative immunofluorescence (QIF) staining. In NSCLC, FGL1 protein was found localized in tumor cells (the pan-cytokeratin-positive) with minimal expression in the stromal compartment (the pan-cytokeratinnegative) (Figure 6a) and no expression in paired normal lung tissues (Figure S6b). Tissue FGL1 levels showed a continuous distribution in this cohort and ~15% of specimens from NSCLC patients showed elevated expression (Figure 6b) which was associated with a significantly decreased 5-year overall survival (Figure 6c). Interestingly, there was no association between FGL1 and B7-H1 (PD-L1) expression levels, but high FGL1 in tumor tissue was significantly associated with high LAG3 levels (Figure S6c). In addition, we also found significantly higher plasma FGL1 levels in NSCLC patients compared to healthy donors in two independent cohorts (also see Table S3): #2 (n=18) from University of Navarra, Pamplona, Spain (Figure 6d) and #3 (n=56) from Fujian Medical University, Fuzhou, China (Figure S6d). Of note, there was no difference in plasma FGL1 levels among NSCLC patients with or without metastasis as well as liver injury (Figure S6e). Furthermore, in cohort #2, we found a positive association of tumor FGL1 QIF scores and plasma FGL1 levels (data not shown). Our findings indicate that FGL1 is upregulated in human cancers, especially in NSCLC.
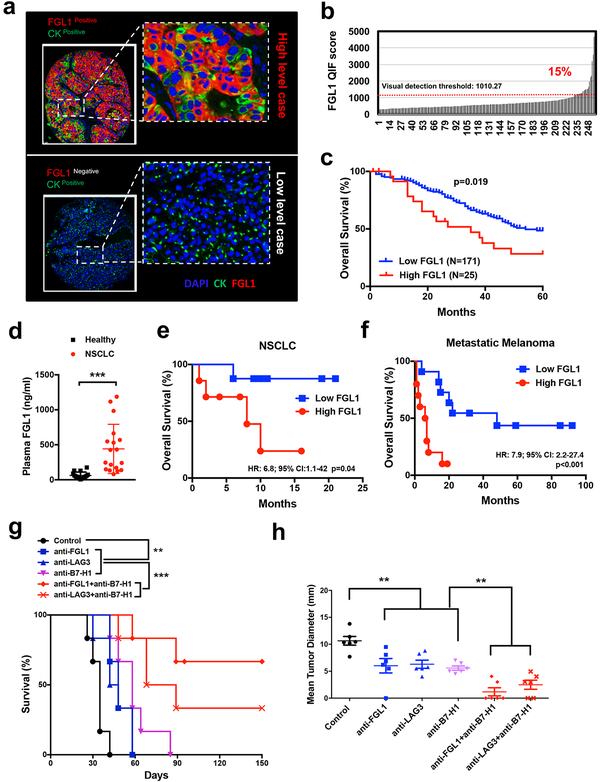
(a) Representative immunofluorescence staining of FGL1, DAPI (for nuclear counterstain), and pancytokeratin (CK) in FGL1 positive or negative NSCLC cancer sections.
(b-c) FGL1 expression as indicated by quantitative immunofluorescence (QIF) staining in NSCLC cancer tissues from cohort #1 (see also Table S3). (b) Distribution of FGL1 expression, (c) association of high or low FGL1 expression with overall survival of the patients. The QIF visual detection threshold (1010.27) was used as a cutoff as indicated by dotted line in (b).
(d) The baseline plasma FGL1 levels were determined by ELISA in cohort #2 (see also Table S3) of NSCLC cancer patients (n=18) and healthy donors (n=16). Data were presented as the mean ± SEM. ***, p<0.001 by Student’s t-test.
(e-f) Kaplan-Meier plots of overall survival stratified by median baseline plasma FGL1 levels in NSCLC (cut-point: 336.5 ng/ml) and melanoma (cut-point: 114 ng/ml) patients treated with single-agent antiPD-1 therapy in NSCLC (d, cohort #2, n=18) and melanoma (e, cohort #4, n=21), see also Table S3 and S4.
(g-h) B6 mice were inoculated s.c. with MC38 cells (0.5×106/mouse) at day 0, followed by the treatment with anti-FGL1, anti-LAG3, or control mAb (n=6 per group) every four days from day 6 to day 18. In some groups, mice were also treated with a single dose of anti-B7-H1 (10B5) at day 6. (g) Survival of the mice is shown. Survival analysis was conducted by Log-rank test, **, p<0.01; ***, p<0.001. The presented data is representative of at least two independent experiments. (h) Tumor sizes are shown as the mean tumor diameter ± SEM at day 22. **, p<0.01 by Student’s t-test.
High plasma FGL1 is associated with poor outcomes in patients with anti-PD therapy
To test if FGL1 acts independently from the B7-H1/PD-1 pathway to suppress tumor immunity, we evaluated the association between the baseline plasma FGL1 levels and the efficacy of the B7H1/PD-1 blockade therapy (anti-PD therapy) in metastatic NSCLC patients. In Cohort #2 (also see Table S3 and S4), we found that higher plasma FGL1 levels were associated with worse overall survival in NSCLC patients treated with anti-PD therapy (hazard ratio (HR)=6.8, 95% confidence interval (CI)=1.1–42 and p-value=0.04) (Figure 6e). Similar results were observed in an independent cohort (Cohort #4, from Yale University, also see Table S3 and S4) of metastatic melanoma patients (n=21) treated with anti-PD-1 mAbs (HR=7.9; 95% CI=2.2–27.4 and p-value <0.001) (Figure 6f). Our results suggest that the FGL1/LAG3 interaction is independent from the B7-H1/PD-1 pathway and could potentially contribute to the resistance of anti-PD therapy in human cancers.
We further tested the role of the anti-FGL1/anti-LAG3 in the presence of the B7-H1/PD-1 pathway blockade using the MC38 tumor model. Mice were inoculated s.c. with MC38, and established tumors at day 6 were treated with the mAbs. When applied individually, anti-FGL1, anti-LAG3 or antiB7-H1 mAb slowed tumor growth and minimally prolonged survival (Figure 6g). However, anti-FGL1 or anti-LAG3 mAb in combination with anti-B7-H1 mAb significantly improved survival (Figure 6g) and decreased tumor burden (Figure 6h) compared to single mAb treatment. A significant proportion of mice (>30% of mice) treated with the combination therapy were free of tumor for over 150 days (Figure 6g). Our results suggest that the FGL1/LAG3 pathway is an independent tumor immune evasion mechanism, and that blockade of this interaction may synergize with anti-PD therapy.
DISCUSSION
In this study, we have identified and characterized FGL1 as a major ligand of LAG3 that is responsible for its T cell inhibitory function in a receptor-ligand interdependent manner both in vitro and in vivo. Genetic ablation or mAbs blocking the FGL1/LAG3 interaction enhanced T cell responses and promoted anti-tumor immunity. With limited expression in the majority of normal tissues, FGL1 is upregulated in several human cancers and is associated with a poor prognosis and therapeutic outcome. Together, our findings support the FGL1/LAG3 pathway as an immune escape mechanism and a potential target for cancer immunotherapy.
Physiological functions of FGL1 are not well understood. Soluble FGL1 protein can be detected in the blood plasma of healthy donors at the nanogram per milliliter (ml) level, while FGL1 mRNA can only be detected in liver and pancreas across a large panel of normal tissues (Figure S5a), suggesting that FGL1 may be produced by the liver and/or pancreas and subsequently released into the bloodstream. In addition to the reported function in hepatocyte regeneration and metabolism, our findings reveal for the first time a prominent role of FGL1 in the negative regulation of inflammatory immune responses. FGL1-KO mice spontaneously developed several autoimmune symptoms including dermatitis and antidsDNA autoantibodies (Figure S3d–f). However, these symptoms developed only in the aged but not in newborn or young adult mice, indicating that endogenous FGL1 is not a major regulator for selftolerance but may suppress environmentally induced inflammation. Interestingly, liver is considered an immune-privileged organ, as allogeneic liver transplants can survive longer without immune suppressive agents (Horst et al., 2016). The underlying mechanism for this immune privilege, however, is largely unknown. We did not observe significant liver inflammation in aged FGL1-KO mice (data not shown), perhaps due to a lack of LAG3 expression on resting T cells in the liver. While FGL1 may play a local role in maintaining the tolerogenic environment of the liver, its secretion as a soluble factor allows for potential cross-talk between the liver and other peripheral tissues that may help fine-tune systemic inflammation. This normal physiological function of FGL1 may be hijacked by several solid tumors that increase FGL1 expression to suppress local anti-tumor immunity. In this context, immune evasion may be mediated by high levels of FGL1 in the tumor microenvironment through the interaction with LAG3 specifically expressed on tumor-infiltrating T cells. Our results in mouse tumor models indicate a preferential activation of T cell immunity in the tumor microenvironment upon FGL1/LAG3 blockade while the effect of this blockade in systemic immune suppression is minimal (Wang et.al, unpublished observation), suggesting a major role for FGL1 in immune suppression of the tumor microenvironment.
Our results support FGL1 as a major ligand for the T cell inhibitory function of LAG3. First, FGL1/LAG3 represents a high affinity interaction that is specific and physiological, as indicated by fusion protein binding experiments involving primary T cells from WT or Lag3 deficient mice. Second, FGL1 mAb has similar effects to anti-LAG3 on the stimulation of T cell responses and antitumor effect in our in vitro and in vivo experiments. Furthermore, anti-FGL1 mAb has no antitumor effect in LAG3 deficient mice while anti-LAG3 mAb likewise loses efficacy in Fgl1 deficient mice. Finally, our preliminary studies indicate that adult Fgl1 deficient mice are also prone to the induction of autoimmune diseases (Wang et al, unpublished data), a phenotype similar to LAG3 deficient mice (Bettini et al., 2011; Woo et al., 2012).
To date, at least four different proteins have been reported to interact with LAG3 including MHCII, galectin-3, LSECtin, and α-synuclein. Galectin-3 and LSECtin have potential roles in T cell regulation, while α-synuclein is possibly involved in the neurological function of LAG3 (Kouo et al., 2015; Mao et al., 2016; Xu et al., 2014). The interaction modality of galectin-3 and LSECtin to LAG3 are less known, but both molecules have previously been shown to have several other binding partners (Kizuka et al., 2015; Li et al., 2009; Liu et al., 2004; Stillman et al., 2006; Tang et al., 2010). It remains to be shown whether their roles in the suppression of T cell-responses and antitumor immunity are dependent on LAG3. Although MHC-II is the first identified ligand for LAG3, the detailed biochemistry and affinity of this interaction is still unclear. Given that FGL1 does not compete with MHC-II for LAG3 binding (Figure S1f), this opens the possibility for the existence of a FGL1/MHC-II/LAG3 trimolecular complex -open questions include the signaling outcome of FGL1 versus MHC-II upon interaction with LAG3 and how this complex could contribute to T cell suppression. A detailed stoichiometry analysis may be required to understand how soluble FGL1 triggers cell surface LAG3 to transmit signals for T cell suppression, which is currently unknown. We found that FGL1 could form oligomers, and these oligomeric forms of FGL1 bound to LAG3 much better than the dimeric form (Figure S1b, c), implicating that oligomeric FGL1 may be required for T cell suppression. The presence of native oligomeric FGL1 may also explain our results in the Octet assay showing a high affinity interaction of purified soluble FGL1 with LAG3. Thus, increased avidity of FGL1, most likely through oligomerization, but potentially through other mechanisms such as attachments to the extracellular matrix, may facilitate its interaction with LAG3 in vivo. Currently, several MHC-II blocking anti-LAG3 mAbs are being evaluated in clinical trials for the treatment of advanced human cancer. Preliminary data of these trials showed minimal or modest effect as a single agent (Ascierto and McArthur, 2017; Ascierto et al., 2017). Based on our findings, a possible interpretation for the clinical results could be that these mAbs block the MHC-II/LAG3 interaction but do not block FGL1/LAG3 binding. Thus, these mAbs may still allow FGL1 to transmit inhibitory signals to LAG3, leading to an incomplete blockade of LAG3-mediated immune suppression. Our findings warrant careful re-evaluation of therapeutic strategies that aim to block the immune inhibitory function of LAG3.
Our findings support that the FGL1/LAG3 pathway maybe an important immune evasion mechanism and could contribute to current cancer immunotherapy efforts for several reasons. Our studies indicate that FGL1 is a major ligand for LAG3 to suppress T-cell responses and constitute a new target for immune modulation. Furthermore, upregulation of FGL1 on tumor cells but not in normal tissues (Figure S5) may allow for a highly tumor-selective targeting of antibody therapy. In addition, tumor model studies using FGL1-KO mice demonstrate that FGL1 has a potent immune suppressive effect on anti-tumor immunity that is dependent on LAG3. Moreover, FGL1 may be a potential biomarker to predict the outcome of anti-PD therapy, since high plasma FGL1 levels are associated with a worse response to anti-PD therapy in NSCLC and melanoma patients (Figure 6e, f). Lastly, FGL1 blockade also synergizes with anti-B7-H1 blockade in animal models (Figure 6g, h), suggesting that FGL1 and anti-PD dual-blockade may be an alternative treatment for patients who are resistant to antiPD therapy. In summary, our findings identify a functional interaction of the LAG3 pathway and reveal a possible mechanism that tumors may employ for immune evasion, with important implications for developing next generation cancer immunotherapies.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Lieping Chen ([email protected]).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Female C57BL/6 (B6) mice, Rag1−/− (herein referred to as Rag1-KO) and OT-1 Rag1-KO mice at 6–8 weeks old were purchased from Charles River (Wilmington, MA) and Taconic Biosciences (Rensselaer, NY), respectively. Fgl1 gene conditional knockout mice were obtained from European Mouse Mutant Archive (EMMA), and homologous targeting of this mouse strain was performed in a genetically engineered B6 ES cell (Agouti JM8A). Fgl1 whole body knockout mice (Fgl1−/−, herein referred to as FGL1-KO) were generated by crossing Fgl1 gene conditional knockout mice with CMV-Cre mice (Jackson Laboratory). The Lag3 whole body knockout mice (Lag3−/−, herein referred to as LAG3-KO) were described previously (Workman and Vignali, 2003). All mouse protocols were in accordance with NIH guidelines and approved by the Animal Care and Use Committee of Yale University School of Medicine.
Plasmids, fusion proteins and antibodies
The human cDNA library constructs coding ~5600 full-length plasma membrane proteins and ~1000 secreted proteins were purchased from Genecopoeia (Rockville, MD), Open Biosystems (Huntsville, AL) or were individually cloned by Chen laboratory at Yale. All of the genes used for the GSRA were cloned into a mammalian expression vector. Human and mouse fusion proteins were generated by tagging the extracellular domain with human Ig Fc or 3xFLAG, expressed in 293T cells, and purified by affinity column, or purchased from R&D systems (Minneapolis, MN). The domain deletion plasmids for FGL1 and LAG3 were produced by PCR or GeneArt synthesis (Thermo Fisher, Waltham, MA). Antibodies used in flow cytometry and in vitro studies were purchased from BD Biosciences (San Jose, CA), BioLegend (San Diego, CA), or Thermo Fisher (Waltham, MA). Mouse CD8 or CD4 in vivo depletion antibodies were purchased from BioXcell (West Lebanon, NH). The hybridoma (clone C9B7W) producing anti-murine LAG3 mAb was described previously (Workman 2002b). The methods to produce and characterize the panels of rat anti-mouse FGL1 and mouse anti-human FGL1 were described previously by immunization with relevant fusion protein (Chen et al., 1992; Dong et al., 2002). The 177m03/177R4 antibodies were also used for flow cytometry and immunohistochemistry. The 177R8 antibody was used for the detection of FGL1 in the mouse liver by western blot.
Human subjects
All four patient cohorts are summarized in Table S3. Cohort #1 (Yale University) included tumor samples in the form of tissue microarrays (TMAs) from 275 NSCLC patients who were not treated with immunotherapy. Cohort #2 (University of Navarra) included plasma samples collected from 18 NSCLC patients who were treated with anti-PD-1 mAbs. Cohort #3 (Fujian Medical University) included plasma samples from 56 NSCLC patients who were not treated with immunotherapy. Cohort #4 (Yale University) included plasma samples from 21 melanoma patients treated with anti-PD-1 mAbs. The treatment characteristics of patients in cohort#2 and #4 are summarized in Table S4 and the plasma samples were collected before anti-PD-1 treatment. The study protocols were approved by the Institutional Review Boards from each participating institution and all patients signed written informed consents. Human peripheral bloods were obtained by venipuncture and centrifuged to isolate plasma that were stored at −80°C.
METHOD DETAILS
The genome-scale receptor array (GSRA) system
The GSRA system was modified from our previous report (Yao et al, 2011). Other than ~5,600 genes encoding cell transmembrane proteins, over 1,000 genes encoding secreted proteins were also included in the GSRA system. The gene selection criteria include the expression profiles and homology with known immune-related gene families (bioinformatics analysis using BioGPS, Immgen, HPM, UCSC genome browser and NCBI databases). The selected genes coding secreted proteins were fused with an artificial transmembrane domain (TM) from FIBCD1 (TM insertion into the N-terminal) or B7-H6 (TM insertion into the C-terminal) to ensure membrane expression. In each plate, cDNA library plasmids were diluted individually in OptiMEM (4ug/ml) before transferring into 384-well plates (VWR) with 200ul plasmid solution in each well. Two uls of plasmid solution from each well were further transferred into 1536-well plates (Greiner) by a robotic liquid handling system (PlateMate, ThermoFisher) and stored at −80°C. For the assay, 2 ul OptiMEM containing Lipofectamine 3000 per well was added to the plates with a Multidrop Combi robotic dispenser (Thermo Fisher), followed by the addition of 2,000 293T.2A cells (a 293T subline overexpressing DAP10, DAP12, FcRγ and CD3Z) in 4 ul DMEM medium per well. After incubation at 37°C for 18 hrs, 2 ng of LAG3-Ig (human or mouse) and 3 ng of anti-Fc secondary antibody were added into each well. These plates were read 8 hrs later in the Applied Biosystems 8200 cellular detection system (CDS) and analyzed by the CDS 8200 software. Human Fc Receptors were served as internal positive controls within each plate.
Protein-protein interaction analysis
Recombinant proteins were purified by HiTrap Protein A column (GE Healthcare) or anti-FLAG M2 affinity gel (Sigma). In some experiments, different protein fractions of size exclusion chromatography (SEC) by SEC650 column (BioRad) were collected, and then performed for a size exclusion chromatography with Multi-Angle Light Scattering analysis (SEC-MALS) for absolute molecular mass and size measurements using Agilent 1200 system (Agilent Technologies) equipped with Superdex 200 column (GE Healthcare). Protein interactions were measured and analyzed by an Octet Red instrument (PALL, NY). The Octet Protein A sensor was dipped into solution containing mouse or human LAG3-Ig fusion protein (10ug/ml) and subsequently loaded with various concentrations of FLAG tagged FGL1 proteins (two-fold serial dilutions) or their different SEC peak fractions (10ug/ml). Protein association and disassociation was monitored and analyzed by Octet software, and the Kd value was calculated using 1:1 binding ratio.
T cell function assays
T cell costimulation assay was described previously (Chapoval et al., 2001; Sica et al., 2003). Briefly, anti-mouse CD3 mAb (eBiosciences) at suboptimal concentrations were pre-coated in 96-well flat plates. Splenocytes from wild-type B6 mice or LAG3-KO mice at 3×105/well were added in the presence of FGL1-Ig or control Ig at 5ug/ml and cultured at 37°C for 3 days. In the last 16 hrs of culture, 3H-TdR was added and counted with a MicroBeta Trilux liquid scintillation counter (PerkinElmer, Waltham, MA). For antigen-specific stimulation, 3A9 T cell hybridoma (with low endogenous LAG3 expression) or 3A9 over-expressing mouse LAG3 gene (3A9-LAG3) was incubated with LK35.2 cell line as antigen-presenting cells in the presence of HEL peptide (1.5ug/ml) and recombinant FGL1 or control protein at the indicated concentrations. Anti-FGL1(177R4) or anti-LAG3 antibodies (5ug/ml) were also included at the beginning of the culture. The IL-2 levels in the supernatant 24 hrs after exposure to antigens were analyzed by CBA kit (BD Pharmingen).
Syngeneic mouse tumor models
Female 8–10-week-old LAG3-KO, FGL1-KO mice or wild-type littermates (WT) were inoculated s.c. with MC38 (5×105/mouse) or Hepa1–6 (1×106/mouse) at day 0 and subsequently treated intraperitoneally (i.p.) with 100 ug of anti-FGL1(177R4) or anti-LAG3 (C9B7W) mAbs on days 6, 10, 14 and 18, with saline or rat-Ig as controls. Anti-B7-H1 (10B5) (Dong et al., 2002; Hirano et al., 2005) at 100ug was injected i.p. at day 6. Tumor growth was monitored by electronic caliper twice a week and shown as the mean tumor diameter: (length + width)/2(Chen et al., 1992; Chen et al., 1994).
Mouse tissue digestion
Mouse tumor tissues were mechanically dissociated with the GentleMACS Dissociator (Miltenyi Biotec) in the presence of RPMI 1640 with 1mg/ml Collagenase IV plus 0.1mg/ml DNase I (Sigma-Aldrich) for 30 min at 37°C. Cell suspension was washed with RPMI containing 10% FBS (GIBCO) and filtered through 70-μm cell strainer (BD Falcon).
Mass cytometry sample preparation
Cells from tumor tissue were re-suspended in PBS with 0.5% BSA and 0.02% NaN3 and incubated with mAb against mouse CD16/32 for 10 min at room temperature to block Fc receptors. Subsequently metal-labeled mAbs cocktail against cell surface molecules were added and further incubated for 20 minutes on ice. Antibodies were either purchased pre-conjugated from Fluidigm or conjugated in-house using mass cytometry antibody conjugation kits (Fluidigm) according to the manufacturer’s instructions. Cells were stained for viability with 5mM cisplatin in FBS (Fluidigm) for 1 minutes on ice. After the treatment with the Fixation/Permeabilization Buffer (Thermo Fisher), cells were further incubated with the metal-labeled mAbs cocktail against intracellular proteins. Cells were then washed and stained with 1 ml of 1:4000 191/193Ir DNA intercalator (Fluidigm) diluted in PBS containing 1.6% Paraformaldehyde (EMS), and stored at 4°C until acquisition.
Mass cytometry sample acquisition
At the moment of acquisition, cells were washed once with PBS with 0.5% BSA and 0.02% NaN3, once with ddH2O, and then re-suspended in ddH2O containing bead standards (Fluidigm) to approximately 106 cells per ml. Samples were subsequently acquired on a CyTOF2 machine (Fluidigm) at an event rate of <400 events/second.
Quantitative multiplex immunofluorescence (QIF)
Simultaneous and quantitative multiplex immunofluorescence protocol was developed as previously described (Schalper et al., 2015) using specific antibodies for indicated antigens carefully titrated and validated individually. Briefly, fresh histology sections from the cases were deparaffinized and subjected to antigen retrieval using EDTA buffer (pH=8.0) and boiled for 20 min at 97°C in a pressure-boiling container (PT module, Lab Vision). Slides were then incubated with dual endogenous peroxidase block (DAKO #S2003, Carpinteria, CA) for 10
min at 97°C in a pressure-boiling container (PT module, Lab Vision). Slides were then incubated with dual endogenous peroxidase block (DAKO #S2003, Carpinteria, CA) for 10 min at room temperature and subsequently with a blocking solution containing 0.3% bovine serum albumin in 0.05% Tween solution for 30 minutes. Slides were stained with 4’, 6-Diamidino-2-Phenylindole (DAPI) for visualization of all cell nuclei, and simultaneously stained with anti-FGL1 (clone 177m03) as well as antibody to pan-cytokeratin (CK, clone AE1/AE3, Abcam). In some experiments, the slides were co-stained with antibodies to LAG-3 (clone 17B4, Abcam) or B7-H1(clone 405–9A11, Cell signaling). Secondary antibodies and fluorescent reagents were later included for the detection of the signal of each marker, including anti-mouse Envision (K4001, DAKO) with Cy5-tyramide (Perkin-Elmer), anti-mouse IgG1 antibody (Abcam) with Cy3 plus (Perkin-Elmer), goat anti-rabbit (Abcam) with biotinylated tyramide/Streptavidine-Alexa750 conjugate (Perkin-Elmer). Residual horseradish peroxidase activity between incubations with secondary antibodies was eliminated by exposing the slides twice for 10 minutes to a solution containing benzoic hydrazide and hydrogen peroxide.
min at room temperature and subsequently with a blocking solution containing 0.3% bovine serum albumin in 0.05% Tween solution for 30 minutes. Slides were stained with 4’, 6-Diamidino-2-Phenylindole (DAPI) for visualization of all cell nuclei, and simultaneously stained with anti-FGL1 (clone 177m03) as well as antibody to pan-cytokeratin (CK, clone AE1/AE3, Abcam). In some experiments, the slides were co-stained with antibodies to LAG-3 (clone 17B4, Abcam) or B7-H1(clone 405–9A11, Cell signaling). Secondary antibodies and fluorescent reagents were later included for the detection of the signal of each marker, including anti-mouse Envision (K4001, DAKO) with Cy5-tyramide (Perkin-Elmer), anti-mouse IgG1 antibody (Abcam) with Cy3 plus (Perkin-Elmer), goat anti-rabbit (Abcam) with biotinylated tyramide/Streptavidine-Alexa750 conjugate (Perkin-Elmer). Residual horseradish peroxidase activity between incubations with secondary antibodies was eliminated by exposing the slides twice for 10 minutes to a solution containing benzoic hydrazide and hydrogen peroxide.
Measurement of plasma FGL1 levels
Two pairs of anti-FGL1 antibodies were used to measure FGL1 levels in the plasma of human (177m01&177m03) or mouse (177R1&177R4) samples by the sandwiched enzyme-linked immunosorbent assay (ELISA). The assay diluent (PBS with 10%FBS) served as negative control.
QUANTIFICATION AND STATISTICAL ANALYSIS
Mass cytometry data analysis
All mass cytometry files were normalized and manually gated in FlowJo (version X 10.0.7r2) or Cytobank (Santa Clara, CA) by DNA, event length, live/dead discrimination, CD45, and 4 bead channels to exclude dead, debris, doublets, and non-immune cells and beads. Data were arcsinh transformed with a cofactor of 5 before applying the downstream analyses. X-shift clustering analysis (Samusik et al., 2016) in Java was performed to pooled samples to automatically identify underlying immune subsets. Heat-maps were generated with selected markers, which had been normalized by dividing the 99% percentile of cluster mean value for each marker. Cell frequency in each cluster was calculated as the assigned cell events dividing the total cell events in the same sample after manual gating. Clusters with less than 1% cell frequency for all samples were not displayed in frequency plot or heat-maps in order to increase the clarity of visualization. t-Distributed Stochastic Neighbor Embedding (t-SNE) dimension reduction was performed using R package (van der Maaten and Hinton, 2008) to WT/FGL1-KO tumors and PBMC datasets. Each sample was randomly sampled for up to 5,000 cells to ensure the total displayed cell events in WT and FGL1-KO scenarios were equivalent. For analysis of antibody treatment data, FCS files were manually pre-gated on Ir193 DNA+CD45+ events, excluding cisplatin+ dead cells, doublets and DNA-negative debris, and analyzed by Cytobank (Santa Clara, CA).
Tissue fluorescence measurement and scoring
Quantitative measurement of the fluorescent signal was performed using the AQUA® method that enables objective and sensitive measurement of targets within user-defined tissue compartments. Briefly, the QIF score of each marker expression was calculated by dividing the corresponding antibody staining pixel intensities by the area of cytokeratin (CK) positive tumor compartment or the DAPI positive total tissue compartment. Scores were normalized to the exposure time and bit depth at which the images were captured, allowing scores collected at different exposure times to be comparable.
Bioinformatics and statistics
The FGL1 expression pattern in normal human or mouse tissues as well as leukocytes were analyzed in BioGPS and HPM databases. The mRNA microarray data on solid cancers and the counterpart normal tissues (254 datasets in total covering 14 types of human cancers) were collected from Oncomine (Thermo Fisher). FGL1 upregulation or downregulation frequency in the datasets of each cancer (fold change ≥ 3 for upregulation, or <0.3 for downregulation, and p-value < 0.05 as cutoff) were further analyzed by the program R. Moreover, FGL1 expression in individual cancer versus counterpart normal tissues was also evaluated using TCGA cancer databases. The original microarray data was normalized by cancer browser (https://genome-cancer.ucsc.edu/) and then analyzed by the program R. Student’s ttest, two-way ANOVA, and Log-rank test on GraphPad Prism 7.0 was used for statistical analysis, and p-values reflect comparison to the control samples. P-values were reported as follows: NS, not significant, *, p<0.05; **, p<0.01; ***, p<0.001, ****, p<0.0001. The error bars in figures represent standard error of the mean (SEM).
DATA AND SOFTWARE AVAILABILITY
The CyTOF FCS files associated with the datasets in this paper have been uploaded in Mendeley database with the following preview link: https://data.mendeley.com/datasets/8gvfpy83jn/draft?a=e1bb6f8d-a1f8-481f-a465-142dabd0f24b
Supplemental Excel Table: Supplemental Table S1. Gene list in the GSRA system, related to Figure 1.
Supplemental Excel Table: Supplemental Table S2. The list of top 200 upregulated genes in the TCGA database. Lung adenocarcinoma versus counterpart normal tissues are presented, related to Figure 6.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Mouse CD145 (clone 30-F11), 141-Pr | Fluidigm | Cat#: 3089005B |
| Anti-Mouse CD11c (clone N418), 142-Nd | Fluidigm | Cat#: 3142003B |
| Anti-Mouse CD69 (clone H1.2F3), 143-Nd | Fluidigm | Cat#: 3143004B |
| Anti-Mouse CD45R (clone RA3–6B2), 144-Nd | Fluidigm | Cat#: 3144011B |
| Anti-Mouse CD4 (clone RM4–5), 145-Nd | Fluidigm | Cat#: 3145002B |
| Anti-Mouse F4/80 (clone BM8), 146-Nd | Fluidigm | Cat#: 3146008B |
| Anti-Mouse FAS (clone SA367H8), 148-Nd | Biolegend | Cat#: 152602; RRID: AB_2629777 |
| Anti-Mouse CD19 (clone 6D5), 149-Sm | Fluidigm | Cat#: 3149002B |
| Anti-Mouse CD25 (clone PC61), 150-Nd | Biolegend | Cat#: 102002; RRID: AB_312851 |
| Anti-Mouse Ly6-G (clone 1A8), 151-Eu | Fluidigm | Cat#: 3151010B |
| Anti-Mouse CD3e (clone 145–2C11), 152-Sm | Fluidigm | Cat#: 3152004B |
| Anti-Mouse CD8a (clone 53–6.7), 153-Eu | Fluidigm | Cat#: 3153012B |
| Anti-Mouse KLRG-1 (clone 2F1), 154-Sm | Thermo Fisher | Cat#: 16–5893–82; RRID: AB_469131 |
| Anti-Mouse CD28 (clone 37.51), 155-Gd | Thermo Fisher | Cat#: 16–0281–81; RRID: AB_468920 |
| Anti-Mouse B7-H1 (clone 10B5), 156-Gd | In-house | |
| Anti-Mouse Foxp3 (clone FJK-16s), 158-Gd | Fluidigm | Cat#: 3158003A |
| Anti-Mouse PD-1 (clone 29F.1A12), 159-Tb | Fluidigm | Cat#: 3159024B |
| Anti-Mouse TBET (clone 4B10), 160-Gd | Fluidigm | Cat#: 3160010B |
| Anti-Mouse Ly6C (clone HK1.4), 162-Dy | Fluidigm | Cat#: 3162014B |
| Anti-Mouse CD62L (clone MEL-14), 164-Nd | Fluidigm | Cat#: 3164003B |
| Anti-Mouse 4–1BB (clone 2A), 166-Er | In-house | |
| Anti-Mouse KI-67 (clone B56), 168-Er | Fluidigm | Cat#: 3168007B |
| Anti-Mouse TIM-3 (clone RMT3–23), 169-Tm | Biolegend | Cat#: 119702; RRID: AB_345376 |
| Anti-Mouse NK1.1 (clone PK136), 170-Er | Fluidigm | Cat#: 3170002B |
| Anti-Mouse CD44 (clone IM7), 171-Yb | Fluidigm | Cat#: 3171003B |
| Anti-Mouse CD11b (clone Ml/70), 172-Yb | Fluidigm | Cat#: 3172012B |
| Anti-Mouse Granzyme B (clone GB11), 173-Yb | Fluidigm | Cat#: 3173006B |
| Anti-Mouse LAG-3 (clone C9B7W), 174-Yb | Fluidigm | Cat#: 3174019B |
| Anti-Mouse CD127 (clone A7R34), 175-Lu | Fluidigm | Cat#: 3175006B |
| Anti-Mouse EOMES (clone Dan11mag), 176-Yb | Thermo Fisher | Cat#: 14–4875–80; RRID: AB_11043546 |
| Anti-Mouse MHC class II (clone M5/114.15.2), 209-Bi | Fluidigm | Cat#: 3209006B |
| Anti-Mouse Pan-cytokeratin (CK, clone AE1/AE3) | Abcam | Cat#: ab27988 |
| Anti-Mouse LAG-3 (clone 17B4) | Abcam | Cat#: ab40466 |
| Anti-Mouse B7H1 (clone 405.9A11) | CST | Cat#: 29122 |
| Anti-mouse CD3, functional grade | BD | Cat#: 553057 |
| Secondary Anti-Mouse Envision | DAKO | Cat#: K4001 |
| Secondary Goat Anti-Rabbit | Abeam | Cat#: ab6721 |
| Secondary Anti-Mouse IgG1 | Abeam | Cat#: ab97240 |
| Anti-mouse FGL1 (clone 177R4) | This paper | N/A |
| Anti-mouse FGL1 (clone 177R1) | This paper | N/A |
| Anti-human FGL1 (clone 177 m03) | This paper | N/A |
| Anti-human FGL1 (clone 177 m01) | This paper | N/A |
| Anti-Mouse LAG3 (clone C9B7W), APC | Biolegend | Cat#: 125210 |
| Anti-FLAG APC | Biolegend | Cat#: 637308 |
| Alexa Fluor® 647 Goat Anti-Human IgG (H+L) | Invitrogen | Cat#: A-21445 |
| Anti-mouse IgG APC | Biolegend | Cat#: 405308 |
| TruStain fcX™ (anti-mouse CD16/32) Antibody | Biolegend | Cat#: 101320 |
| Anti-mouse IgG HRP | Sigma | Cat#: A9044–2ML |
| Bacterial and Virus Strains | ||
| Lenti-virus packaging kit | GeneCopoeia | Cat#: LT001 |
| GeneCopoeia GCI-L3 chemically competent cells | GeneCopoeia | Cat#: STK-300–10 |
| pLenti CMV Puro DEST (w118–1) | Addgene | Cat#: 17452 |
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase from Clostridium Histolyticum, Type IV | Sigma-Aldrich | Cat#: C5138 |
| DNase, Type I | Sigma-Aldrich | Cat#: D4527 |
| Cell-ID Intercalator-Ir—125 μM | Fluidigm | Cat#: 201192A |
| Cell-ID Cisplatin | Fluidigm | Cat#: 201064 |
| 16% Paraformaldehyde Aqueous Solution, EM Grade | EMS | Cat#: 15710 |
| TSA Plus Cy3 System | Perkin Elmer | Cat#: NEL744001KT |
| TSA Cy5-tyramide | Perkin Elmer | Cat#: SAT705A001EA |
| Biotinylated Tyramide | Perkin Elmer | Cat#: SAT700001EA |
| Streptavidin Alexa-750 | Thermo Fisher | Cat # S-21384 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher | Cat # D1306 |
| I-Ak HEL peptide | MBL international | Cat# TS-M708-P |
| OVA 257–264 peptide | GenScript | Cat#: RP10611 |
| Lipofectamine 2000 | Invitrogen | Cat#:11668019 |
| Critical Commercial Assays | ||
| Mass cytometry antibody conjugation kits | Fluidigm | Cat#: 201300 |
| Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent | Thermo Fisher | Cat#: 00–5521–00 |
| EQTM Four Element Calibration Beads | Fluidigm | Cat#: 201078 |
| Permeabilization Buffer (10X) | Thermo Fisher | Cat#: 00–8333–56 |
| BD CBA mouse Inflammation/Th1,2,17 kit | BD Biosciences | Cat#:552364;560485 |
| Mouse anti-dsDNA IgG specific ELISA kit | Alpha diagnostic International | Cat#: 5120 |
| Octet Protein A Biosensor | PALL | https://www.fortebio.com/biosensors-Protein-A.html |
| Deposited Data | ||
| CyTOF data for MC38 tumor from WT/FGL1-KO mice | This paper | Mendeley Data, see methods. |
| CyTOF data for PBMCs from WT/FGL1-KO mice | This paper | Mendeley Data, see methods. |
| Experimental Models: Cell Lines | ||
| 3A9 | ATCC | Cat#: CRL3293 |
| 3A9-LAG3 | Dr. Dario Vignali | Workman et al., 2002b |
| LK35.2 | ATCC | Cat#: HB98 |
| MC-38 | NCI | Chen et al., 1994 |
| Hepal-6 | ATCC | Cat#: CRL1830 |
| 10B5(anti-B7-Hl/PD-Ll) Hybridoma | Chen Lab | Hirano et al., 2005 |
| Recombinant DNA | ||
| cDNA library | See methods | N/A |
| Software and Algorithms | ||
| CyTOF Normalization | Finck et al., 2013 | https://github.com/nolanlab/bead-normalization |
| X-Shift | Amusik et al., 2016 | http://web.stanford.edu/$samusik/vortex/ |
| Octet analysis software | PALL | https://www.fortebio.com/octet-software.html |
| R version 3.2 | N/A | https://www.r-project.org/ |
| FlowJo X 10.0.7r2 | Flowjo, LLC | https://www.flowjo.com/ |
| AQUA (automated quantitative analysis) | HistoRx | https://medicine.yale.edu/pathology/ypts/sts/servicemenu/qifaqua.aspx |
Supplementary Material
1
Figure S1. Characterization of the FGL1/LAG3 interaction, related to Figure 1
(a) Schematic representation of the FGL1 constructs with an artificial transmembrane domain (FGL1TM). The coil-coil domain (CCD) and fibrinogen domain (FD) of FGL1 (23–312aa) was fused with the FIBCD intracellular domain (IC) and transmembrane (TM) (1–57aa). SP, signal peptide; ECD, ectodomain.
(b) The purified FLAG-tagged mouse FGL1 showed three peaks in the SEC650 size exclusion column, the typical oligomeric state (peak1–2) and a dimeric peak 3 (validated by SEC-MALS, ~77.6 kD).
(c) Octet analysis of mouse FGL1/LAG3 interaction. The Octet Protein A sensor was loaded with mouse LAG3-Ig and then tested for binding kinetics with oligomeric (peak 1–2) or dimeric form (peak 3) of FLAG-tagged mouse FGL1 (10ug/ml) as well as the control PBS buffer.
(d) Octet analysis of human FGL1/LAG3 interaction. The Octet Protein A sensor was loaded with human LAG3-Ig and then tested for binding kinetics with FLAG-tagged human FGL1 (starting from 100ug/ml, two-fold serial dilutions).
(e) Schematic representation of Ig tagged full length FGL1, coiled-coil domain (CCD) or fibrinogen domain (FD) truncated FGL1 (left panel), and binding to mouse LAG3+ 293T.2A cells by CDS (right panel). Control Ig was included as a negative control.
(f) The binding of recombinant mouse MHC-II(I-Ab) fusion protein (100ng/ml) to mouse LAG3+ 293T.2A cells was shown in the presence of 10ug/ml FGL1-Ig (orange) or control Ig (blue). Mock transfected 293T.2A cells were also stained as a negative control (red). Data are representative of at least two independent experiments.
2
Figure S2. Anti-FGL1 potentiates antigen-specific T cell responses in vivo, related to Figure 2
(a) Staining of 293T.2A cells with mock (red) or FGL1-TM (blue) transfection by anti-FGL1 mAb (clone 177R4) and analyzed by flow cytometry.
(b) Binding of mouse FLAG-tagged FGL1 (100ng/ml) to mouse LAG3+293T.2A cells in the presence of anti-FGL1, anti-LAG3, or control mAb at the indicated concentrations. Data were acquired and quantified by CDS software and are expressed as the mean ± SEM.
(c-d) Splenocytes from OT-1/Rag1-KO mice were transferred i.v. into B6 mice at 2×106/mouse, followed by the treatment with OVA peptide (500ug/mouse) one day later. Injections of control, antiFGL1, or anti-LAG3 mAb (n=4 per group) were performed at day 1 and day 2. (c) Schematic representation of the experimental design. (d) Plasma cytokine levels at day 4 were measured by CBA. Data are presented as the mean ± SEM. *, p<0.05; NS, not significant by Student’s t-test.
3
Figure S3. Autoimmunity phenotypes in FGL1-KO mice, related to Figure 3
(a) Fgl1 mRNA expression in mouse tissues (left) and hematopoietic cells (embedded) from BioGPS microarray database.
(b) The levels of FGL1 in the plasma of 3-month-old WT or FGL1-KO mice (n=5).
(c) Western blot analysis of FGL1 protein in the liver from wild type littermates (WT) and FGL1-KO mice by anti-FGL1 mAb with β-actin as the internal control.
(d) Lesions of skin dermatitis (left) and the disease frequency (right) observed in 12 to 16-month-old FGL1-KO mice versus WT mice.
(e) Skin tissue H&E staining from representative FGL1-KO and WT mice.
(f) The plasma levels of anti-double-stranded (ds) DNA antibodies in 12 to 16-month-old female or male FGL1-KO and WT mice. Data are presented as the mean ± SEM. *, p<0.05 by Student’s t-test.
4
Figure S4. The efficacy of FGL1 silencing in vivo, related to Figure 4 and and55.
(a-b) B6 mice were inoculated s.c. with Hepa1–6 cells at 1×106/mouse (a), or MC38 cells at 0.5×106/mouse (b) as described in Figure 4c, followed by the treatment with anti-FGL1, anti-LAG3, or control mAb (n=5 per group). Some groups of mice (b) were additionally treated with either anti-CD8 (middle panel) or anti-CD4 (right panel) depleting mAb twice a week from day 1 after tumor inoculation. Tumor growth is shown as mean tumor diameter ± SEM. *, p<0.05; NS, not significant by Two-way ANOVA.
(c-d) FGL1-KO or WT mice were inoculated with MC38 cells at day 0 and euthanized at day 17 as in Figure 5. The weight of tumor or spleen, as well as the total numbers of tumor-draining (TDLN) or nontumor-draining (LN) lympho-node are shown (c). In addition, the percentage of CD45+ leukocytes in the tumor single cell population, and absolute number of CD45+ cells per mg tumor tissue is shown (d). *, p<0.05; NS, not significant by Student’s t test.
(e-f) B6 mice were inoculated s.c. with MC38 cells at 0.5×106/mouse, followed by the treatment with anti-FGL1, anti-LAG3, or control mAb (n=5 per group) as described in Figure 4c. Mice were euthanized at day 18. The absolute numbers of CD45+, CD8+ and CD4+ cells per mg of tumor tissues (e), as well as the activation/functional phenotypes of CD8+ or CD4 tumor-infiltrating T cells (f) are shown. Data are presented as the mean ± SEM. *, p<0.05; ** p<0.01; *** p<0.001 by Student’s t test.
5
Figure S5. FGL1 is upregulated in some human cancers, related to Figure 6
(a) FGL1 expression in human tissues as indicated in BioGPS microarray database (mRNA) and in human proteome map (HPM) mass spectrometry database (embedded).
(b) Frequency of datasets (studies) with FGL1 gene up or downregulation (fold change ≥ 3 or < 0.3, p < 0.05 as cutoff) in human cancers compared to the counterpart normal tissues from the Oncomine database.
(c) FGL1 mRNA expression in the indicated human cancers versus the counterpart normal tissues from TCGA cancer database, see also Table S2. **, p<0.01; ****, p<0.0001 by Student’s t-test.
6
Figure S6. Upregulation of FGL1 protein in NSCLC and its association with disease prognosis, related to Figure 6
(a) Validation of human FGL1 protein expression by immunofluorescence assay with different dilutions of anti-FGL1 antibody on 293T cells transfected with FGL1-TM. Mock-transfected 293T cells were used as negative controls.
(b) Representative immunofluorescence staining of FGL1, DAPI, and cytokeratin (CK) in a NSCLC cancer section and counterpart normal tissue.
(c) Comparison of B7-H1 or LAG3 quantitative immunofluorescence (QIF) scores in FGL1 high or low NSCLC cancer sections (cohort #1, related to Figure 6b, c).
(d) Plasma FGL1 levels in cohort #3 (see also Table 3) of 56 NSCLC patients and 29 healthy donors tested by FGL1 specific ELISA.
(e) Plasma FGL1 levels in cohort #3 NSCLC patients grouped by the status of metastasis or liver injury (as indicated by plasma ALT levels).
Data are presented as the mean ± SEM. **, p<0.01; ***, p<0.001; NS, not significant by Student’s t-test. Survival analysis was conducted by Log-rank test.
8
Table S2. The list of top 200 upregulated genes in the TCGA database. Lung adenocarcinoma versus counterpart normal tissues are presented, related to Figure 6.
9
Table S3. Demographic information of the four patient cohorts, related to Figure 6.
Table S4. Summary of clinical response of the cohort #2 and #4 patients to anti-PD therapy, related to Figure 6.
Acknowledgements
We thank Beth Cadugan for editing the manuscript; Jun Zhao for data analysis, and other members in the laboratories of Yale for helpful discussions and technical assistance, as well as Drs. Tim Zheng, Anne Vogt, Birgit Fogal, Timothy Fenn, Sven Mostboeck, Karl-Heinz Heider, David Blair from Boehringer Ingelheim for their helpful discussion. This research was supported partially by National Institutes of Health grants P30 CA16359, P50 CA196530, a sponsored research funding from Boehringer Ingelheim and an endowment from United Technologies Corporation to L.C. M.F.S. is supported by a Miguel Servet contract from Instituto de Salud Carlos III, Fondo de Investigación Sanitaria (Spain). D.A.A.V. is supported by National Institutes of Health (P01 AI108545) and NCI Comprehensive Cancer Center Support CORE grant (CA047904).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
LC is a consultant/advisory board member and receives consulting fees from Pfizer, Vcanbio, and GenomiCare; is a scientific founder of NextCure and TAYU Biotech and has sponsored research grants from Boehringer Ingelheim and NextCure. There is a patent application pending related to this work.
References
- Anderson AC, Joller N, and Kuchroo VK (2016). Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 44, 989–1004. [Europe PMC free article] [Abstract] [Google Scholar]
- Andrews LP, Marciscano AE, Drake CG, and Vignali DA (2017). LAG3 (CD223) as a cancer immunotherapy target. Immunol Rev 276, 80–96. [Europe PMC free article] [Abstract] [Google Scholar]
- Annunziato F, Manetti R, Cosmi L, Galli G, Heusser CH, Romagnani S, and Maggi E (1997). Opposite role for interleukin-4 and interferon-gamma on CD30 and lymphocyte activation gene-3 (LAG-3) expression by activated naive T cells. Eur J Immunol 27, 2239–2244. [Abstract] [Google Scholar]
- Annunziato F, Manetti R, Tomasevic I, Guidizi MG, Biagiotti R, Gianno V, Germano P, Mavilia C, Maggi E, and Romagnani S (1996). Expression and release of LAG-3-encoded protein by human CD4+ T cells are associated with IFN-gamma production. FASEB J 10, 769–776. [Abstract] [Google Scholar]
- Ascierto PA, and McArthur GA (2017). Checkpoint inhibitors in melanoma and early phase development in solid tumors: what’s the future? J Transl Med 15, 173. [Europe PMC free article] [Abstract] [Google Scholar]
- Ascierto PA, Melero I, Bhatia S, Bono P, Sanborn RE, Lipson EJ, Callahan MK, Gajewski T, Gomez-Roca CA, Hodi FS, et al. (2017). Initial efficacy of anti-lymphocyte activation gene-3 (anti–LAG-3; BMS-986016) in combination with nivolumab (nivo) in pts with melanoma (MEL) previously treated with anti–PD-1/PD-L1 therapy. Journal of Clinical Oncology 35, 9520–9520. [Google Scholar]
- Baixeras E, Huard B, Miossec C, Jitsukawa S, Martin M, Hercend T, Auffray C, Triebel F, and Piatier-Tonneau D (1992). Characterization of the lymphocyte activation gene 3-encoded protein. A new ligand for human leukocyte antigen class II antigens. J Exp Med 176, 327–337. [Europe PMC free article] [Abstract] [Google Scholar]
- Bettini M, Szymczak-Workman AL, Forbes K, Castellaw AH, Selby M, Pan X, Drake CG, Korman AJ, and Vignali DA (2011). Cutting edge: accelerated autoimmune diabetes in the absence of LAG-3. J Immunol 187, 3493–3498. [Europe PMC free article] [Abstract] [Google Scholar]
- Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, and Wherry EJ (2009). Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10, 29–37. [Europe PMC free article] [Abstract] [Google Scholar]
- Bruniquel D, Borie N, Hannier S, and Triebel F (1998). Regulation of expression of the human lymphocyte activation gene-3 (LAG-3) molecule, a ligand for MHC class II. Immunogenetics 48, 116–124. [Abstract] [Google Scholar]
- Camisaschi C, Casati C, Rini F, Perego M, De Filippo A, Triebel F, Parmiani G, Belli F, Rivoltini L, and Castelli C (2010). LAG-3 expression defines a subset of CD4(+)CD25(high)Foxp3(+) regulatory T cells that are expanded at tumor sites. J Immunol 184, 6545–6551. [Abstract] [Google Scholar]
- Cemerski S, Zhao S, Chenard M, Laskey J, Cui L, Shukla R, … De Waal Malefyt R (2015). T cell activation and anti-tumor efficacy of anti-LAG-3 antibodies is independent of LAG-3 – MHCII blocking capacity. Journal for Immunotherapy of Cancer 3(Suppl 2), 183. [Google Scholar]
- Chapoval AI, Ni J, Lau JS, Wilcox RA, Flies DB, Liu D, Dong H, Sica GL, Zhu G, Tamada K, et al. (2001). B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol 2, 269–274. [Abstract] [Google Scholar]
- Chen L, Ashe S, Brady WA, Hellstrom I, Hellstrom KE, Ledbetter JA, McGowan P, and Linsley PS (1992). Costimulation of antitumor immunity by the B7 counterreceptor for the T lymphocyte molecules CD28 and CTLA-4. Cell 71, 1093–1102. [Abstract] [Google Scholar]
- Chen L, McGowan P, Ashe S, Johnston J, Li Y, Hellstrom I, and Hellstrom KE (1994). Tumor immunogenicity determines the effect of B7 costimulation on T cell-mediated tumor immunity. J Exp Med 179, 523–532. [Europe PMC free article] [Abstract] [Google Scholar]
- Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, Nyman J, Marjanovic ND, Kowalczyk MS, Wang C, et al. (2018). Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558, 454–459. [Europe PMC free article] [Abstract] [Google Scholar]
- DeLong JH, Hall AO, Konradt C, Coppock GM, Park J, Harms Pritchard G, and Hunter CA (2018). Cytokine- and TCR-Mediated Regulation of T Cell Expression of Ly6C and Sca-1. J Immunol 200, 1761–1770. [Europe PMC free article] [Abstract] [Google Scholar]
- Demchev V, Malana G, Vangala D, Stoll J, Desai A, Kang HW, Li Y, Nayeb-Hashemi H, Niepel M, Cohen DE, et al. (2013). Targeted deletion of fibrinogen like protein 1 reveals a novel role in energy substrate utilization. PLoS One 8, e58084. [Europe PMC free article] [Abstract] [Google Scholar]
- Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al. (2002). Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8, 793–800. [Abstract] [Google Scholar]
- Gagliani N, Magnani CF, Huber S, Gianolini ME, Pala M, Licona-Limon P, Guo B, Herbert DR, Bulfone A, Trentini F, et al. (2013). Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med 19, 739–746. [Abstract] [Google Scholar]
- Grosso JF, Goldberg MV, Getnet D, Bruno TC, Yen HR, Pyle KJ, Hipkiss E, Vignali DA, Pardoll DM, and Drake CG (2009). Functionally distinct LAG-3 and PD-1 subsets on activated and chronically stimulated CD8 T cells. J Immunol 182, 6659–6669. [Europe PMC free article] [Abstract] [Google Scholar]
- Grosso JF, Kelleher CC, Harris TJ, Maris CH, Hipkiss EL, De Marzo A, Anders R, Netto G, Getnet D, Bruno TC, et al. (2007). LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J Clin Invest 117, 3383–3392. [Europe PMC free article] [Abstract] [Google Scholar]
- Hara H, Yoshimura H, Uchida S, Toyoda Y, Aoki M, Sakai Y, Morimoto S, and Shiokawa K (2001). Molecular cloning and functional expression analysis of a cDNA for human hepassocin, a liverspecific protein with hepatocyte mitogenic activity. Biochim Biophys Acta 1520, 45–53. [Abstract] [Google Scholar]
- Hirano F, Kaneko K, Tamura H, Dong H, Wang S, Ichikawa M, Rietz C, Flies DB, Lau JS, Zhu G, et al. (2005). Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res 65, 1089–1096. [Abstract] [Google Scholar]
- Horst AK, Neumann K, Diehl L, and Tiegs G (2016). Modulation of liver tolerance by conventional and nonconventional antigen-presenting cells and regulatory immune cells. Cell Mol Immunol 13, 277–292. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, Hipkiss EL, Ravi S, Kowalski J, Levitsky HI, et al. (2004). Role of LAG-3 in regulatory T cells. Immunity 21, 503–513. [Abstract] [Google Scholar]
- Huard B, Mastrangeli R, Prigent P, Bruniquel D, Donini S, El-Tayar N, Maigret B, Dreano M, and Triebel F (1997). Characterization of the major histocompatibility complex class II binding site on LAG-3 protein. Proc Natl Acad Sci U S A 94, 5744–5749. [Europe PMC free article] [Abstract] [Google Scholar]
- Huard B, Prigent P, Pages F, Bruniquel D, and Triebel F (1996). T cell major histocompatibility complex class II molecules down-regulate CD4+ T cell clone responses following LAG-3 binding. Eur J Immunol 26, 1180–1186. [Abstract] [Google Scholar]
- Huard B, Prigent P, Tournier M, Bruniquel D, and Triebel F (1995). CD4/major histocompatibility complex class II interaction analyzed with CD4- and lymphocyte activation gene-3 (LAG-3)-Ig fusion proteins. Eur J Immunol 25, 2718–2721. [Abstract] [Google Scholar]
- Huard B, Tournier M, Hercend T, Triebel F, and Faure F (1994). Lymphocyte-activation gene 3/major histocompatibility complex class II interaction modulates the antigenic response of CD4+ T lymphocytes. Eur J Immunol 24, 3216–3221. [Abstract] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. (2014). A draft map of the human proteome. Nature 509, 575–581. [Europe PMC free article] [Abstract] [Google Scholar]
- Kizuka Y, Kitazume S, Sato K, and Taniguchi N (2015). Clec4g (LSECtin) interacts with BACE1 and suppresses Abeta generation. FEBS Lett 589, 1418–1422. [Abstract] [Google Scholar]
- Kouo T, Huang L, Pucsek AB, Cao M, Solt S, Armstrong T, and Jaffee E (2015). Galectin-3 Shapes Antitumor Immune Responses by Suppressing CD8+ T Cells via LAG-3 and Inhibiting Expansion of Plasmacytoid Dendritic Cells. Cancer Immunol Res 3, 412–423. [Europe PMC free article] [Abstract] [Google Scholar]
- Li CY, Cao CZ, Xu WX, Cao MM, Yang F, Dong L, Yu M, Zhan YQ, Gao YB, Li W, et al. (2010). Recombinant human hepassocin stimulates proliferation of hepatocytes in vivo and improves survival in rats with fulminant hepatic failure. Gut 59, 817–826. [Abstract] [Google Scholar]
- Li Y, Hao B, Kuai X, Xing G, Yang J, Chen J, Tang L, Zhang L, and He F (2009). C-type lectin LSECtin interacts with DC-SIGNR and is involved in hepatitis C virus binding. Mol Cell Biochem 327, 183–190. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu W, Tang L, Zhang G, Wei H, Cui Y, Guo L, Gou Z, Chen X, Jiang D, Zhu Y, et al. (2004). Characterization of a novel C-type lectin-like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J Biol Chem 279, 18748–18758. [Abstract] [Google Scholar]
- Liu Z, and Ukomadu C (2008). Fibrinogen-like protein 1, a hepatocyte derived protein is an acute phase reactant. Biochem Biophys Res Commun 365, 729–734. [Europe PMC free article] [Abstract] [Google Scholar]
- Mao X, Ou MT, Karuppagounder SS, Kam TI, Yin X, Xiong Y, Ge P, Umanah GE, Brahmachari S, Shin JH, et al. (2016). Pathological alpha-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 353. [Europe PMC free article] [Abstract] [Google Scholar]
- Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, Eppolito C, Qian F, Lele S, Shrikant P, et al. (2010). Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci U S A 107, 7875–7880. [Europe PMC free article] [Abstract] [Google Scholar]
- Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, and Skarnes WC (2009). Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods 6, 493–495. [Europe PMC free article] [Abstract] [Google Scholar]
- Pihlgren M, Dubois PM, Tomkowiak M, Sjogren T, and Marvel J (1996). Resting memory CD8+ T cells are hyperreactive to antigenic challenge in vitro. J Exp Med 184, 2141–2151. [Europe PMC free article] [Abstract] [Google Scholar]
- Rotte A, Jin JY, and Lemaire V (2018). Mechanistic overview of immune checkpoints to support the rational design of their combinations in cancer immunotherapy. Ann Oncol 29, 71–83. [Abstract] [Google Scholar]
- Samusik N, Good Z, Spitzer MH, Davis KL, and Nolan GP (2016). Automated mapping of phenotype space with single-cell data. Nat Methods 13, 493–496. [Europe PMC free article] [Abstract] [Google Scholar]
- Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, Herbst RS, and Rimm DL (2015). Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 107. [Europe PMC free article] [Abstract] [Google Scholar]
- Shevach EM (2009). Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645. [Abstract] [Google Scholar]
- Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, Chapoval AI, Flies DB, Bajorath J, and Chen L (2003). B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 18, 849–861. [Abstract] [Google Scholar]
- Stillman BN, Hsu DK, Pang M, Brewer CF, Johnson P, Liu FT, and Baum LG (2006). Galectin-3 and galectin-1 bind distinct cell surface glycoprotein receptors to induce T cell death. J Immunol 176, 778–789. [Abstract] [Google Scholar]
- Tang L, Yang J, Tang X, Ying W, Qian X, and He F (2010). The DC-SIGN family member LSECtin is a novel ligand of CD44 on activated T cells. Eur J Immunol 40, 1185–1191. [Abstract] [Google Scholar]
- Triebel F, Jitsukawa S, Baixeras E, Roman-Roman S, Genevee C, Viegas-Pequignot E, and Hercend T (1990). LAG-3, a novel lymphocyte activation gene closely related to CD4. J Exp Med 171, 1393–1405. [Europe PMC free article] [Abstract] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing Data using t-SNE. J Mach Learn Res 9, 2579–2605. [Google Scholar]
- Walunas TL, Bruce DS, Dustin L, Loh DY, and Bluestone JA (1995). Ly-6C is a marker of memory CD8+ T cells. J Immunol 155, 1873–1883. [Abstract] [Google Scholar]
- Williams JB, Horton BL, Zheng Y, Duan Y, Powell JD, and Gajewski TF (2017). The EGR2 targets LAG-3 and 4–1BB describe and regulate dysfunctional antigen-specific CD8+ T cells in the tumor microenvironment. J Exp Med 214, 381–400. [Europe PMC free article] [Abstract] [Google Scholar]
- Woo SR, Turnis ME, Goldberg MV, Bankoti J, Selby M, Nirschl CJ, Bettini ML, Gravano DM, Vogel P, Liu CL, et al. (2012). Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 72, 917–927. [Europe PMC free article] [Abstract] [Google Scholar]
- Workman CJ, Cauley LS, Kim IJ, Blackman MA, Woodland DL, and Vignali DA (2004). Lymphocyte activation gene-3 (CD223) regulates the size of the expanding T cell population following antigen activation in vivo. J Immunol 172, 5450–5455. [Abstract] [Google Scholar]
- Workman CJ, Dugger KJ, and Vignali DA (2002a). Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J Immunol 169, 5392–5395. [Abstract] [Google Scholar]
- Workman CJ, Rice DS, Dugger KJ, Kurschner C, and Vignali DA (2002b). Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3). Eur J Immunol 32, 2255–2263. [Abstract] [Google Scholar]
- Workman CJ, and Vignali DA (2003). The CD4-related molecule, LAG-3 (CD223), regulates the expansion of activated T cells. Eur J Immunol 33, 970–979. [Abstract] [Google Scholar]
- Workman CJ, and Vignali DA (2005). Negative regulation of T cell homeostasis by lymphocyte activation gene-3 (CD223). J Immunol 174, 688–695. [Abstract] [Google Scholar]
- Xu F, Liu J, Liu D, Liu B, Wang M, Hu Z, Du X, Tang L, and He F (2014). LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res 74, 3418–3428. [Abstract] [Google Scholar]
- Yamamoto T, Gotoh M, Sasaki H, Terada M, Kitajima M, and Hirohashi S (1993). Molecular cloning and initial characterization of a novel fibrinogen-related gene, HFREP-1. Biochem Biophys Res Commun 193, 681–687. [Abstract] [Google Scholar]
- Yan J, Ying H, Gu F, He J, Li YL, Liu HM, and Xu YH (2002). Cloning and characterization of a mouse liver-specific gene mfrep-1, up-regulated in liver regeneration. Cell Res 12, 353–361. [Abstract] [Google Scholar]
- Yao S, Zhu Y, Zhu G, Augustine M, Zheng L, Goode DJ, Broadwater M, Ruff W, Flies S, Xu H, et al. (2011). B7-h2 is a costimulatory ligand for CD28 in human. Immunity 34, 729–740. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2018.11.010
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867418315022/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cell.2018.11.010
Article citations
Influencing immunity: role of extracellular vesicles in tumor immune checkpoint dynamics.
Exp Mol Med, 11 Nov 2024
Cited by: 0 articles | PMID: 39528800
Review
Deciphering LAG-3: unveiling molecular mechanisms and clinical advancements.
Biomark Res, 12(1):126, 18 Oct 2024
Cited by: 0 articles | PMID: 39425148 | PMCID: PMC11487938
Review Free full text in Europe PMC
FBXO family genes promotes hepatocellular carcinoma via ubiquitination of p53.
J Cancer Res Clin Oncol, 150(10):458, 14 Oct 2024
Cited by: 0 articles | PMID: 39397119 | PMCID: PMC11471714
HDAC inhibitor SAHA enhances antitumor immunity via the HDAC1/JAK1/FGL1 axis in lung adenocarcinoma.
J Immunother Cancer, 12(10):e010077, 09 Oct 2024
Cited by: 0 articles | PMID: 39384195 | PMCID: PMC11474878
Multiple myeloma and the potential of new checkpoint inhibitors for immunotherapy.
Ther Adv Vaccines Immunother, 12:25151355241288453, 09 Oct 2024
Cited by: 0 articles | PMID: 39399301 | PMCID: PMC11467827
Review Free full text in Europe PMC
Go to all (441) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protocols & materials
Ximbio (4)
 https://ximbio.com/reagent/159685/anti-fibrinogen-like-protein-1-177r1-monoclonal-antibody
https://ximbio.com/reagent/159685/anti-fibrinogen-like-protein-1-177r1-monoclonal-antibody https://ximbio.com/reagent/157963/anti-fibrinogen-like-protein-1-177-m03-monoclonal-antibody
https://ximbio.com/reagent/157963/anti-fibrinogen-like-protein-1-177-m03-monoclonal-antibody https://ximbio.com/reagent/157964/anti-fibrinogen-like-protein-1-177r4-monoclonal-antibody
https://ximbio.com/reagent/157964/anti-fibrinogen-like-protein-1-177r4-monoclonal-antibody https://ximbio.com/reagent/159684/anti-fibrinogen-like-protein-1-177-m01-monoclonal-antibody
https://ximbio.com/reagent/159684/anti-fibrinogen-like-protein-1-177-m01-monoclonal-antibody
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Binding of LAG-3 to stable peptide-MHC class II limits T cell function and suppresses autoimmunity and anti-cancer immunity.
Immunity, 55(5):912-924.e8, 11 Apr 2022
Cited by: 67 articles | PMID: 35413245
Fibrinogen-like protein 1 (FGL1): the next immune checkpoint target.
J Hematol Oncol, 14(1):147, 15 Sep 2021
Cited by: 59 articles | PMID: 34526102 | PMCID: PMC8444356
Review Free full text in Europe PMC
A novel LAG3 neutralizing antibody improves cancer immunotherapy by dual inhibition of MHC-II and FGL1 ligand binding.
Biomed Pharmacother, 175:116782, 21 May 2024
Cited by: 0 articles | PMID: 38776682
LAG3 ectodomain structure reveals functional interfaces for ligand and antibody recognition.
Nat Immunol, 23(7):1031-1041, 27 Jun 2022
Cited by: 24 articles | PMID: 35761082 | PMCID: PMC10191176
Funding
Funders who supported this work.
Boehringer Ingelheim
Instituto de Salud Carlos III, Fondo de Investigación Sanitaria
NCI Comprehensive Cancer Center Support CORE (1)
Grant ID: P30 CA047904
NCI NIH HHS (3)
Grant ID: P30 CA047904
Grant ID: P50 CA196530
Grant ID: P30 CA016359
NIAID NIH HHS (1)
Grant ID: P01 AI108545
NIH (3)
Grant ID: P01 AI108545
Grant ID: P30 CA16359
Grant ID: P50 CA196530





