Abstract
Free full text

Chromosome Segregation Errors Generate a Diverse Spectrum of Simple and Complex Genomic Rearrangements
Abstract
Cancer genomes are frequently characterized by numerical and structural chromosomal abnormalities. Here we integrated a centromere-specific inactivation approach with selection for a conditionally essential gene, a strategy termed ‘CEN-SELECT’, to systematically interrogate the structural landscape of missegregated chromosomes. We show that single-chromosome missegregation into a micronucleus can directly trigger a broad spectrum of genomic rearrangement types. Cytogenetic profiling revealed that missegregated chromosomes exhibit 120-fold higher susceptibility to developing seven major categories of structural aberrations, including translocations, insertions, deletions, and complex reassembly through chromothripsis coupled to classical non-homologous end joining. Whole-genome sequencing of clonally propagated rearrangements identified random patterns of clustered breakpoints with copy-number alterations resulting in interspersed gene deletions and extrachromosomal DNA amplification events. We conclude that individual chromosome segregation errors during mitotic cell division are sufficient to drive extensive structural variations that recapitulate genomic features commonly associated with human disease.
Alterations in chromosome structure are pervasive in human cancers1,2 and define a disease group known as genomic disorders3–5. These abnormalities can comprise a continuum of complexity, ranging from simple arm-level deletions to intricate networks of rearrangements connecting multiple chromosomes6–10. Tumors frequently harbor one or more forms of structural genomic rearrangements, which can underlie disease pathogenesis by altering DNA copy-numbers11, reshuffling linear DNA sequences10,12–15, and/or perturbing regulatory architecture5,16–20. Cancer genome sequencing has recently uncovered complex structural changes that previously escaped detection by conventional cytogenetics, including chromothripsis – localized rearrangements from the catastrophic shattering of individual chromosomes and subsequent re-stitching in haphazard order21.
Chromosomes are constantly under assault from intrinsic and extrinsic sources of DNA double-strand breaks (DSBs)7,22–25, although these challenges are often counteracted by DNA damage repair mechanisms to maintain genomic integrity. Errors in mitotic cell division have emerged as a potent source of endogenous DSBs inflicted onto missegregated chromosomes26. Improper chromosome segregation causes numerical aneuploidy and has recently been linked to structural anamolies. For instance, missegregated chromosomes can generate unbalanced translocations when damaged in the cytokinetic furrow27, acquire arm-level segmental imbalances28,29, or alternatively, become encapsulated into abnormal structures called micronuclei.
Micronuclei act as a spatially restricted compartment for DSB accumulation during interphase30,31 with fragmentation of the micronucleated chromosome during the following mitosis30,32,33. Although the establishment of a genetically heritable, clonally rearranged human chromosome derived from these events has not been experimentally achieved, chromosomes within micronuclei can acquire localized rearrangement junctions in the next cell cycle34 resembling chromothriptic-like signatures from cancer genomes21. Examples of chromothripsis involving multiple chromosomes and in combination with translocations have been identified in somatic21,35–37 and inherited5,38–42 cases, demonstrating that chromothriptic alterations can include events more diverse than rearrangements restricted to an individual chromosome. Despite its prevalence in disease, it remains unknown whether the spectrum of simple and complex genomic rearrangements can be initiated by a unifying mechanism.
We previously established a somatic human cell-based approach to selectively inactivate the Y chromosome centromere by using a gene replacement strategy to rapidly exchange the centromere-specific histone H3 variant CENP-A with a loss-of-function chimeric mutant, which supports ongoing function of all centromeres except for the Y chromosome33. Centromere inactivation caused chromosome-specific missegregation into micronuclei, triggering whole-chromosome shattering during the subsequent mitosis coupled with efficient chromosome loss33. To explore the structural rearrangement landscape of missegregated chromosomes, we now develop CEN-SELECT – a strategy combining centromere inactivation-induced chromosome shattering with a drug selection marker engineered into the Y chromosome, which otherwise lacks somatically essential genes. The resulting chromosomal byproducts are therefore unbiased from constraints typically imposed by autosomal gene-dosage effects. Implementing this approach with comprehensive cytogenetic and whole-genome sequencing (WGS) analyses, we show that mitotic segregation errors directly generate a broad spectrum of chromosomal aberrations that reconstruct the complex structural features of cancer genomes.
RESULTS
Development of the CEN-SELECT system in diploid human cells
To induce mitotic errors and identify cells maintaining the missegregated chromosome, we combined a Y centromere-specific inactivation strategy33 with a Y chromosome-encoded selection marker, an approach we call CEN-SELECT (Fig. 1a). We started with p53-inactivated DLD-1 colorectal cancer cells that are chromosomally stable (46,XY) and carry one disrupted CENPA allele with the second allele modified to encode CENP-A fused to an EYFP-linked auxin-inducible degron (AID) tag (CENP-AEYFP-AID/–)33. Addition of the auxin hormone indole-3-acetic acid (IAA) activates a constitutively expressed E3 ubiquitin ligase (osTIR1) to induce rapid CENP-AEYFP-AID degradation43,44. CRISPR/Cas9-mediated genome editing was used to insert a neomycin-resistance gene (NeoR) (Supplementary Fig. 1a,b), which confers survival in the presence of the antibiotic G418, at the Y-chromosome AZFa locus located ~2.5Mb from the centromere on proximal Yq11.22145. Correctly targeted clones were identified (Supplementary Fig. 1b,c), and a single-copy, doxycycline (DOX)-inducible wild-type CENP-A (CENP-AWT) or chimeric mutant carrying a histone H3 carboxy-terminal tail substitution (CENP-AC–H3) was integrated at a defined FRT site. Replacement of CENP-AEYFP-AID with chimeric CENP-AC–H3 supports functional kinetochore assembly at the centromere of all chromosomes except for the Y centromere33, which lacks CENP-B binding sequences46. Genetic modifications and cell cloning steps are outlined in Supplementary Fig. 1a.
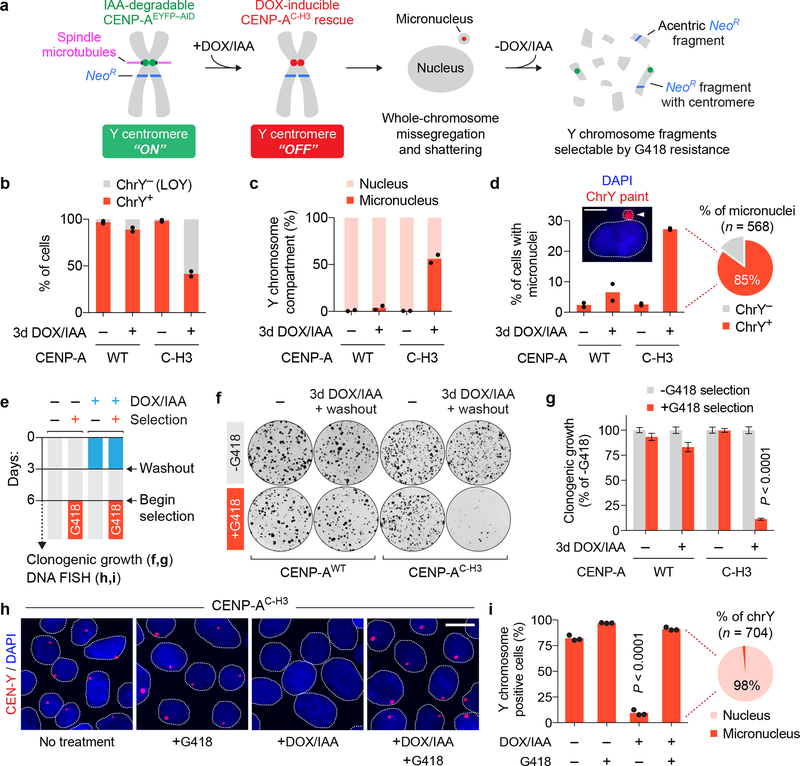
a) Overview of the CEN-SELECT approach, which combines a CENP-A replacement strategy to induce Y chromosome missegregation and micronucleus-mediated shattering with a CRISPR/Cas9-integrated neomycin-resistance gene (NeoR). b-d) Engineered DLD-1 cells carrying a CENP-AWT or CENP-AC–H3 rescue gene were treated with DOX/IAA and quantified by interphase FISH for b) Y chromosome loss (LOY), c) the compartment of the Y chromosome in chrY-positive cells, and d) the proportion of cells with micronuclei. Pie chart shows the fraction of micronuclei carrying a chrY-positive or chrY-negative micronucleus from CENP-AC–H3 rescued cells treated with DOX/IAA. Data represent the mean of n = 2 independent experiments from b,d) 1,081–2,085 cells or c) 861–1,242 Y chromosomes. Scale bar, 5 μm. e) Experimental schematic for panels f-i. f) Representative colony formation plate scans and g) quantification. Bar graph represents the mean ± SEM of n = 9 biological replicates pooled from 3 independent experiments; P-value derived from two-tailed Student’s t-test compared to untreated cells. h) Representative interphase FISH images of CENP-AC–H3 rescue cells and i) quantification. Scale bar, 10 μm. Bar graph represents the mean ± SEM of n = 3 independent experiments from 833–918 cells; P-value derived from two-tailed Student’s t-test compared to untreated cells. Pie chart represents the fraction of Y chromosomes compartmentalized within the nucleus or micronucleus following DOX/IAA treatment and G418 selection.
Over half (59%) of cells underwent expected loss of the Y chromosome (LOY) (Fig. 1b) within three days of centromere inactivation induced by DOX/IAA addition, as determined by interphase fluorescent in situ hybridization (FISH) with Y centromere-targeting DNA probes (CEN-Y). Consistent with increased mitotic errors, the Y chromosome was encapsulated within micronuclei in 56% of cells that retained it – a ~100-fold increase over untreated cells (Fig. 1c,d).
Despite loss of kinetochore function, Y centromere identity is epigenetically preserved by CENP-AC–H3-assembled chromatin33. We next determined whether inactivated Y centromeres could be reactivated following the removal of DOX/IAA. Endogenous CENP-AEYFP-AID returned to its initial steady-state level within 3–4 days of drug washout and accompanied by a corresponding decline in CENP-AC–H3 synthesis (Supplementary Fig. 2a). Reaccumulated CENP-AEYFP-AID incorporated into centromeric chromatin in interphase and mitotic chromosomes, including the Y chromosome (Supplementary Fig. 2b–d), consistent with Y centromere reactivation, and the proportion of cells containing micronuclei approached baseline.
We next used transient Y centromere inactivation coupled with selection for NeoR gene retention (Fig. 1e). While transient replacement of CENP-AEYFP-AID with CENP-AC–H3 alone had no effect on clonogenic growth from single cells, its combination with G418 selection sharply reduced survival by ~89% (Fig. 1f,g). Inspection of G418-resistant populations by FISH revealed that >90% of recovered cells scored positive for retention of the Y chromosome, the vast majority (98%) of which were positioned within the main nucleus (Fig. 1h,i), consistent with centromere reactivation. Additionally, there was no effect on the viability of control cells expressing CENP-AWT treated in parallel (Fig. 1h,i). Thus, CEN-SELECT can experimentally identify cells carrying chromosomes previously exposed to high frequency missegregation into micronuclei.
Release of damaged micronuclear chromatin during mitosis
Micronuclei are intrinsically prone to acquiring DSBs30,31,33. Consistent with this, micronuclei produced by Y centromere inactivation recruited components involved in the DNA damage response, including the MRE11-RAD50–NBS1 (MRN) complex (Supplementary Fig. 3a) that is critical for DSB recognition and signaling. Two ATM kinase substrates, H2AX47 and NBS148,49, were also readily phosphorylated (S139 and S343, respectively) within most micronuclei examined. By contrast, whereas the 53BP1 DNA repair factor50,51 was rapidly recruited to DSB sites induced by chemotherapeutic agents in the major nucleus, micronuclear DSBs failed to actively recruit or retain 53BP1 (Supplementary Fig. 3b), in accordance with previous findings52, suggesting that micronuclear DNA damage developing during interphase can persist unrepaired into the next mitosis.
Live-cell imaging of nuclear and micronuclear membranes fluorescently labeled with lamin A–GFP revealed that 6.4% of micronuclei (n=140, pooled from three experiments) exhibited abnormal nuclear envelope breakdown (NEBD) throughout mitosis (Supplementary Videos 1–2). Micronuclear NEBD kinetics were next assessed by time-lapse microscopy of semi-permeabilized interphase cells treated with mitotic Xenopus egg extracts to trigger NEBD53,54 (Supplementary Fig. 4a–c). Extract-induced nuclear and micronuclear NEBD kinetics were identical in most cells (Supplementary Fig. 4d,e), while in a subset (14%, n=51) the micronuclear envelope only partially disassembled and at reduced rate (Supplementary Fig. 4e–g). Thus, whereas partially-disassembled micronuclei can persist for another cell cycle, most micronuclei enter mitosis with timely NEBD, promoting further chromosome missegregation55 and/or the reincorporation of damaged chromatin into daughter cell nuclei after mitotic exit30.
Mitotic errors drive widespread structural rearrangements
To test whether chromosome segregation errors directly provoke gross chromosomal rearrangements, Y chromosome paint probes were hybridized to metaphase spread preparations. A majority (59±8%, 92/153 metaphases) of cells examined from CEN-SELECT harbored at least one type of Y-specific structural change (Fig. 2a) with different alterations occurring in each cell (described below). Structural rearrangements were undetectable from 94 control metaphases rescued with CENP-AWT and at <1% frequency (1/122 metaphases) in G418-resistant cells without centromere inactivation (Fig. 2a), consistent with a low spontaneous rearrangement rate.
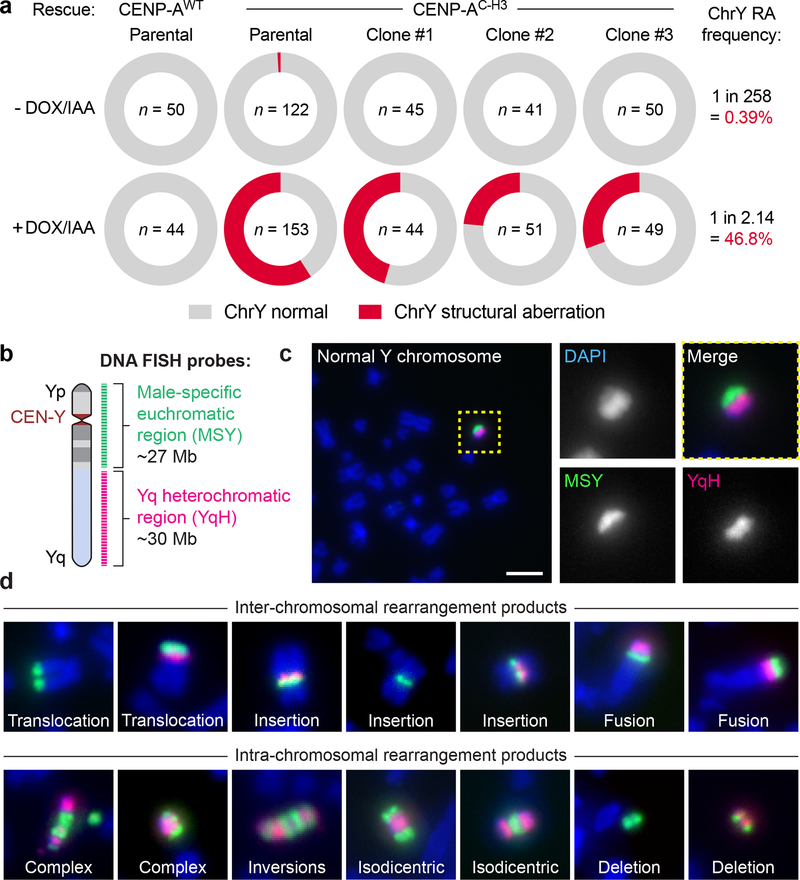
a) Measurements of Y chromosome rearrangement frequencies in parental cells and 3 independent clonal lines following 0d or 3d DOX/IAA treatment and G418 selection. Metaphase spreads were subjected to DNA fluorescence in situ hybridization (FISH) using Y chromosome-specific paint probes. n = number of metaphase spreads examined. Parental CENP-AC–H3 frequencies were pooled from 3 independent experiments. RA, rearrangement. b) Schematic of multi-colored DNA FISH probes used to characterize structural anomolies of the Y chromosome. c-d) Representative FISH images of c) a normal Y chromosome without visible defects from control metaphases (scale bar, 5 μm) and d) examples of derivative Y chromosomes from 3d DOX/IAA-treated, G418-resistant metaphases. See Supplementary Note for a description of each rearrangement type.
To confirm that rearrangements did not result from selection for rare cells with pre-existing abnormalities, three independent clones were isolated that carried a cytogenetically normal Y chromosome (Supplementary Fig. 1a). Each clone underwent efficient LOY following centromere inactivation (Supplementary Fig. 1d). Y chromosome rearrangement frequencies were elevated in each clone following CEN-SELECT, supporting that the majority of rearrangements were generated de novo rather than selection for pre-existing alterations (Fig. 2a). Across the parental cells and clones, CEN-SELECT produced Y chromosome rearrangements at a frequency >2 orders-of-magnitude higher than G418 selection without centromere inactivation (139/297 metaphases versus 1/258 metaphases, respectively) (Fig. 2a). CEN-SELECT did not affect the low rearrangement frequency (1.4%, 2/144 metaphases) of a control autosome (chromosome 14), as determined using chromosome-paint probes, and rearrangements between the Y and chromosome 14 were observed at a frequency (4%, 1/24 Y-translocations) expected by chance (~4.4% in diploid cells). One clone (#1) was selected for subsequent efforts as it was completely haploid for the Y under untreated conditions (n=45 metaphases) and had a Y chromosome rearrangement frequency closely matching the parental clone after CEN-SELECT.
Rearrangements recapitulate a diverse SV landscape
To visualize the types of structural rearrangements generated by CEN-SELECT, we designed a metaphase FISH-based assay utilizing two DNA probe sets that distinctly labeled both halves of the Y chromosome (Fig. 2b). A set of green paint probes hybridized to the euchromatic portion of the male-specific region of the Y (MSY) spanning Yp and proximal Yq56, and another set of red probes (41H10) hybridized to the repetitive, heterochromatic array on distal Yq (YqH). These probe sets produced a characteristic morphology when hybridized to control metaphases, enabling routine discrimination between normal (Fig. 2c) and derivative Y chromosomes.
Cytogenetic inspection of 200 Y chromosomes exhibiting microscopic abnormalities revealed seven major categories of diverse inter- and intra-chromosomal structural alterations (Fig. 2d,,3a),3a), including end-fusions, translocations, insertions, complex rearrangements, inversions (Supplementary Fig. 5), isodicentrics, and deletions (see Supplementary Note for detailed descriptions of each type). Derivative chromosomes consisting of multiple rearrangement types were also observed, exemplified by seven cases of complex aberrations combined with translocations (Fig. 3a).
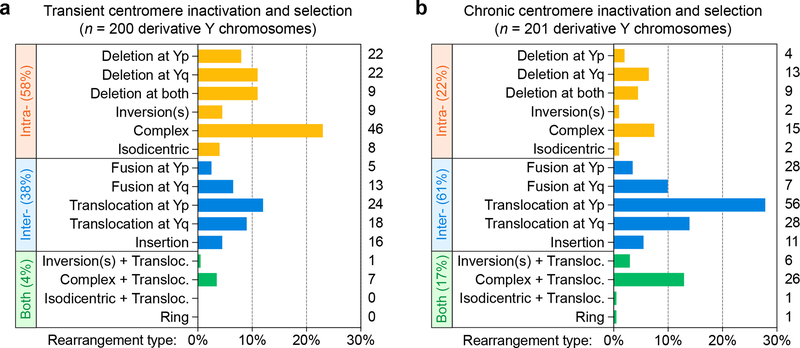
a-b) The distribution of structural rearrangement types quantified from metaphase spreads using MSY/YqH FISH probes following a) transient centromere inactivation induced by 3d DOX/IAA treatment, washout, and G418 selection, or b) prolonged centromere inactivation induced by continuous passage in DOX/IAA and G418 (detailed in Supplementary Fig. 6). The number of each case detected is depicted on the right of each graph.
To determine whether chronic centromere inactivation would influence the spectrum of rearrangement types, cells were continuously passaged with G418 alone or under CEN-SELECT conditions (Supplementary Fig. 6a). Whereas cell-doubling rates remained steady under G418 selection, CEN-SELECT cells displayed reduced growth rates during early passages (Supplementary Fig. 6b,c), reflecting competition between proliferating Y chromosome-positive cells and LOY cells undergoing cell death. Proliferation rate accelerated after p4, approaching baseline growth as Y chromosome-positive cells began outcompeting LOY cells (Supplementary Fig. 6c,d). FISH analysis revealed a passage-dependent reduction in Y chromosome fragmentation with a corresponding enrichment in Y chromosome rearrangements that eventually relocated to the nucleus (Supplementary Fig. 6d,e). At the sixth passage, >97% of metaphases (n=43) harbored 1–3 copies of a normal or rearranged Y chromosome. Cytogenetic profiling of 201 derivative chromosomes obtained from this final passage detected heterogeneous rearrangement types similar to transient CEN-SELECT (Fig. 3a), but with a notable shift towards more inter-chromosomal rearrangements (Fig. 3b). A possible ring chromosome was also detected (1/401 derivative chromosomes), as well as another uncharacterized rearrangement (Supplementary Fig. 6f). Thus, mitotic chromosome segregation errors trigger a wide spectrum of rearrangements that frequently recapitulate cancer-associated structural alterations.
Rearrangement frequency and specificity through NHEJ repair
We next compared rearrangements generated by CEN-SELECT to two established sources of structural aberrations: DNA damage induced by ionizing radiation (IR) (Supplementary Fig. 7a,b) and random chromosome segregation errors (Supplementary Fig. 7c,d) caused by spindle assembly checkpoint (SAC) inactivation27,57. Following the G418 selection procedure, frequencies of de novo structural abnormalities affecting four autosomes or both sex chromosomes were measured from 1,968 metaphases (Supplementary Fig. 7e–g). IR caused an expected dose- and chromosome length-dependent increase in rearrangements (Fig. 4a, Supplementary Fig. 7h), whereas SAC inactivation produced modest levels of structural aberrations that weakly correlated with chromosome size (Fig. 4a). In contrast, CEN-SELECT generated Y chromosome-specific rearrangements at a frequency comparable to rearrangements on a single autosome from sublethal IR doses (Fig. 4a), demonstrating that CEN-SELECT achieves efficient rearrangements specifically on chromosomes undergoing increased missegregation rates.
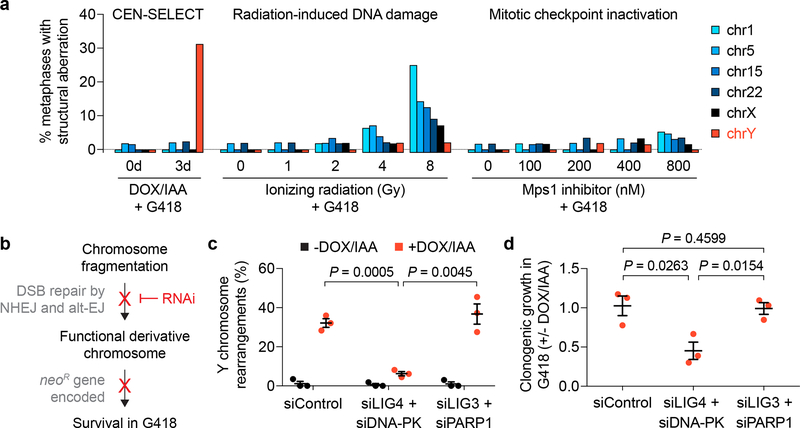
a) DLD-1 cells carrying a CENP-AC–H3 rescue gene were treated as indicated in Supplementary Fig. 7e, followed by metaphase spread preparation and hybridization to the indicated chromosome paint probes. Metaphases were examined for structural rearrangements affecting each chromosome. Bar graph represents n = 42–65 metaphases scored per chromosome per condition, analysing a total of 1,968 metaphase spreads (exact sample sizes provided in Supplementary Fig. 7g). b) Schematic of experimental hypothesis tested in panels c and d. NHEJ, non-homologous end joining; alt-EJ, alternative end joining. c-d) DLD-1 CENP-AC–H3 rescue cells were treated with or without 3d DOX/IAA and transfected with the indicated siRNAs simultaneous with DOX/IAA washout for an additional 3d. Cells were then re-plated into G418 medium for c) 10d selection followed by metaphase FISH using MSY/YqH probes (102–159 metaphase spreads per condition) or d) 14d at single-cell density for colony formation assays. Data in c and d represent the mean ± SEM of n = 3 independent experiments; P-values were derived from two-tailed Student’s t-test comparing groups as indicated.
With the exception of terminal deletions, the observed rearrangement products required at least one DNA joining event between two broken chromosome ends. To test the involvement of the two distinct DSB end-joining pathways [classical non-homologous end joining (NHEJ) and alternative end joining (alt-EJ)], each was transiently suppressed by RNAi-mediated co-depletion of LIG4/DNA-PK or LIG3/PARP1, respectively26 (Fig. 4b). Cytogenetic assessment revealed that inhibition of NHEJ, but not alt-EJ, markedly reduced Y chromosome rearrangement frequencies in the majority of G418-resistant cells (Fig. 4c) – 94% of which harbored a Y chromosome without visible defects (149/159 metaphases). A complementary approach confirmed this, with NHEJ-deficiency conferring clonogenic sensitivity to G418 selection (Fig. 4d), consistent with the failure to reassemble a functional NeoR-encoding Y chromosome. These data support classical NHEJ as the major DSB repair mechanism facilitating rearrangement formation on missegregated chromosomes in human cells.
Sequencing of stable and complex genomic rearrangement events
The structural rearrangement landscape of derivative chromosomes produced by CEN-SELECT was next determined at high-resolution by WGS. Single-cell derived subclones were isolated (Fig. 5a) from sustained CEN-SELECT populations that harbored a rearranged Y chromosome within the main nucleus (Supplementary Fig. 6d–e). Metaphase FISH revealed that 22/40 clones carried distinct, clonal Y chromosome rearrangements (Fig. 5b) spanning the diverse spectrum observed in the bulk population (Fig. 3b). Five clones had inter-chromosomal translocations involving the Y with complex patterns of intra-chromosomal rearrangements exhibiting overlapping FISH signals between the MSY/YqH regions (Fig. 5c). Metaphase FISH determined that >95% of cells within each clone propagated the same derivative chromosome even after long-term continuous passage without G418 (Fig. 5d,e), demonstrating that, in the absence of selective pressure, these chromosomes were stably heritable without further changes.
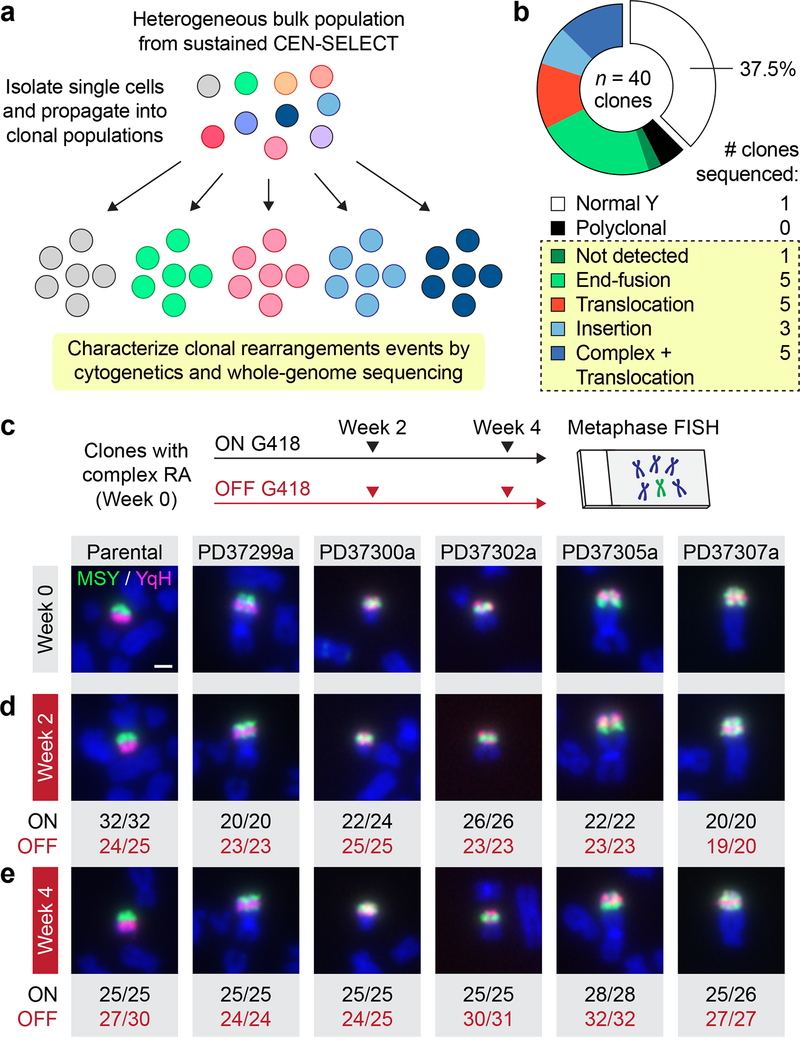
a) Schematic of approach used to generate clonally propagated Y chromosome rearrangements from a bulk cell population. b) Frequency of Y chromosome rearrangement types obtained from single cell-derived clones. The boxed section indicates clonal rearrangements, and the number of clones subjected to whole-genome sequencing is shown on the right. c-d) Experimental schematic and representative metaphase FISH images from the indicated clones, which were passaged in parallel cultures with (ON) or without (OFF) G418 selection for c) 0 weeks, d) 2 weeks, and e) 4 weeks. Scale bar, 2 μm. Values below the image represent the number of metaphases positive for the depicted derivative chromosome over the total number of metaphases examined. RA, rearrangements.
To identify de novo structural variants (SVs) and DNA copy-number alterations, the parental DLD-1 clone and 20 derived clones (Fig. 5b) were subjected to paired-end WGS (35.7x mean depth). A control clone carrying a normal Y chromosome (PD37303a) is shown in Fig. 6a, and a summary of cytogenetics and WGS data for each clone is provided in Supplementary Table 1. The frequency of Y chromosome-specific SVs per Mb sequence was enriched ~21-fold compared to the mean of individual autosomes, and the total number of SVs otherwise scaled by chromosome size (Supplementary Fig. 8a,b). We note that these measurements likely underestimate the number of Y chromosome SVs since rearrangements involving reference assembly gaps, including the repetitive YqH region, were excluded from SV analyses, although such junctions were evident by MSY copy-number changes without an associated SV call. The rearrangement partner was identified by WGS for seven clones – each validated by metaphase FISH (Supplementary Fig. 9).
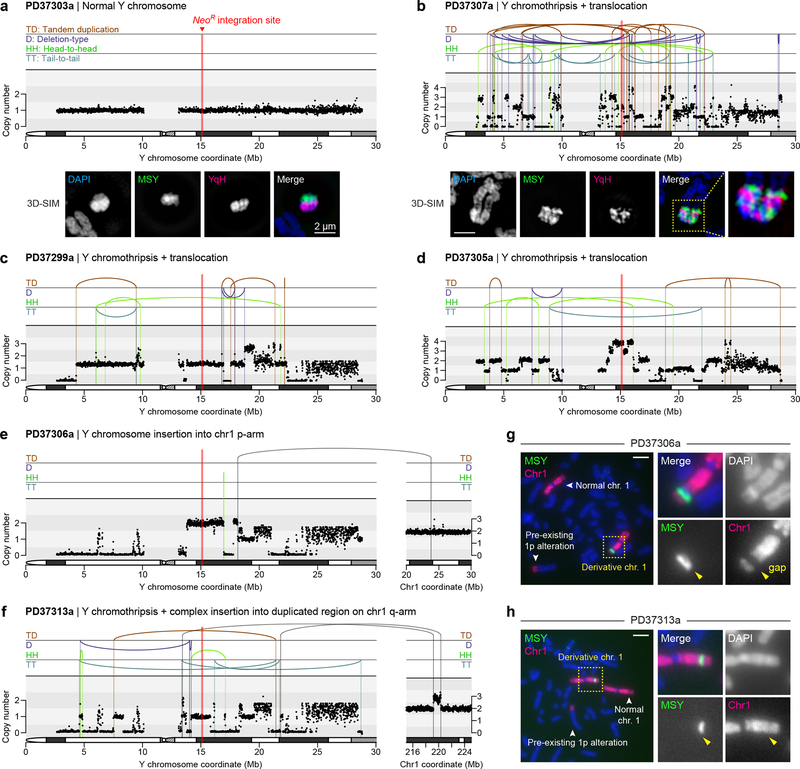
a-b) DNA copy-number profiles from WGS (top) and representative metaphase FISH images hybridized to the indicated probes captured by super-resolution microscopy (bottom) from a) clone PD37303a with a normal Y chromosome and b) clone PD37307a with 83 breakpoints detected across the mappable Y chromosome region. 3D-SIM, 3D structured illumination microscopy. c-f) DNA copy-number profiles of additional clones carrying a chromothriptic Y chromosome coupled to c,d) translocations, e) a simple insertion into chromosome 1p, or f) a complex insertion at a duplicated region on chromosome 1q. X-axes of Y chromosome plots are clipped at 30 Mb to exclude the Yq heterochromatic region. g-h) Metaphase FISH images using MSY and chromosome 1 painting probes on clones g) PD37306a and h) PD37313a. Scale bar, 5 μm.
Reconstructing 71 Y-specific intra-chromosomal rearrangement breakpoints revealed that the majority of junctions lacked microhomology beyond a frequency expected by chance from random simulations (Supplementary Fig. 8c; Online Methods). Among these junctions, seven contained 3bp of microhomology and two had ≥4bp (Supplementary Fig. 8d), consistent with predominant NHEJ-dependent ligation with minimal alt-EJ involvement. Junctions with ≥2bp of non-templated insertions were also observed within 32% of junctions (23/71) with a median length of 13.5bp and maximum length of 94bp (Supplementary Fig. 8d). Junction sequences with microhomology tracts and long insertions are provided in Supplementary Table 2. Additionally, measurements of intra-DSB distance between consecutive breakpoints indicated that a majority (76%, n=228) of chromosome fragments were <1Mb in size with a median length of 300kb (Supplementary Fig. 8e).
Chromothripsis coupled to inter-chromosomal rearrangements
WGS identified 13 clones with intra-chromosomal rearrangements of the Y chromosome, at least nine of which harbored the canonical signatures of chromothripsis (Fig. 6b–d; Supplementary Fig. 10), including oscillating copy-number states and clustered breakpoint patterns randomly distributed across the four possible orientations (Supplementary Fig. 8a). Similar complex rearrangements were not observed on other chromosomes. Super-resolution visualization of metaphase chromosomes revealed that complex rearrangements produced intricately-linked FISH patterns with interspersed euchromatic and heterochromatic segments (Fig. 6b). Copy-number profiling further identified short deletions interspersed along the chromosome, a hallmark of chromosome shattering followed by the loss of DNA fragments, and copy-number increases at multiple loci (Fig. 6b–d; Supplementary Fig. 10), indicative of DNA replication errors and/or co-fragmentation of sister chromatids. Oscillations across three, sometimes more, copy-number levels were also observed, patterns suggesting the simultaneous involvement of both sister chromatids in the chromothriptic event58. Indeed, as previously shown33, centromere-inactivated chromosomes occasionally underwent non-dysjunction during mitosis and entrapped both chromatids into the same micronucleus.
Sequencing of two distinct clones resolved independent simple or complex events involving interstitial insertion of Y chromosome fragments into different chromosome 1 arms (Fig. 6e,f). FISH analysis revealed that one (PD37306a) had distinct separation between Y and chromosome 1p sequences, as indicated by an apparent gap using a chromosome 1 painting probe (Fig. 6g). By contrast, another (PD37313a) carried co-localized chromosome 1 and MSY FISH signals (Fig. 6h). PD37313a was determined by WGS to contain a series of shuffled Y chromosome sequences juxtaposed between a de novo duplicated segment located on chromosome 1q (Fig. 6f), consistent with chromothriptic fragments inserted into a tandem duplication site, which could arise from stalled replication forks59.
Hypermutation hotspots were also observed near five chromothriptic junctions (Supplementary Fig. 8f), indicative of kataegis co-occurring with chromosomal rearrangements60. These regions comprised strictly C>T mutations with an intermutation distance of <1kb. Kataegic events occurred in proximity to Y-Y breakpoints associated with an SV call (130bp–6.5kb) or binned copy-number change (61–109kb) and were otherwise not observed elsewhere throughout the genome. Interphase DNA bridges61 containing Y chromosome sequences were undetectable across all CEN-SELECT passages (Supplementary Fig. 6d), supporting micronucleus-mediated origins underlying these kataegis signatures.
We further identified complex Y chromosome rearrangements from an independent cohort of 3/18 male osteosarcomas interrogated by WGS62, including chromothripsis in a 10-year-old child (PD7193a) (Supplementary Fig. 11a–c). The architecture of these rearrangements resembled those generated by CEN-SELECT, including complex intra- and inter-chromosomal rearrangements accompanied by oscillating copy-number patterns. Similar molecular features were also observed at reconstructed breakpoint junctions (Supplementary Fig. 11d–f). These examples provide proof-of-principle evidence that such rearrangements occur in the context of human cancer, although the degree to which mitotic errors contributes to somatic rearrangements in different cancer types warrants further investigation.
Consequences of chromothriptic DNA copy-number alterations
We next examined how structural rearrangements provoked gene inactivation by mapping gene copy-number states along the Y chromosome. Analysis of 213 well-mapped Y-linked genes and pseudogenes revealed widespread deletions scattered across regions of the initially missegregated chromosome, including five clones with >65% gene loss and eight harboring less frequent (0.5–37%) deletions (Fig. 7a,b). The genomic loci of two Y-encoded genes broadly expressed in somatic cells, KDM5D and EIF1AY, were inspected at greater resolution. Among the 20 sequenced clones, five carried a structural deletion in either gene and three exhibited co-deletions in both (Supplementary Fig. 12a,b). RT-PCR analysis verified that these genes were not expressed in the corresponding clones (Supplementary Fig. 12c) and provided further evidence that these derivative chromosomes were transcriptionally functional beyond NeoR expression. Thus, in contrast to whole-chromosome aneuploidy that alters gene-dosage across the missegregated chromosome, complex rearrangements arising from chromosome missegregation deletes a subset of interspersed genes through copy-number alterations (Fig. 7c).
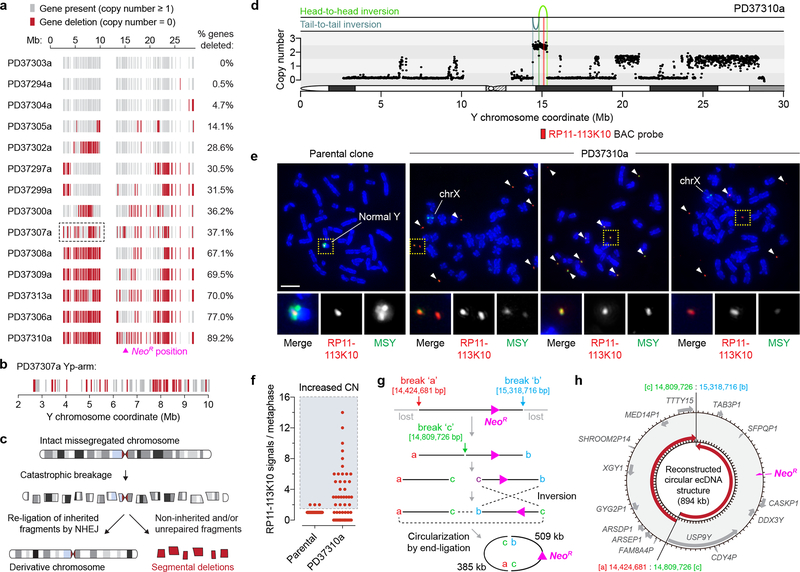
a) Each grey vertical line represents an individual gene or pseudogene depicted at its chromosomal start position, and red lines represent a copy-number of zero. Clones are ranked from fewest to most gene deletions. b) Magnification of clone PD37307a (boxed region in a) exhibiting oscillating patterns of gene retention and deletion within an 8 Mb segment. c) Schematic of chromosome shattering and reassembly events resulting in a derivative chromosome harboring rearrangements with interspersed deletions. d) DNA copy-number profile of clone PD37310a showing extensive Y chromosome loss except for the region harboring the selection marker accompanied by two inversions. e) Images of metaphase spreads prepared from the parental or PD37310a clone and hybridized to MSY (green) and RP11–113K10 BAC (red) probes recognizing the region shown in d. Arrows denote extrachromosomal DNA fragments hybridizing to the RP11–113K10 probe, and regions of the X chromosome partially hybridize to MSY probes due to X-Y sequence homology. Scale bar, 5 μm. f) Quantification of e. Each data point represents an individual metaphase spread derived from the parental clone (n = 48) or PD37310a clone (n = 56). CN, copy-number. g) Schematic depicting the predicted steps leading to the generation of the extrachromosomal DNA (ecDNA) element through the circular reassembly of two broken DNA fragments. h) Reconstructed ecDNA sequence from WGS. Genes and pseudogenes in the corresponding region are shown, and red arrows depict 5’ to 3’ orientation.
Clone PD37310a was resistant to G418 selection despite lacking a cytogenetically-detectable Y chromosome when hybridized to painting probes. WGS revealed that the majority of the Y was lost except for a ~0.9Mb segment surrounding the NeoR marker at Yq11.221 (Fig. 7d). Cytogenetics with FISH probes targeting this locus (RP11–113K10) showed that this region spatially manifested as small fragments of extrachromosomal DNA (ecDNA) (Fig. 7e) reminiscent of cancer-associated double-minute chromosomes63,64. PD37310a cells, but not parental controls, heterogeneously maintained multiple copies of this sequence (Fig. 7f), demonstrating that chromosome shattering induced by mitotic errors could elicit ecDNA production – consistent with observations in chromothriptic tumors21,65. The head-to-head and tail-to-tail orientation of the two rearrangement junctions (Fig. 7d) suggested that the ecDNA structure was produced from three DSBs that generated two fragments (509kb and 385kb in length), followed by an inversion of one fragment and subsequent re-ligation between the four available ends (Fig. 7g,h).
DISCUSSION
Here we show that chromosomal instability can initiate stable and diverse structural rearrangements that span the spectrum of complexity observed in cancers and genomic disorders, expanding the catalogue of possible genomic rearrangement types caused by cell division errors. Missegregated chromosomes are frequently encapsulated into micronuclei that assemble dysfunctional nuclear envelopes31,66. The ensuing loss of nucleocytoplasmic compartmentalization throughout interphase triggers micronucleus-specific DSBs that persist unrepaired into mitosis. These DSBs subsequently resolve into highly fragmented chromatin during mitotic entry30,32,33 that reintegrates into daughter cell genomes after mitotic exit30,34.
Following the completion of mitosis, inherited chromosome fragments are deposited into a functional G1-phase nucleus to initiate the reassembly process by activating the DNA damage response. Because NHEJ is temporally active during early cell cycle phases, we propose that the NHEJ machinery engages these fragments for re-ligation, triggering both intra- and inter-chromosomal rearrangements depending on the availability and/or spatial proximity of compatible DNA ends. We show that inhibiting NHEJ suppresses chromosome rearrangement formation (Fig. 4c), and the breakpoint junctions of re-ligated fragments do not utilize sequence homology beyond that expected from random chance (Supplementary Fig. 8c,d). Microhomology-mediated repair through alt-EJ, however, may be involved in processing remaining DSBs that persist into S-/G2-phases when end-resection is operative67. Whereas recombination-dependent DSB repair preserves sequence integrity at breakpoint junctions, end joining-mediated repair can generate mutagenic indels. Although the mechanisms linking rearrangements with kataegis remain unclear, our data suggest that some breakpoints (Supplementary Fig. 8f) are likely reassembled through an intermediate step consisting of exposed single-stranded DNA processed by APOBEC enzymes68.
Use of CEN-SELECT has generated functional and genetically stable chromosomes that have undergone chromothripsis and which persist over multiple cell cycles even without continued selection. Importantly, apart from the requirement during selection to retain NeoR, the sparse gene content and lack of essential genes on the Y chromosome eliminates constraints on the possible ‘solution space’ of SV configurations that can arise, allowing for an unbiased view of the rearrangement landscape driven by mitotic errors. For instance, complex reassembly events retained multiple segments of Y-sequences despite no selective pressure for its maintenance (Figure 6, Supplementary Figure 10). At a minimum, the reconstitution of a stable derivative chromosome requires the re-stitching of fragments that include an epigenetically-active centromere with two end-capping telomeres. Because the majority of DNA fragments produced from micronucleus-mediated chromosome shattering are acentric33, retention of any essential chromosomal element(s) may be facilitated through rearrangements with a normally segregating, non-homologous chromosome harboring a spontaneous DSB, thereby reacquiring the capacity for its faithful inheritance.
Here we exploited unique features of the Y chromosome to reconstruct the mechanisms and consequences of genomic aberrations69. Although a contribution from underlying experimental factors (Y-specific features, resistance gene position) or genetic background (TP53 status, MMR-deficiency) cannot be formally excluded, we anticipate that the observations reported here are generalizable across chromosomes. We provide evidence that chromothriptic rearrangements can focally delete genes located on missegregated chromosomes (Fig. 7a,b), findings with implications for tumor suppressor gene loss in cancer. Such deletions likely arise through the unequal distribution of DNA fragments during mitosis34 and from inherited fragments that either failed to participate in the chromosome rearrangement event (Fig. 7c) or stochastically reassembled into an unstable configuration. Conversely, copy-number increases can be generated through DNA replication errors (e.g., template-switching), or in more extreme cases, the ligation of fragment ends to produce autonomously-replicating, circular ecDNA structures lacking centromeres and telomeres, which act as a frequent mode of oncogene amplification in cancer63,64. Indeed, the ecDNA carried by clone PD37310a co-amplified 14 additional genes within the vicinity of the selection marker (Fig. 7h). The short fragments generated by chromosome shattering (Supplementary Fig. 8e) may be conducive for ecDNA production to drive gene amplification events associated with tumorigenesis and/or chemotherapeutic-resistance.
Rearrangements arising from chromosome shattering are likely frequent in chromosomally unstable cells exhibiting high rates of chromosome segregation errors, especially those deficient in the p53 checkpoint28,29,65. The majority of random chromosome products formed from these events, however, is probably deleterious or confers no positive advantage. In rarer instances, chromosome shattering may trigger a series of rearrangements in which a ‘perfect storm’ of genetic lesions are created through DNA copy-number alterations, disruptive breakpoints, and/or mutations to enable clonal and oncogenic selective advantages70. Such catastrophic events can operate as a punctuated mutational mechanism to rapidly remodel the cancer genome26,71.
METHODS
Cell culture and reagents
T-REx Flp-In DLD-1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher) containing 10% tetracycline-free fetal bovine serum (FBS) (Omega Scientific) and 100 U/mL penicillin-streptomycin. hTERT-immortalized RPE-1 cells were cultured in DMEM/F12 (Thermo Fisher) supplemented with 10% FBS, 100 U/mL penicillin-streptomycin, 2 mM L-glutamine, and 0.348% sodium bicarbonate. All cells were maintained at 37°C under 5% CO2 and atmospheric oxygen.
Doxycycline (DOX) and indole-3-acetic acid (IAA) were purchased from Sigma, dissolved in cell culture-grade water, and used at 1 μg/mL and 500 μM, respectively. Where indicated, DOX/IAA washout was performed by two washes in PBS followed by addition of growth medium. The Mps1 inhibitor reversine (Santa Cruz) was dissolved in DMSO and used at the indicated concentrations. Geneticin (G418 Sulfate) and hygromycin (Thermo Fisher Scientific) were used at selection concentrations of 300 μg/mL and 200 μg/mL, respectively. DLD-1 siRNA transfections were conducted with Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) using characterized siRNAs (GE Healthcare Dharmacon) as previously described33.
Genome engineering and modifications
T-REx Flp-In DLD-1 cells, which were previously modified to carry a heterozygous CENP-A EYFP-AID fusion tag (CENP-AEYFP-AID/–) and stably express osTIR133, were used as the starting material for all DLD-1 experiments. To integrate a selectable marker into the Y chromosome by CRISPR editing, target sequences for guide RNAs were selected using CRISPR Design Tool (see URLs). An oligo (5’-AACACTTCTCTAGCACGATT-3’) encoding a guide RNA targeting the Y chromosome AZFa locus (target coordinate 12,997,776 using GRCh38 reference assembly) was cloned into the Bbs1 restriction site of the pSpCas9(BB)-2A-GFP vector (PX458, Addgene #48138) and co-transfected into DLD-1 CENP-AEYFP-AID/– cells with a synthesized pUC57-based donor vector containing an EF1α promoter driving the neomycin resistance gene (NeoR) flanked by 450 bp left and right AZFa homology arms (GenScript). Following transfection using X-tremeGENE 9 DNA Transfection Reagent (Sigma-Aldrich), cells were selected with G418 for 20 days and plated by limiting dilution into 96-well plates. G418-resistant, single cell-derived clones were expanded and screened by PCR for targeted insertion of NeoR into the AZFa locus using a forward primer located outside the left homology arm and a reverse primer located within the EF1α promoter. A forward primer sitting inside the left donor homology arm with the same reverse primer were used as a control, as well as primers amplifying the sY1250 sequence-tagged site (STS). All PCR primer sequences are provided in Supplementary Table 3. Clones identified by PCR were confirmed to harbor a single copy of a morphologically normal-appearing Y chromosome using DNA FISH on metaphase spreads.
Following Y chromosome editing, CENP-A WT or C–H3 rescue constructs were integrated into the Flp-In locus, as previously described33. Briefly, cells were co-transfected with pOG44 and pcDNA5 constructs carrying the indicated CENP-A rescue gene using X-tremeGENE 9 DNA Transfection Reagent (Sigma-Aldrich), and successfully targeted cells with correct integration were selected with hygromycin.
To generate cells with fluorescently-labeled nuclear envelopes, DLD-1 and RPE-1 H2B–mRFP cells were transduced with retroviruses packaged in 293GP cells to stably express pBABE-puro-GFP-wt-lamin A (Addgene #17662). Fluorescent cells were isolated by flow cytometry on a BD FACSAria II Cell Sorter (UCSD Stem Cell Core).
Cell growth measurements
For colony formation assays, trypsinized cells were pelleted, diluted as needed, and seeded at low density into 10-cm2 or 6-well plates for up to two weeks. Cells were fed with fresh growth medium each week. Cells were fixed with 100% methanol at room temperature, incubated in 0.5% crystal violet, 25% methanol solution with gentle agitation for 30 minutes, and washed with water. To account for expected ~10-fold differences in clonogenic growth in Fig. 4d, all DOX/IAA-treated conditions were seeded at a 10-fold higher cell density than control non-DOX-IAA-treated cells to yield a comparable number of colonies at time of fixation. For quantification of cell proliferation rates, 1×106 cells were seeded into 10-cm2 plates and total cell numbers were counted using a hemacytometer every 5 days. 1×106 cells were re-seeded into new 10-cm2 plates and repeated for up to 30 days.
Nuclear envelope disassembly assay using Xenopus egg extracts
Interphase and CSF-arrested extracts were prepared from Xenopus laevis eggs as described72. Crude extracts were supplemented with 250 mM sucrose and aliquots were snap frozen and stored at −80°C. All animal protocols complied with ethical regulations set forth and approved by UC San Diego and the Institutional Animal Care and Use Committee (IACUC). To generate RPE-1 cells with micronuclei by nocodazole washout, RPE-1 H2B–mRFP and lamin A–GFP cells were arrested in mitosis for 6 hours with 100 ng/mL nocodazole (Sigma), harvested by mitotic shake-off, and released into fresh growth medium overnight in 4-well chamberslides. For nuclear envelope disassembly assays, a slightly modified protocol adapted from Mühlhäusser et al.54 and Shankaran et al.73 were closely followed. Briefly, chamberslides were incubated on ice for 5 minutes and washed with cold permeabilization buffer (20 mM Hepes, pH 7.4, 110 mM KoAc, 5 mM MgOAc, 0.5 mM EGTA, 250 mM sucrose) for 5 minutes. Cells were then incubated on ice for 5 minutes in permeabilization buffer containing 40 μg/mL digitonin (Promega) while frozen egg extracts were allowed to thaw on ice. Cells were washed three times with cold permeabilization buffer for 5 minutes each. 500 μL of thawed egg extracts were added per chamber and cells were imaged on a DeltaVision RT (GE Healthcare) microscope system using a 60x objective with 20 × 0.2-μm z-sections at 4–6 minute intervals. Time-lapse images were deconvolved and maximum intensity quick projections videos were generated using softWoRx (Applied Precision) program. GFP fluorescent intensities were measured using ImageJ.
Metaphase spread preparation
Subconfluent cell cultures were incubated with 100 ng/mL KaryoMAX (Thermo Fisher Scientific) diluted in fresh growth medium for 4 hours. Cells were trypsinized, pelleted, and slowly resuspended in 0.075 M KCl solution dropwise with gentle vortexing, followed by incubation at 37°C for 6 minutes and addition of 1:6 total volume of pre-chilled Carnoy’s fixative (3:1 methanol:acetic acid). Cells were then pelleted, resuspended in pre-chilled Carnoy’s fixative, and centrifuged. Cells were resuspended in pre-chilled Carnoy’s fixative and stored at −20°C. To prepare metaphase spreads, 10 μL of fixed cell suspensions were dropped onto glass slides and air-dried.
Interphase and metaphase DNA fluorescence in situ hybridization (FISH)
Chromosome paint and centromere enumeration probes were purchased from MetaSystems. Yq heterochromatin (YqH, 41H10) and RP11–113K10 BAC probes were obtained from BACPAC Resources Program, CHORI (see URLs). 41H10 BAC DNA was isolated from 200 mL bacterial cultures using the Plasmid Midi Kit (Qiagen) and labeled with Cy3 label using nick-translation protocol for DNA Polymerase I/Dnase I (Life Technologies). RP11–113K10 BAC DNA was isolated from 50 mL bacterial cultures using the BACMAX DNA Purification Kit (Epicentre), sonicated (Branson Ultrasonics Sonifier S-450), and labeled with TM-rhodamine using a Label IT Nucleic Acid Labeling Kit (Mirus Bio). Labeled BAC probes were resuspended in FITC-labeled Y chromosome paint probes (Metasystems).
To perform FISH on interphase cells, unsynchronized cells cultured in chamberslides were washed with PBS and fixed with Carnoy’s fixative for 15 minutes at room temperature. Slides were dehydrated in 80% ethanol and air-dried.
FISH probes were combined as indicated, applied to interphase cells or metaphase spreads on glass slides, and sealed with a coverslip. Samples and probes were co-denatured at 75°C for 2 minutes, followed by sealing with rubber cement and overnight hybridization at 37°C in a humidified chamber. Slides were washed in 0.4x SSC at 72°C for 2 minutes and in 2x SSC, 0.05% Tween-20 at room temperature for 30 seconds. Slides were rinsed in water, stained with DAPI, and mounted in ProLong Gold anti-fade mounting solution.
For quantification of chromosomal rearrangement frequencies, randomly selected metaphases were imaged, manually inspected using the indicated DNA FISH probes, and scored for de novo structural chromosomal abnormalities when compared to the parental DLD-1 karyotype.
Immunofluorescence and immuno-FISH
For immunofluorescence, cells seeded on coverslips or chamberslides were fixed with 4% formaldehyde diluted in PBS for 10 minutes and washed with PBS. Cells were then permeabilized with 0.3% Triton X-100, PBS for 5 minutes, washed with PBS, and blocked with Triton Block (0.1% Triton X-100, 2.5% FBS, 0.2 M glycine, PBS). Primary antibodies were diluted in Triton Block and applied to cells for 1 hour at room temperature or overnight at 4°C, followed by three 10-minute washes with 0.1% Triton X-100, PBS. The following primary antibodies were used at 1:1,000 dilution in Triton Block: CENP-A (ab13939, Abcam), GFP (A10262, Invitrogen), phospho-S139 γH2AX (clone JBW301, 05–636, Millipore), and 53BP1 (NB100–304, Novus Biologicals). The following primary antibodies were obtained from GeneTex and used at 1:500 dilution in Triton Block: MRE11 (GTX70212), RAD50 (GTX70228), NBS1 (GTX70222), and phospho-S343 NBS1 (GTX61779). Fluorescence-conjugated secondary antibodies (Thermo Fisher Scientific) were diluted 1:500 in Triton Block and applied to cells for 1 hour at room temperature, followed by three 10-minute washes with 0.1% Triton X-100, PBS and staining with DAPI. Cells were mounted in ProLong Gold anti-fade mounting solution and imaged as described.
For immunofluorescence combined with DNA FISH, the immunofluorescence procedure was performed as described followed by fixation in Carnoy’s fixative for 10 minutes at room temperature and dehydration in 80% ethanol for 5 minutes. Samples were then processed for FISH as described.
Fixed and live-cell microscopy
Metaphase FISH images were captured on a DeltaVision Elite (GE Healthcare) microscope system with a 60x objective and 5 × 0.5-μm z-sections. Maximum intensity quick projections were generated using softWoRx. Immunofluorescence images were captured at 30 × 0.2-μm z-sections, followed by deconvolution and maximum intensity quick projections using softWoRx. Structured illumination microscopy imaging was performed on metaphase spreads using a DeltaVision OMX SR (GE Healthcare) microscope system with a 100x objective (Olympus PlanApo 1.4 NA) and EMCCD 10 Mhz camera mode. Raw images were reconstructed and aligned for super-resolution using softWoRx with the following Wiener filters: 525 nm, 0.020; 442 nm, 0.020; 600 nm, 0.015.
For live-cell imaging experiments, 1×104 cells/well were seeded into CELLSTAR μClear 96-well plates (Greiner Bio-One) and imaged on a Yokogawa CQ1 confocal image cytometer with a 40x objective. To determine mitotic timing and the formation of micronuclei with mitotic checkpoint inactivation, cells were seeded 16 hours prior to staining with 500 nM SiR-DNA (Spirochrome) and treatment with reversine (Santa Cruz) at the indicated concentrations, followed by image acquisition at 5-minute intervals with 5 × 2-μm z-sections per field. To monitor nuclear envelope breakdown in cells stably expressing lamin A–GFP, micronuclei were induced by 3d DOX/IAA treatment, followed by image acquisition at 5- or 6-minute intervals with 6 × 1.5-μm z-sections per field. Mean intensity projections were created using ImageJ.
Immunoblotting
Whole-cell extracts were collected in SDS sample buffer and boiled for 10 minutes. Samples were resolved by SDS-PAGE, transferred to PVDF, and blocked with 5% milk in PBST (PBS, 0.1% Tween-20). Primary antibodies against CENP-A (2186, Cell Signaling) and GAPDH (14C10, Cell Signaling) were diluted 1:1,000 and 1:2,000, respectively, in PBST followed by incubation and three 10-minute washes in PBST. Membranes were incubated with HRP-conjugated secondary antibodies diluted 1:4,000 in 5% milk in PBST followed by three 10-minute washes in PBST and exposure to film.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted using a NucleoSpin RNA kit (Machery-Nagel) according to manufacturers’ instructions. cDNA libraries were created from 4 μg of total RNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with random primers. PCR was conducted using Q5 High-Fidelity polymerase (NEB) with the following reaction conditions: 98°C for 3 minutes, 30 cycles of 98°C for 1 minute, 63°C for 1 minute, and 72°C for 1 minute, followed by 72°C extension for 5 minutes. PCR products were resolved by electrophoresis on a 2% agarose gel and visualized with ethidium bromide. All RT-PCR primer sequences are provided in Supplementary Table 3.
Next-generation DNA sequencing and mapping
Genomic DNA was prepared using a Quick-DNA Plus Kit (Zymo) according to manufacturers’ instructions. Sequencing libraries were generated from genomic DNA on robots according to manufacturers’ protocols. DNA was sequenced on an Illumina HiSeq X Ten platform to a targeted whole human genome coverage of 30x. Cluster generation and sequencing were performed using the manufacturers’ pipelines. Average sequence coverage achieved across all samples was 35.7x (range, 31.4–39.8x) with 151 bp paired-ends and insert size of ~500 bp. Mapping to the human genome was performed using the BWA algorithm74 BWA-MEM (Version 0.7.8) and genome build hs37d5.
Copy-number and structural variant calling
Copy-number was called using the ascatNgs algorithm75. The variant calling pipeline of the Cancer Genome Project, Wellcome Sanger Institute was used to call somatic mutations76. The following algorithms with standard settings and no additional post-processing were used: CaVEMan for substitutions and BRASS for rearrangements.
We note that clones harboring Y chromosome end-to-end fusions without additional rearrangements identified by cytogenetics proved uninformative due to the sequence junctions spanning repetitive telomeric sequences. We also note that X-Y translocation calls arose in three clones due to mapping errors, most likely caused by sequence similarities between regions of the X and Y chromosomes. All of these calls mapped to the distal end of Xp, which were confirmed by FISH to be false-positive signals and subsequently removed from the reported sequencing data.
Simulation of random microhomology at breaksites
To simulate the expected frequency of microhomology lengths at breaksites, we used a randomization approach to mimic the sequence context at the actual breaksites identified in our samples. The set of all rearrangement calls were replicated 100x and a random number within the range of −5,000 and 5,000 was added to each breaksite position. The length of homology at the junctions of these simulated breaksite sequences were then measured.
Statistical analysis and reproducibility
Statistical analyses were performed using GraphPad Prism 7.0 software with the tests as described in the figure legends; all P-values were derived from experiments repeated independently at least three times. Error bars represent SEM of at least three independent experiments unless indicated. Figures with representative images were reproduced and obtained from two or more independent experiments.
Reporting summary
Further information is available in the Life Sciences Reporting Summary linked to this article.
Accession Codes
Whole-genome sequencing data have been deposited at the European Genome-phenome Archive (EGA) under accession number EGAD00001004163.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request. Whole-genome sequencing data have been deposited at the European Genome-phenome Archive (EGA) under accession number EGAD00001004163.
URLs
CRISPR Design Tool, http://crispr.mit.edu; BACPAC Resources Program, CHORI, https://bacpacresources.org
Supplementary Material
1
2
3
4
5
6
7
ACKNOWLEDGEMENTS
We thank H. Skaletsky for advice on targeting the Y chromosome, P. Rao and T. Senaratne for assistance with initial cytogenetic experiments, P. Mischel for helpful discussions, J. Santini and M. Erb for assistance with super-resolution imaging, the UCSD School of Medicine Microscopy Core (P30 NS047101) and A. Shiau for shared use of equipment, and the UCSD Animal Care Program for irradiator use. This work was supported by the US National Institutes of Health (K99 CA218871 to P.L. and R35 GM122476 to D.W.C.), Wellcome Trust (WT088340MA to P.J.C. and 110104/Z/15/Z to S.B.), Hope Funds for Cancer Research (HFCR-14-06-06 to P.L.), Swiss National Science Foundation (P2SKP3–171753 and P400PB-180790 to S.F.B.), St. Baldricks Foundation (Robert J. Arceci Innovation Award to S.B.), NIHR UCLH Biomedical Research Centre, UCL Experimental Cancer Centre, and RNOH NHS Trust (to A.M.F., who is a NIHR Senior Investigator), and Howard Hughes Medical Institute (to D.C.P.) D.W.C. receives salary support from the Ludwig Institute for Cancer Research.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Editorial Summary:
Cytogenetic and whole-genome sequencing analyses using CEN-SELECT show that mitotic segregation errors generate a broad spectrum of chromosomal aberrations that recapitulate the complex structural features of cancer genomes.
REFERENCES
METHODS-ONLY REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1038/s41588-019-0360-8
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc6441390?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/s41588-019-0360-8
Article citations
High prevalence of chromosome 17 in breast cancer micronuclei: a means to get rid of tumor suppressors?
Hum Cell, 38(1):5, 22 Oct 2024
Cited by: 0 articles | PMID: 39438374
Chromothripsis: an emerging crossroad from aberrant mitosis to therapeutic opportunities.
J Mol Cell Biol, 16(4):mjae016, 01 Sep 2024
Cited by: 0 articles | PMID: 38710586 | PMCID: PMC11487160
Review Free full text in Europe PMC
The Fanconi anemia pathway induces chromothripsis and ecDNA-driven cancer drug resistance.
Cell, 187(21):6055-6070.e22, 23 Aug 2024
Cited by: 0 articles | PMID: 39181133
Chromosomal instability as a driver of cancer progression.
Nat Rev Genet, 29 Jul 2024
Cited by: 3 articles | PMID: 39075192
Review
Patterns of Aneuploidy and Signaling Consequences in Cancer.
Cancer Res, 84(16):2575-2587, 01 Aug 2024
Cited by: 1 article | PMID: 38924459 | PMCID: PMC11325152
Review Free full text in Europe PMC
Go to all (104) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
European Genome-Phenome Archive
- (2 citations) European Genome-Phenome Archive - EGAD00001004163
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Chromothripsis from DNA damage in micronuclei.
Nature, 522(7555):179-184, 27 May 2015
Cited by: 615 articles | PMID: 26017310 | PMCID: PMC4742237
Processes shaping cancer genomes - From mitotic defects to chromosomal rearrangements.
DNA Repair (Amst), 107:103207, 10 Aug 2021
Cited by: 3 articles | PMID: 34425515
Review
Deciphering the complexity of simple chromosomal insertions by genome sequencing.
Hum Genet, 140(2):361-380, 29 Jul 2020
Cited by: 12 articles | PMID: 32728808
Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements.
Nat Med, 18(11):1630-1638, 07 Nov 2012
Cited by: 157 articles | PMID: 23135524 | PMCID: PMC3616639
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R00 CA218871
Grant ID: K99 CA218871
NIGMS NIH HHS (1)
Grant ID: R35 GM122476
NINDS NIH HHS (1)
Grant ID: P30 NS047101
National Institute for Health Research (NIHR) (1)
Grant ID: NF-SI-0616-10112
Swiss National Science Foundation (1)
Grant ID: 180790
Wellcome Trust (3)
Grant ID: 110104/Z/15/Z
Grant ID: WT088340MA
Grant ID: 103858/Z/14/Z





