Abstract
Free full text

A Nationwide Screen of Carbapenem-Resistant Klebsiella pneumoniae Reveals an Isolate with Enhanced Virulence and Clinically Undetected Colistin Heteroresistance
Associated Data
ABSTRACT
The convergence of hypervirulence and multidrug resistance in Klebsiella pneumoniae is a significant concern. Here, we report the first screen for hypermucoviscosity, a trait associated with increased virulence, using a U.S. surveillance collection of carbapenem-resistant (CR) K. pneumoniae isolates. We identified one hypermucoviscous isolate, which carried a gene encoding the KPC-3 carbapenemase, among numerous resistance genes. The strain further exhibited colistin heteroresistance undetected by diagnostics. This convergence of diverse resistance mechanisms and increased virulence underscores the need for enhanced K. pneumoniae surveillance.
INTRODUCTION
Several recent reports have described carbapenem-resistant (CR), hypervirulent Klebsiella pneumoniae strains in China (1,–4). Alarmingly, strains described by Gu et al. harbored plasmids containing virulence genes, including those controlling capsule-mediated hypermucoviscosity, and antibiotic resistance genes, including that encoding the KPC-2 carbapenemase (1). Previously, hypervirulent/hypermucoviscous Klebsiella pneumoniae (hvKP) strains were largely antibiotic susceptible. While the majority of infections with hvKP occur in Asia, we were interested in systematically assessing the prevalence of carbapenem-resistant hvKP (CR-hvKP) strains in the United States.
We tested 265 carbapenem-resistant K. pneumoniae clinical isolates collected between 2012 and 2015 from the Emerging Infections Program (EIP) Multi-site Gram-negative Surveillance Initiative (MuGSI) which has a surveillance population of >15 million people in 8 states across the United States. We phenotypically screened isolates using the string test to detect hypermucoviscosity, which has been strongly correlated with hypervirulence. K. pneumoniae isolates collected through MuGSI between 2012 and 2015 were streaked out on blood agar plates (tryptic soy agar [TSA] with 5% sheep blood; Remel, Thermo Fisher Scientific). The string test was performed to detect hypermucoviscosity, using a 1-μl inoculation loop on a single colony. A positive string test is >5mm in length. One isolate collected in 2014 from New York, termed CDC98, was positive by the string test (Fig. S1 and Movie S1). Subsequent testing by six independent scientists who were blind to the strains being tested confirmed CDC98 as being string test positive compared to three nonhypermucoviscous negative-control strains (Fig. S2).
Whole-genome Illumina sequencing revealed that CDC98 is sequence type 258 (ST258), capsule type KL107, and contains numerous resistance genes, including kpc3. ST258 is the current global pandemic strain of K. pneumoniae, and although the reasons for its success as a global pathogenic strain are uncertain, it is likely to have a fitness and selective advantage, as well as success, due to its antibiotic-resistance elements (5, 6). Additionally, we discovered by sequencing that the isolate harbors a previously unreported hybrid of plasmids pKPN-498 and pUHKPC45-117, encoding antibiotic resistance [SHV-12, sulI, catA1, mph(A), LEN-4, dfrA12, and aadA2] and virulence (sil and pco) genes. Interestingly, the isolate does not harbor the virulence plasmid pLVPK (or its close derivatives, collectively referred to here as pLVPKs) found in most Asian hvKP strains or the previously identified regulators of hypermucoviscosity and hypervirulence, rmpA and rmpA2 (7). The genes rmpA and rmpA2 have been thought to be responsible for the hypermucoviscous phenotype by direct regulation of the cps locus, although examples have been published where hypermucoviscosity was observed in the absence of these genes (8,–13). In addition to the putative virulence genes encoded on the hybrid plasmid, CDC98 possesses chromosomal copies of the siderophore receptor genes iutA and iroN, which are found on pLVPKs in many Asian hvKP isolates.
The level of virulence of CDC98 was difficult to predict, since it is hypermucoviscous yet lacks pLVPK. We directly assessed this using a murine in vivo infection model; C57BL/6 mice were infected intraperitoneally with 106 CFU of the indicated K. pneumoniae strains. Mice were provided with food and water ad libitum and monitored for signs of illness or weight loss. Mice exhibiting weight loss below 80% of starting weight were euthanized. CDC98 was indeed more virulent than three nonhypermucoviscous ST258 K. pneumoniae isolates from the same study site (New York; Fig. 1). However, while CDC98 displays significantly increased virulence compared to the other K. pneumoniae isolates tested, this level of virulence is far lower than that observed in canonical hypervirulent K. pneumoniae strains (14).
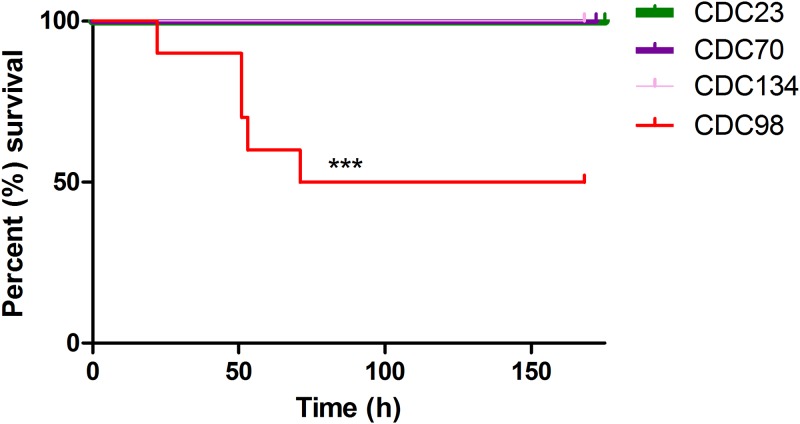
CDC98 displays increased virulence compared to that of nonhypermucoviscous K. pneumoniae isolates. Mice were infected intraperitoneally with CDC98 or the indicated nonhypermucoviscous K. pneumoniae isolates. Results shown are the combination of two independent experiments (n =
= 10 total). Comparison of survival curves was performed with the Mantel-Cox test (P
10 total). Comparison of survival curves was performed with the Mantel-Cox test (P =
= 0.0004).
0.0004).
As the sequencing of CDC98 indicated the presence of multiple antibiotic-resistance genes, antibiotic susceptibility testing was performed using the MicroScan WalkAway 96 plus instrument (Beckman Coulter, Inc., Brea, CA) and Neg Breakpoint Combo 44 panel, and the resultant antibiogram of this strain revealed extensive drug resistance to multiple classes of antibiotics, including carbapenems (Table S1). In the case of multidrug-resistant strains, such as CDC98, last-line antibiotics are often required for treatment. One such drug is the cationic polymyxin antibiotic colistin, which disrupts the membranes of Gram-negative bacteria.
Additional susceptibility testing by population analysis profile (PAP), whereby serial dilutions of an overnight bacterial culture are plated on increasing concentrations of an antibiotic on agar plates, revealed CDC98 to be heteroresistant to colistin (Fig. 2A). Heteroresistance describes a phenomenon wherein a subpopulation of bacteria in an otherwise homogenous population displays antibiotic resistance. Importantly, previous work has shown that colistin heteroresistance mediates antibiotic treatment failure in mice, and it has been postulated that this may in part contribute to unexplained antibiotic treatment failure in the clinic (15). Diagnostic testing for colistin susceptibility by Etest (Fig. 2B) or broth microdilution (BMD; MIC <
< 0.5
0.5 μg/ml) classified CDC98 as susceptible, indicating that the heteroresistance was not detected.
μg/ml) classified CDC98 as susceptible, indicating that the heteroresistance was not detected.
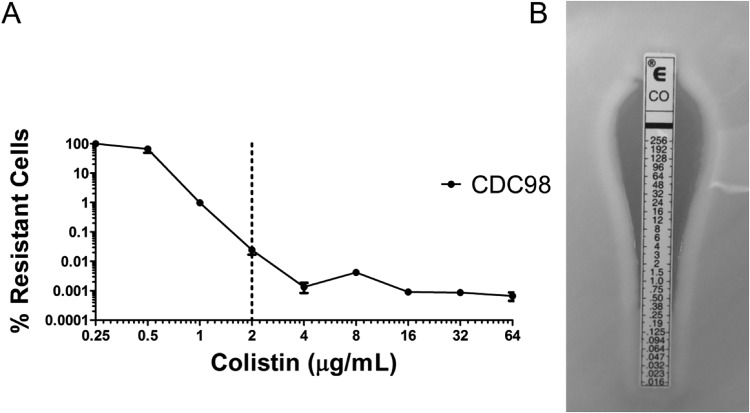
CDC98 displays heteroresistance to the last-line antibiotic colistin. (A) Population analysis profile (PAP) of CDC98 reveals a subpopulation of colistin-resistant cells. The dotted line represents the CLSI epidemiological cutoff value (ECV) for determining K. pneumoniae susceptibility or resistance to colistin (16). (B) CDC98 was designated by Etest as susceptible to colistin.
This is the first report of a systematic survey of carbapenem-resistant Klebsiella pneumoniae isolates for hypermucoviscosity in the United States. Discrepancies between Asian CR-hvKP strains (such as strain K1; [1]) and CDC98 suggest that a distinct lineage of carbapenem-resistant K. pneumoniae may be emerging in the United States, as CDC98 exhibits phenotypic similarities to contemporary hypervirulent K. pneumoniae strains while containing significant genetic differences (Table S2). These findings are further supported by a recent U.S. isolate exhibiting rmpA and/or rmpA2-independent increased capsular polysaccharide production and enhanced virulence, which was not as virulent as classical hvKP strains (8). Together, these results are indicative of the convergent evolution of distinct lineages of K. pneumoniae in Asia and the United States, leading to acquisition of enhanced virulence and antibiotic resistance. This convergence, taken together with emerging antibiotic resistance mechanisms, like heteroresistance, that can often go undetected by current diagnostics, pose a new and worrisome threat to public health.
Recent discussion in the field has suggested that using hypermucoviscosity as the singular criterion for identifying hypervirulence is insufficient. Hypermucoviscous isolates with enhanced virulence, such as CDC98, can lack the common genomic biomarkers for hypervirulence (rmpA, rmpA2, K1 capsular serotype, etc.). Therefore, while our findings reveal that CR-hvKP isolates have been relatively rare, both phenotypic and genotypic screening will be critical to gain a comprehensive understanding of CR-hvKP in the United States.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chui-Yoke Chin, Emily Crispell, David Hufnagel, Sree Sai Jaggavarappu, and Hannah Ratner for critical reading of the manuscript. We also thank the Emerging Infections Program and Centers for Disease Control and Prevention staff involved in the Multi-site Gram-negative Surveillance Initiative for the collection, cataloging, and characterization of relevant clinical isolates, including Sandra Bulens and Maria Karlsson. We thank the Genomics Resource Center at the University of Maryland School of Medicine for Illumina sequencing.
D.S.W. is supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award, VA Merit Award I01 BX002788, and NIH grant AI141883. This work was supported in part by the Emerging Infections Program and the National Center for Emerging and Zoonotic Infectious Diseases at the U.S. Centers for Disease Control and Prevention.
The findings and conclusions in this paper are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
We declare no competing financial interests.
Experiments, data collection, and analysis were performed by J.W., V.I.B., A.B.C., L.R., E.M.B., and S.W.S. Data interpretation was conducted by J.W., V.I.B., A.C., I.K.J., L.R., E.M.B., S.W.S., and D.S.W. The manuscript was prepared by J.W. and D.S.W. The study was planned and directed by D.S.W., who had full access to all data presented in the study and final responsibility to submit for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00107-19.
REFERENCES
Articles from Antimicrobial Agents and Chemotherapy are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aac.00107-19
Read article for free, from open access legal sources, via Unpaywall:
https://aac.asm.org/content/aac/63/5/e00107-19.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/aac.00107-19
Article citations
In vivo evolution to hypermucoviscosity and ceftazidime/avibactam resistance in a liver abscess caused by Klebsiella pneumoniae sequence type 512.
mSphere, 9(9):e0042324, 22 Aug 2024
Cited by: 0 articles | PMID: 39171923 | PMCID: PMC11423586
A novel small RNA PhaS contributes to polymyxin B-heteroresistance in carbapenem-resistant Klebsiella pneumoniae.
Emerg Microbes Infect, 13(1):2366354, 10 Jul 2024
Cited by: 0 articles | PMID: 38979571 | PMCID: PMC11238654
The Rapid Emergence of Hypervirulent Klebsiella Species and Burkholderia pseudomallei as Major Health Threats in Southeast Asia: The Urgent Need for Recognition as Neglected Tropical Diseases.
Trop Med Infect Dis, 9(4):80, 08 Apr 2024
Cited by: 0 articles | PMID: 38668541 | PMCID: PMC11054678
Review Free full text in Europe PMC
Prevalence of multidrug-resistant hypervirulent Klebsiella pneumoniae without defined hypervirulent biomarkers in Anhui, China: a new dimension of hypervirulence.
Front Microbiol, 14:1247091, 05 Oct 2023
Cited by: 2 articles | PMID: 37869673 | PMCID: PMC10585048
Genomic Analysis of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae in a Chinese Tertiary Hospital.
Infect Drug Resist, 16:6385-6394, 27 Sep 2023
Cited by: 1 article | PMID: 37789842 | PMCID: PMC10543750
Go to all (19) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Inactivation of mgrB gene regulator and resistance to colistin is becoming endemic in carbapenem-resistant Klebsiella pneumoniae in Greece: A nationwide study from 2014 to 2017.
Int J Antimicrob Agents, 55(4):105930, 01 Mar 2020
Cited by: 22 articles | PMID: 32130981
Rapid Increase in Prevalence of Carbapenem-Resistant Enterobacteriaceae (CRE) and Emergence of Colistin Resistance Gene mcr-1 in CRE in a Hospital in Henan, China.
J Clin Microbiol, 56(4):e01932-17, 26 Mar 2018
Cited by: 36 articles | PMID: 29386265 | PMCID: PMC5869811
Carbapenem-Resistant Klebsiella pneumoniae Exhibiting Clinically Undetected Colistin Heteroresistance Leads to Treatment Failure in a Murine Model of Infection.
mBio, 9(2):e02448-17, 06 Mar 2018
Cited by: 52 articles | PMID: 29511071 | PMCID: PMC5844991
A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae.
J Glob Antimicrob Resist, 25:26-34, 02 Mar 2021
Cited by: 95 articles | PMID: 33667703
Review
Funding
Funders who supported this work.
BLRD VA (1)
Grant ID: I01 BX002788
Burroughs Wellcome Fund
HHS | NIH | National Institute of Allergy and Infectious Diseases (1)
Grant ID: AI141883
NIAID NIH HHS (1)
Grant ID: R01 AI141883
U.S. Department of Veterans Affairs (1)
Grant ID: BX002788
 b,c,d,e
b,c,d,e




