Abstract
Free full text

Risk assessment studies on succinate dehydrogenase inhibitors, the new weapons in the battle to control Septoria leaf blotch in wheat
SUMMARY
Chemical control of Septoria leaf blotch, caused by Mycosphaerella graminicola, is essential to ensure wheat yield and food security in most European countries. Mycosphaerella graminicola has developed resistance to several classes of fungicide and, with the efficacy of azoles gradually declining over time, new modes of action and/or improvements in host varietal resistance are urgently needed to ensure future sustainable disease control. Several new‐generation carboxamide fungicides with broad‐spectrum activity have recently been introduced into the cereal market. Carboxamides inhibit succinate dehydrogenase (Sdh) of the mitochondrial respiratory chain (complex II) but, because of their single‐site specificity, these fungicides may be prone to resistance development. The objective of this study was to assess the risk of resistance development to different Sdh inhibitor (SDHI) fungicides in M. graminicola. UV mutagenesis was conducted to obtain a library of carboxin‐resistant mutants. A range of SDHI resistance‐conferring mutations was found in Sdh subunits B, C and D. Pathogenicity studies with a range of Sdh variants did not detect any fitness costs associated with these mutations. Most of the amino acid residues identified (e.g. B‐S221P/T, B‐H267F/L/N/Y, B‐I269V and D‐D129E/G/T) are directly involved in forming the cavity in which SDHI fungicides bind. Docking studies of SDHI fungicides in structural models of wild‐type and mutated Sdh complexes also indicated which residues were important for the binding of different SDHI fungicides and showed a different binding for fluopyram. The predictive power of the model was also shown. Further diagnostic development, enabling the detection of resistant alleles at low frequencies, and cross‐resistance studies will aid the implementation of anti‐resistance strategies to prolong the cost‐effectiveness and lifetime of SDHI fungicides.
graminicola. UV mutagenesis was conducted to obtain a library of carboxin‐resistant mutants. A range of SDHI resistance‐conferring mutations was found in Sdh subunits B, C and D. Pathogenicity studies with a range of Sdh variants did not detect any fitness costs associated with these mutations. Most of the amino acid residues identified (e.g. B‐S221P/T, B‐H267F/L/N/Y, B‐I269V and D‐D129E/G/T) are directly involved in forming the cavity in which SDHI fungicides bind. Docking studies of SDHI fungicides in structural models of wild‐type and mutated Sdh complexes also indicated which residues were important for the binding of different SDHI fungicides and showed a different binding for fluopyram. The predictive power of the model was also shown. Further diagnostic development, enabling the detection of resistant alleles at low frequencies, and cross‐resistance studies will aid the implementation of anti‐resistance strategies to prolong the cost‐effectiveness and lifetime of SDHI fungicides.
INTRODUCTION
The control of fungal foliar cereal diseases is mainly dependent on the deployment of resistant host cultivars and the programmed application of fungicides. Septoria leaf blotch (SLB), caused by the ascomycete Mycosphaerella graminicola (anamorph Septoria tritici), has been the most important foliar disease of winter wheat in the UK since the 1980s (Bearchell et al., 2005; Hardwick et
al., 2005; Hardwick et al., 2001). Current commercial wheat cultivars are susceptible or moderately resistant to the disease in the UK, because of the relatively low priority of disease resistance in breeding programmes, in combination with the availability of effective systemic fungicides over the last three decades. However, this approach may change as a result of fungicide resistance evolution and the introduction of new European Union regulations on the use of plant protection products (Directive 2009/128/EC and Regulation 1107/2009) impacting on product (re‐)registration and discovery. As a result of resistance development to benzimidazole fungicides (Griffin and Fischer, 1985) and quinone outside inhibitors (QoIs) (Fraaije et
al., 2001). Current commercial wheat cultivars are susceptible or moderately resistant to the disease in the UK, because of the relatively low priority of disease resistance in breeding programmes, in combination with the availability of effective systemic fungicides over the last three decades. However, this approach may change as a result of fungicide resistance evolution and the introduction of new European Union regulations on the use of plant protection products (Directive 2009/128/EC and Regulation 1107/2009) impacting on product (re‐)registration and discovery. As a result of resistance development to benzimidazole fungicides (Griffin and Fischer, 1985) and quinone outside inhibitors (QoIs) (Fraaije et al., 2005), in 1984 and 2002, respectively, the control of SLB is currently dependent on the use of systemic demethylation inhibitors (DMIs; azole fungicides) and the protective multi‐site inhibitor chlorothalonil.
al., 2005), in 1984 and 2002, respectively, the control of SLB is currently dependent on the use of systemic demethylation inhibitors (DMIs; azole fungicides) and the protective multi‐site inhibitor chlorothalonil.
Fungicide programmes, typically two to four sprays of fungicide mixtures during the growing season, contribute to profitable farming by increasing yields by around 2 tonnes/ha, equivalent to 25% of the average UK yield. They have also been shown to reduce greenhouse gas emissions significantly by a more optimal use of nitrogen fertilizer in the absence of disease (Berry et al., 2008). The sensitivity of M.
al., 2008). The sensitivity of M. graminicola populations to azole fungicides has been declining gradually over time, already affecting field performance for some products (Clark, 2006). As a result of the continued evolution of novel CYP51 (sterol 14α‐demethylase) variants, further shifts in sensitivity towards different azoles are expected (Brunner et
graminicola populations to azole fungicides has been declining gradually over time, already affecting field performance for some products (Clark, 2006). As a result of the continued evolution of novel CYP51 (sterol 14α‐demethylase) variants, further shifts in sensitivity towards different azoles are expected (Brunner et al., 2008; Cools and Fraaije, 2008; Cools et
al., 2008; Cools and Fraaije, 2008; Cools et al., 2011; Leroux and Walker, 2011) and new modes of action are urgently needed to ensure future sustainable disease control.
al., 2011; Leroux and Walker, 2011) and new modes of action are urgently needed to ensure future sustainable disease control.
Several new‐generation carboxamide fungicides have been introduced recently [e.g. boscalid (2005), isopyrazam (2010) and bixafen (2011)] or are expected to be launched shortly into the UK cereal market (e.g. penthiopyrad) (Fig. 1). Carboxamides inhibit succinate dehydrogenase (Sdh) of the mitochondrial respiratory chain (complex II). In contrast with early‐generation Sdh inhibitors (SDHIs), such as carboxin introduced in 1969, the new compounds have a wide spectrum of activity and some curative properties. So far, resistance to SDHI fungicides has not been found in field isolates of cereal pathogens. However, carboxin‐resistant laboratory mutants of M.
1). Carboxamides inhibit succinate dehydrogenase (Sdh) of the mitochondrial respiratory chain (complex II). In contrast with early‐generation Sdh inhibitors (SDHIs), such as carboxin introduced in 1969, the new compounds have a wide spectrum of activity and some curative properties. So far, resistance to SDHI fungicides has not been found in field isolates of cereal pathogens. However, carboxin‐resistant laboratory mutants of M. graminicola have been reported (Skinner et
graminicola have been reported (Skinner et al., 1998). These mutants carry amino acid substitutions H267L and H267Y in the Sdh iron sulphur protein subunit B (SdhB) and are cross‐resistant to boscalid (B. A. Fraaije, unpublished). Similar or identical mutations at this codon and mutations at other positions within SdhB, and subunits SdhC and SdhD, have also been found to confer resistance to SDHI fungicides in laboratory mutants and field isolates of a range of fungi, including Alternaria alternata (2008, 2009), Aspergillus oryzae (Shima et
al., 1998). These mutants carry amino acid substitutions H267L and H267Y in the Sdh iron sulphur protein subunit B (SdhB) and are cross‐resistant to boscalid (B. A. Fraaije, unpublished). Similar or identical mutations at this codon and mutations at other positions within SdhB, and subunits SdhC and SdhD, have also been found to confer resistance to SDHI fungicides in laboratory mutants and field isolates of a range of fungi, including Alternaria alternata (2008, 2009), Aspergillus oryzae (Shima et al., 2009), Botrytis cinerea (Leroux et
al., 2009), Botrytis cinerea (Leroux et al., 2010; Stammler et
al., 2010; Stammler et al., 2008) and Corynespora cassiicola (Miyamoto et
al., 2008) and Corynespora cassiicola (Miyamoto et al., 2010). Interestingly, for some inhibitors (e.g. fluopyram), a lack of cross‐resistance to other SDHIs was identified when resistant isolates were tested (see Avenot and Michailides, 2010; Ishii et
al., 2010). Interestingly, for some inhibitors (e.g. fluopyram), a lack of cross‐resistance to other SDHIs was identified when resistant isolates were tested (see Avenot and Michailides, 2010; Ishii et al., 2011; White et
al., 2011; White et al., 1978).
al., 1978).
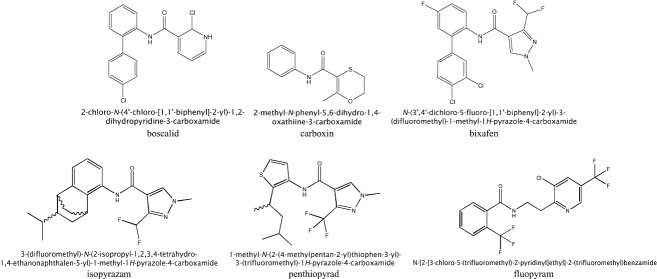
Structural formulae and systematic names of the studied succinate dehydrogenase inhibitor (SDHI) fungicides.
To prevent or delay resistance development to SDHI fungicides in cereal pathogens, the Fungicide Resistance Action Committee (FRAC) has recommended the use of SDHI fungicides preventatively in mixtures with a maximum of two sprays per season. Mixing partners should ideally have a different mode of action and, on their own, provide satisfactory disease control. For the control of SLB, azoles are the preferred mixing partner, and it is expected that most SDHI products will enter the UK wheat market as co‐formulations with azoles.
The objective of this study was to assess the risk of resistance development to different SDHI fungicides in M. graminicola. We measured the baseline sensitivities to SDHIs and other fungicides in old and recent M.
graminicola. We measured the baseline sensitivities to SDHIs and other fungicides in old and recent M. graminicola field isolates from different geographical origins. UV mutagenesis was conducted to obtain carboxin‐resistant mutants. These mutants were further characterized for levels of resistance and cross‐resistance patterns using in
graminicola field isolates from different geographical origins. UV mutagenesis was conducted to obtain carboxin‐resistant mutants. These mutants were further characterized for levels of resistance and cross‐resistance patterns using in vitro fungicide sensitivity assays to a range of SDHIs. For a selection of mutants, pathogenicity tests were carried out to establish their fitness in
vitro fungicide sensitivity assays to a range of SDHIs. For a selection of mutants, pathogenicity tests were carried out to establish their fitness in planta. In addition, in
planta. In addition, in silico studies on the binding of fungicides to the Sdh complex of M.
silico studies on the binding of fungicides to the Sdh complex of M. graminicola were carried out. Docking studies of SDHIs in structural models of wild‐type and mutated Sdh complexes established which residues were important for binding, and the predictive power of the model was also investigated. Knowledge from this study can be used to devise and test anti‐resistance strategies to prolong the cost‐effectiveness and lifetime of SDHIs.
graminicola were carried out. Docking studies of SDHIs in structural models of wild‐type and mutated Sdh complexes established which residues were important for binding, and the predictive power of the model was also investigated. Knowledge from this study can be used to devise and test anti‐resistance strategies to prolong the cost‐effectiveness and lifetime of SDHIs.
RESULTS
Fungicide baseline sensitivity testing of M. graminicola field strains
graminicola field strains
Table 1 shows the fungicide sensitivities of 30 field strains representing past and recent field populations sampled from different geographical regions. Carboxin was the least effective compound amongst the SDHIs tested, with recorded EC50 values between 0.608 and 5.52
1 shows the fungicide sensitivities of 30 field strains representing past and recent field populations sampled from different geographical regions. Carboxin was the least effective compound amongst the SDHIs tested, with recorded EC50 values between 0.608 and 5.52 µg/mL and an average value of 1.84
µg/mL and an average value of 1.84 µg/mL. Boscalid (EC50 between 0.161 and 1.14
µg/mL. Boscalid (EC50 between 0.161 and 1.14 µg/mL; average, 0.482
µg/mL; average, 0.482 µg/mL) was less effective in
µg/mL) was less effective in vitro than isopyrazam (EC50 between 0.020 and 1.1
vitro than isopyrazam (EC50 between 0.020 and 1.1 µg/mL; average, 0.262
µg/mL; average, 0.262 µg/mL) and bixafen (EC50 between 0.019 and 0.664
µg/mL) and bixafen (EC50 between 0.019 and 0.664 µg/mL; average, 0.219
µg/mL; average, 0.219 µg/mL). When sensitivity data from strains isolated before 2005 were compared with the sensitivities of those isolated after 2005 (coinciding with the introduction of boscalid in the UK), there were no differences in EC50 values for the four SDHIs and chlorothalonil. The sensitivity data for epoxiconazole and prothioconazole‐desthio showed a clear trend of decreasing sensitivity for strains isolated more recently in north‐west Europe, a region with a high Septoria disease pressure, compared with older strains. This change in sensitivity has been correlated with the evolution of more complex CYP51 variants harbouring multiple amino acid alterations (Stammler et
µg/mL). When sensitivity data from strains isolated before 2005 were compared with the sensitivities of those isolated after 2005 (coinciding with the introduction of boscalid in the UK), there were no differences in EC50 values for the four SDHIs and chlorothalonil. The sensitivity data for epoxiconazole and prothioconazole‐desthio showed a clear trend of decreasing sensitivity for strains isolated more recently in north‐west Europe, a region with a high Septoria disease pressure, compared with older strains. This change in sensitivity has been correlated with the evolution of more complex CYP51 variants harbouring multiple amino acid alterations (Stammler et al., 2008).
al., 2008).
Table 1
Characterization of Mycosphaerella graminicola field strains.
| Isolate | CYP51 amino acid alterations* | Origin | Year | Fungicide sensitivity (EC50 in µg/mL)† | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Carboxin | Boscalid | Bixafen | Isopyrazam | Epoxiconazole | Prothioconazole‐desthio | Chlorothalonil | ||||
| IPO323 | None | The Netherlands | 1981 | 1.50 | 0.970 | 0.273 | 0.418 | <0.0005 | 0.0018 | 0.167 |
| IPO88004 | None | Ethiopia | 1988 | 1.43 | 0.208 | 0.031 | 0.107 | <0.0005 | 0.0009 | 0.145 |
| A12/3B/8 | None | USA | 1988 | 1.99 | 0.210 | 0.055 | 0.053 | <0.0005 | 0.0015 | 0.191 |
| IPO90012 | None | Mexico | 1990s | 2.05 | 0.300 | 0.086 | 0.073 | <0.0005 | 0.0013 | 0.119 |
| IPO001 | None | The Netherlands | 1991 | 1.27 | 0.203 | 0.054 | 0.078 | <0.0005 | 0.0015 | 0.141 |
| RD14 | Y137F | UK | 1993 | 0.622 | 0.271 | 0.094 | 0.119 | 0.0057 | 0.0060 | 0.040 |
| ST16 | L50S, G460D | UK | 1995 | 1.61 | 0.603 | 0.381 | 0.539 | 0.0018 | 0.0023 | 0.145 |
| IPO92006 | L50S, S188N, N513K | Portugal | 1990s | 3.75 | 0.615 | 0.393 | 0.169 | <0.0005 | 0.0015 | 0.218 |
| CTRL01‐1 | Y137F, S524T | UK | 2001 | 3.13 | 0.438 | 0.130 | 0.176 | 0.0536 | 0.0042 | 0.097 |
| LARS15 | L50S, I381V, Y459D | UK | 2003 | 1.40 | 0.433 | 0.175 | 0.247 | 0.0549 | 0.0105 | 0.109 |
| G303 | L50S, S188N, A379G, I381V, DEL, N513K | UK | 2003 | 2.25 | 1.14 | 0.664 | 0.591 | 0.0749 | 0.0134 | 0.176 |
| IRE13 | L50S, V136A, Y461S | Ireland | 2003 | 0.868 | 0.197 | 0.030 | 0.042 | 0.0268 | 0.0127 | 0.069 |
| IRE30 | L50S, V136A, Y461S | Ireland | 2003 | 1.75 | 0.522 | 0.090 | 0.130 | 0.0399 | 0.0110 | 0.162 |
| POL27 | L50S, I381V, Y461H | Poland | 2004 | 1.27 | 0.524 | 0.207 | 0.397 | 0.127 | 0.0144 | 0.155 |
| NZ75 | Y137F | New Zealand | 2004 | 1.20 | 0.292 | 0.057 | 0.074 | 0.0056 | 0.0011 | 0.235 |
| BD04 | G460D | Spain | 2006 | 0.711 | 0.655 | 0.127 | 0.435 | 0.0056 | 0.0023 | 0.127 |
| MM20 | None | Spain | 2006 | 3.42 | 0.394 | 0.383 | 0.368 | <0.0005 | 0.0018 | 0.210 |
| V212‐2 | V136A, S188N, DEL | UK | 2006 | 2.69 | 0.415 | 0.215 | 0.184 | 0.0712 | 0.0118 | 0.141 |
| 4439 | None | Finland | 2007 | 0.765 | 0.604 | 0.312 | 0.410 | <0.0005 | 0.0006 | 0.165 |
| T2 | D107V, I381V, N513K, S524T | France | 2008 | 0.608 | 0.446 | 0.179 | 0.294 | 0.0578 | 0.0063 | 0.113 |
| NZ11‐1 | L50S, S188N, N513K | New Zealand | 2008 | 3.56 | 0.454 | 0.078 | 0.130 | <0.0005 | 0.0019 | 0.212 |
| I43‐2 | L50S, V136A, Y461S, S524T | Ireland | 2009 | 1.10 | 0.630 | 0.391 | 0.206 | 0.244 | 0.0514 | 0.104 |
| R35‐3 | L50S, I381V, Y461H | UK | 2009 | 1.71 | 0.375 | 0.183 | 0.279 | 0.0910 | 0.0193 | 0.254 |
| R39‐1 | L50S, S188N, A379G, I381V, DEL, N513K | UK | 2009 | 5.52 | 0.795 | 0.662 | 1.10 | 0.278 | 0.0262 | 0.325 |
| BC1 | L50S, I381V, Y461H | UK | 2009 | 1.64 | 0.269 | 0.095 | 0.210 | 0.0818 | 0.0155 | 0.316 |
| BC4 | L50S, I381V, Y461H | UK | 2009 | 1.42 | 0.472 | 0.408 | 0.328 | 0.151 | 0.0270 | 0.329 |
| TAG71‐3 | L50S, D134G, V136A, Y461S, S524T | UK | 2009 | 2.05 | 0.161 | 0.019 | 0.020 | 0.453 | 0.156 | 0.336 |
| TAG1‐9 | L50S, V136A, I381V, Y461H, S524T | UK | 2010 | 1.35 | 0.498 | 0.271 | 0.206 | 1.50 | 0.180 | 0.081 |
| TAG74‐3 | L50S, S188N, I381V, DEL, N513K | UK | 2010 | 1.46 | 0.305 | 0.115 | 0.123 | 1.05 | 0.167 | 0.151 |
| ROS1.40.1 | L50S, V136A, I381V, Y461S, S524T | UK | 2010 | 1.24 | 0.522 | 0.402 | 0.130 | 0.658 | 0.152 | 0.146 |
Clear linear cross‐sensitivity patterns were observed for bixafen and isopyrazam, bixafen and boscalid, and isopyrazam and boscalid, with R
2 values of 0.65 (P < 0.001), 0.61 (P < 0.001) and 0.46 (P < 0.001) (n= 30), respectively. The R
2 values for carboxin and bixafen, carboxin and isopyrazam and carboxin and boscalid were lower, at 0.17 (P= 0.022), 0.17 (P= 0.025) and 0.03 (P= 0.385) (n= 30), respectively. As expected, of the remaining fungicide pairs tested, a clear linear cross‐resistance relationship was only measured between the two DMI fungicides epoxiconazole and prothioconazole‐desthio (R
2= 0.85; P < 0.001; n= 21) after discarding the ‘wild‐type’ strains (strains with no alterations in CYP51 or variants carrying L50S, S188N and N513K) (Leroux et al., 2007).
al., 2007).
Sdh sequence variation among M. graminicola field strains
graminicola field strains
DNA sequence comparison between IPO323, IRE30 and five other strains (BC1, BC4, R35‐3, R39‐1 and V212‐2) showed that the Sdh subunits were generally well conserved (GenBank accession numbers JF916683–916700). For SdhB, only changes resulting in the replacement of lysine (aag) by arginine (agg) at position 48 (B‐K48R) and cysteine (tgc) by arginine (cgc) at codon 276 (B‐C276R) were found in strains BC1 and V212‐2, respectively. Strain R35‐3 had a proline (ccc) residue [instead of serine (tcc)] at position 51 in SdhC (C‐S51P), and isolate R39‐1 had two changes in this subunit, a valine (gtc) residue [instead of isoleucine (atc)] at codon 29 (C‐I29V) and a glycine (ggc) residue [instead of arginine (cgc)] at codon 54 (C‐R54G). Two SdhC changes were also found in strain V212‐2, with two asparagine (aac) residues replaced by threonine (acc) at codons 33 (C‐N33T) and 34 (C‐N34T). For SdhD, strain R35‐3 carried a glycine (ggt) residue [instead of alanine (gct)] at codon 10 (D‐A10G), whereas IRE30 harboured a unique proline (ccg) residue [instead of arginine (cgg)] at codon 47 (D‐R47P). SdhC was most variable with regard to additional nucleotide changes leading to synonymous substitutions and sequence variation within intron regions (data not shown).
Docking of carboxin and other SDHIs in the quinone binding site of the M. graminicola Sdh complex
graminicola Sdh complex
Docking studies ((2,2, ,3)3) showed carboxin positioned in a similar manner as in the crystal structure of the avian Sdh complex (Huang et al., 2006). SdhA did not form part of the quinone binding site. SdhB residues P220, S221, W223, W224, H267 and I269 form the major part of the binding pocket. SdhC (L71, W80, S83, A84 and R87) and SdhD (D129 and Y130) form the remainder of the fungicide binding cavity. The docking study also indicates that the binding energy for the carboxin–Sdh complex is relatively high, suggesting relatively poor binding when compared with the other fungicides (Table
al., 2006). SdhA did not form part of the quinone binding site. SdhB residues P220, S221, W223, W224, H267 and I269 form the major part of the binding pocket. SdhC (L71, W80, S83, A84 and R87) and SdhD (D129 and Y130) form the remainder of the fungicide binding cavity. The docking study also indicates that the binding energy for the carboxin–Sdh complex is relatively high, suggesting relatively poor binding when compared with the other fungicides (Table 2). This probably reflects the relatively small size of this molecule, reducing extensive protein–fungicide interactions. Boscalid and bixafen contain an additional phenyl ring compared with carboxin, which is reflected in the lower binding energies. The predicted binding energies of carboxin, boscalid, bixafen and isopyrazam (Table
2). This probably reflects the relatively small size of this molecule, reducing extensive protein–fungicide interactions. Boscalid and bixafen contain an additional phenyl ring compared with carboxin, which is reflected in the lower binding energies. The predicted binding energies of carboxin, boscalid, bixafen and isopyrazam (Table 2) are perfectly in line with the respective baseline fungicide sensitivity levels (Table
2) are perfectly in line with the respective baseline fungicide sensitivity levels (Table 1). The chlorinated phenyl rings of boscalid and bixafen occupy the same binding pocket as the phenyl moiety of docked carboxin (Fig.
1). The chlorinated phenyl rings of boscalid and bixafen occupy the same binding pocket as the phenyl moiety of docked carboxin (Fig. 3A–C). However, the amide moiety is bound in another position, as is the second phenyl moiety of these fungicides, which is located at the surface of the protein. The pyrazole and pyridine moieties of bixafen and boscalid are found in the same pocket as the nonaromatic ring of carboxin. Binding seems to be stronger for bixafen, suggesting that the pyrazole moiety interacts favourably when compared with the pyridine moiety of boscalid, as both fungicides adopt similar binding modes when docked into the Sdh complex. Docking of isopyrazam (Fig.
3A–C). However, the amide moiety is bound in another position, as is the second phenyl moiety of these fungicides, which is located at the surface of the protein. The pyrazole and pyridine moieties of bixafen and boscalid are found in the same pocket as the nonaromatic ring of carboxin. Binding seems to be stronger for bixafen, suggesting that the pyrazole moiety interacts favourably when compared with the pyridine moiety of boscalid, as both fungicides adopt similar binding modes when docked into the Sdh complex. Docking of isopyrazam (Fig. 3D) and penthiopyrad (Fig.
3D) and penthiopyrad (Fig. 3E) yields similar binding energies when compared with bixafen (Table
3E) yields similar binding energies when compared with bixafen (Table 2). However, the binding mode more closely resembles the binding of carboxin. The pyrazole moieties occupy the same pocket as the phenyl group of carboxin, whereas their amide and hydrophobic moieties bind in the same location as the amide and the nonaromatic ring of carboxin. To compare the effectiveness of fluopyram in
2). However, the binding mode more closely resembles the binding of carboxin. The pyrazole moieties occupy the same pocket as the phenyl group of carboxin, whereas their amide and hydrophobic moieties bind in the same location as the amide and the nonaromatic ring of carboxin. To compare the effectiveness of fluopyram in silico, we also docked this novel fungicide. The predicted fluopyram binding is largely similar to the binding of carboxin, as the phenyl ring and the amide moiety are in the same location as the nonaromatic ring and amide of carboxin (Fig.
silico, we also docked this novel fungicide. The predicted fluopyram binding is largely similar to the binding of carboxin, as the phenyl ring and the amide moiety are in the same location as the nonaromatic ring and amide of carboxin (Fig. 3F). However, the other moiety of fluopyram, a benzamide group, is separated from the amide bond by two carbons. This facilitates extended binding of this fungicide, as the benzamide moiety is favourably stacked in between two tryptophans, SdhB W223 and SdhC W80 (Fig.
3F). However, the other moiety of fluopyram, a benzamide group, is separated from the amide bond by two carbons. This facilitates extended binding of this fungicide, as the benzamide moiety is favourably stacked in between two tryptophans, SdhB W223 and SdhC W80 (Fig. 2). There is no such spacer between the amide and the analogous phenyl group in carboxin. This suggests very tight binding for fluopyram, as the binding energy, −138.6
2). There is no such spacer between the amide and the analogous phenyl group in carboxin. This suggests very tight binding for fluopyram, as the binding energy, −138.6 kJ/mol, is the lowest of all the docked SDHIs (Table
kJ/mol, is the lowest of all the docked SDHIs (Table 2).
2).
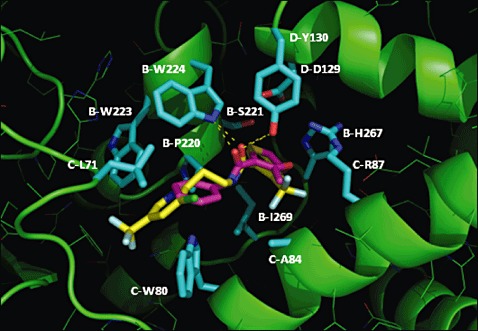
Docking of carboxin and fluopyram in the quinone binding site. The succinate dehydrogenase (Sdh) structural model is shown with the carbon atoms in green, with the residues forming the binding pocket with the carbon atoms in cyan. Carboxin and fluopyram are shown with the carbon atoms in magenta and yellow, respectively. Hydrogen bonds are indicated as broken lines.
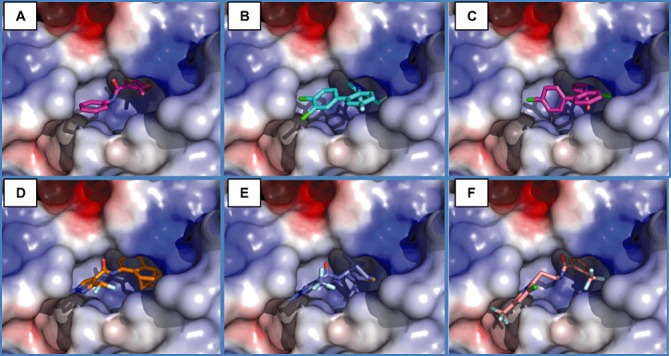
Binding of succinate dehydrogenase inhibitors (SDHIs) to the quinone binding site of the succinate dehydrogenase (Sdh) complex. Carboxin (A), bixafen (B), boscalid (C), isopyrazam (D), penthiopyrad (E) and fluopyram (F). The electrostatic potential of the protein surface is coloured from red (negative charge) to blue (positive charge).
Table 2
Lowest binding energies obtained by docking of fungicides in a structural model of the wild‐type (WT) succinate dehydrogenase (Sdh) complex from Mycosphaerella graminicola and fungicide resistance‐related Sdh variants.
| Inhibitor* | Binding energies for each complex (kJ/mol)† | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | S221P SdhB | R265P SdhB | H267L SdhB | H267Y SdhB | H267N SdhB | I269V SdhB | N86K SdhC | I127V SdhD | D129E SdhD | D129G SdhD | |
| Boscalid | −113.0 | −114.9 | −114.2 | −111.8 | −112.7 | −112.5 | −112.8 | −114.8 | −114.6 | −116.1 | −110.5 |
| Carboxin | −92.0 | −92.5 | −90.2 | −90.0 | −93.1 | −89.1 | −88.7 | −90.3 | −90.5 | −91.1 | −84.0 |
| Bixafen | −121.9 | −129.4 | −122.6 | −121.8 | −122.2 | −125.1 | −120.0 | −124.8 | −122.7 | −121.7 | −122.1 |
| Isopyrazam | −122.8 | −115.5 | −119.8 | −115.2 | −113.0 | −126.4 | −123.4 | −118.9 | −123.4 | −118.6 | −136.2 |
| Penthiopyrad | −122.4 | −120.5 | −122.2 | −120.2 | −119.2 | −122.6 | −121.1 | −124.1 | −122.2 | −121.1 | −125.6 |
 kJ/mol) are presented in bold.
kJ/mol) are presented in bold.Identification of Sdh mutations in carboxin‐resistant UV mutants
In total 124 mutants, 98 derived from IPO323 (Table 3) and 26 from IRE30 (Table
3) and 26 from IRE30 (Table 4), were isolated and characterized. Mutants in the collection had a wide range of nonsynonymous mutations (Fig.
4), were isolated and characterized. Mutants in the collection had a wide range of nonsynonymous mutations (Fig. 4) and corresponding SDHI sensitivity profiles. Some rare mutants had multiple mutations in the same or in different Sdh subunit encoding genes. Figure
4) and corresponding SDHI sensitivity profiles. Some rare mutants had multiple mutations in the same or in different Sdh subunit encoding genes. Figure 4 shows the position of nine key amino acids that were substituted in SDHI‐insensitive mutants of M.
4 shows the position of nine key amino acids that were substituted in SDHI‐insensitive mutants of M. graminicola, and how conserved are these residues in SdhB, SdhC and SdhD sequences from A.
graminicola, and how conserved are these residues in SdhB, SdhC and SdhD sequences from A. alternata, As.
alternata, As. oryzae, B.
oryzae, B. cinerea, C.
cinerea, C. cassiicola and chicken (Gallus gallus). SdhB‐C137R is unique for M.
cassiicola and chicken (Gallus gallus). SdhB‐C137R is unique for M. graminicola and no mutations have been found at or in close proximity to this position in SDHI‐resistant strains of other fungi. This residue is normally part of an FeS cluster, relatively far from the quinone binding site, and it is expected that such a mutation would result in a nonfunctional protein. SdhB codon S221 (S221P/T) is one position away from codon 225 in B.
graminicola and no mutations have been found at or in close proximity to this position in SDHI‐resistant strains of other fungi. This residue is normally part of an FeS cluster, relatively far from the quinone binding site, and it is expected that such a mutation would result in a nonfunctional protein. SdhB codon S221 (S221P/T) is one position away from codon 225 in B. cinerea, where substitutions P225L/F/T have been reported for field strains (Leroux et
cinerea, where substitutions P225L/F/T have been reported for field strains (Leroux et al., 2010; Stammler et
al., 2010; Stammler et al., 2008). Codon 230 in B.
al., 2008). Codon 230 in B. cinerea, where N230I has been linked to SDHI resistance (Leroux et
cinerea, where N230I has been linked to SDHI resistance (Leroux et al., 2010), is positioned next to W224 in M.
al., 2010), is positioned next to W224 in M. graminicola, a residue predicted to form part of the quinone binding site (Fig.
graminicola, a residue predicted to form part of the quinone binding site (Fig. 2). SdhB codons 265 (R265P), 267 (H267F/L/N/Y) and 269 (I269V) are clustered together, and mutations resulting in H267Y/L/N have also been reported in a range of other fungi (Avenot and Michailides, 2010). SdhC N80K, reported for Coprinus cinereus (Ito et
2). SdhB codons 265 (R265P), 267 (H267F/L/N/Y) and 269 (I269V) are clustered together, and mutations resulting in H267Y/L/N have also been reported in a range of other fungi (Avenot and Michailides, 2010). SdhC N80K, reported for Coprinus cinereus (Ito et al., 2004), is identical to N86K in M.
al., 2004), is identical to N86K in M. graminicola. In addition, SdhC L85P and N86S were also found in M.
graminicola. In addition, SdhC L85P and N86S were also found in M. graminicola mutants. SdhC S73P and T90I, reported for C.
graminicola mutants. SdhC S73P and T90I, reported for C. cassiicola (Miyamoto et
cassiicola (Miyamoto et al., 2010) and As.
al., 2010) and As. oryzae (Shima et
oryzae (Shima et al., 2009), respectively, are within three positions of codons 85 and 86 in M.
al., 2009), respectively, are within three positions of codons 85 and 86 in M. graminicola. SdhD codons 127 (I127V) and 129 (D129E/G/T) are clustered together. SdhD D89G and D124E, reported in Paracoccus denitrificans (Matsson et
graminicola. SdhD codons 127 (I127V) and 129 (D129E/G/T) are clustered together. SdhD D89G and D124E, reported in Paracoccus denitrificans (Matsson et al., 1998) and As.
al., 1998) and As. oryzae (Shima et
oryzae (Shima et al., 2009), respectively, are equivalent to D129G and D129E in M.
al., 2009), respectively, are equivalent to D129G and D129E in M. graminicola. G109V in C.
graminicola. G109V in C. cassiicola (Miyamoto et
cassiicola (Miyamoto et al., 2010), H133R in A.
al., 2010), H133R in A. alternata (Avenot et
alternata (Avenot et al., 2009) and H132R in B.
al., 2009) and H132R in B. cinerea (Leroux et
cinerea (Leroux et al., 2010) are part of the α‐helix that positions the two crucial quinone binding site residues in M.
al., 2010) are part of the α‐helix that positions the two crucial quinone binding site residues in M. graminicola, SdhD D129 and Y130. However, they are too far from the quinone binding site (approximately 12
graminicola, SdhD D129 and Y130. However, they are too far from the quinone binding site (approximately 12 Å) to have a direct effect on fungicide binding.
Å) to have a direct effect on fungicide binding.
Table 3
Overview of key mutations and associated amino acid substitutions in succinate dehydrogenase (Sdh) complexes of IPO323‐derived mutants (n= 98) and their succinate dehydrogenase inhibitor (SDHI) sensitivity profiles.
| Strains/mutant Sdh variants | Corresponding codon changes | Mutant number | Fungicide sensitivity range (EC50 in µg/mL) | |||
|---|---|---|---|---|---|---|
| Carboxin† | Boscalid | Bixafen | Isopyrazam | |||
| IPO323 | 3.50 | 0.718 | 0.216 | 0.316 | ||
| B‐C137R | tgt > cgt | 1 | 159 | >12.5 | 2.01 | 1.93 |
| B‐S221P (M152)* | tcc > ccc | 1 | 11.8 | 0.344 | 0.014 | <0.001 |
| B‐S221P* | tcc > ccc | 5 | 102–190 | >12.5 | 0.761–1.91 | 0.889–2.34 |
| B‐S221T | tcc > act | 2 | 81.7–113 | 0.959–2.0 | 0.465–0.583 | 0.742–1.11 |
| B‐R265P | cga > cca | 6 | 133–175 | 3.26–5.0 | 0.235–1.02 | 0.644–1.83 |
| B‐H267F | cac > ttc | 1 | 58.1 | >12.5 | 2.2 | 3.13 |
| B‐H267L | cac > ctt/ctc | 9 | >200 | >12.5 | >12.5 | 10.0–>12.5 |
| B‐H267N | cac > aac | 5 | 81.0–131 | 0.662–1.33 | 0.284–0.787 | 0.451–2.09 |
| B‐H267Y | cac > tac | 48 | 67.7–193 | >12.5 | 0.482–3.05 | 0.232–4.6 |
| B‐I269V | att > gtt | 7 | 63.5–143 | 1.1–2.86 | 0.397–1.03 | 1.15–1.86 |
| B‐S221P, C‐R54G | tcc > ccc, cgc > ggc | 1 | 102 | 2.65 | 0.462 | 1.08 |
| B‐H267Y, C‐N86S | cac > tac, aac > agc | 1 | 153 | >12.5 | 1.46 | 2.11 |
| B‐D166G, D‐D129G | gac > ggc, gac > gga | 1 | >200 | >12.5 | 4.62 | 5.13 |
| B‐I13V, D‐D129E, D‐V154A | att > gtt, gac > gaa, gtt > gct | 1 | 40.0 | 3.08 | 0.969 | 1.44 |
| C‐L85P | ctc > ccc | 1 | 85.1 | 1.64 | 0.792 | 0.669 |
| C‐N86K | aac > aaa | 1 | 112 | >12.5 | >12.5 | >12.5 |
| D‐I127V | atc > gtc | 1 | 85.1 | 1.86 | 0.433 | 1.27 |
| D‐D129E | gac > gag/gaa | 5 | 85.1–148 | 11.6–>12.5 | 0.412–2.15 | 0.666–4.07 |
| D‐D129T | gac > acc | 1 | >200 | 3.41 | 3.49 | 3.6 |
 4 because of the wider range of fungicide concentrations tested to measure the high resistance levels in the mutants.
4 because of the wider range of fungicide concentrations tested to measure the high resistance levels in the mutants.Table 4
Overview of key mutations and associated amino acid substitutions in succinate dehydrogenase (Sdh) complexes of IRE30‐derived mutants (n= 26) and their succinate dehydrogenase inhibitor (SDHI) sensitivity profiles.
| Strains/mutant Sdh variants* | Corresponding codon changes | Mutant number | Fungicide sensitivity range (EC50 in µg/mL)† | |||
|---|---|---|---|---|---|---|
| Carboxin | Boscalid | Bixafen | Isopyrazam | |||
| IRE30 | 2.82 | 0.389 | 0.070 | 0.088 | ||
| B‐S221P | tcc > ccc | 5 | 57.8–108 | 0.447–1.09 | 0.058–0.243 | 0.096–0.338 |
| B‐H267N | cac > aac | 4 | 64.7–99.3 | 1.11–1.88 | 0.284–0.81 | 0.398–2.09 |
| B‐H267Y | cac > tac | 14 | 69.0–155 | >12.5 | 0.777–2.6 | 0.882–4.04 |
| B‐P155L, B‐H267Y | cca > cta, cac > tac | 1 | 23.6 | 0.878 | 0.682 | 0.486 |
| C‐L85P, D‐V96A | ctc > ccc, gtc > gcc | 1 | 53.1 | 3.33 | 0.874 | 1.75 |
| D‐D129E | gac > gag | 1 | 149 | >12.5 | 1.25 | 2.4 |
 4 because of the wider range of fungicide concentrations tested to measure the high resistance levels in the mutants.
4 because of the wider range of fungicide concentrations tested to measure the high resistance levels in the mutants.
Positions of key amino acid alterations in different succinate dehydrogenase (Sdh) subunits from succinate dehydrogenase inhibitor (SDHI)‐resistant mutants of Mycosphaerella graminicola. Partial Sdh subunit sequences are presented for M. graminicola (Mg), Alternaria alternata (Aa), Aspergillus oryzae (Ao), Botrytis cinerea (Bc), Corynespora cassiicola (Cc) and Gallus gallus (Gg). Positions of key amino acid residues are marked with a star. Shaded residues are amino acid residues that have been altered and linked with SDHI resistance. Bold amino acids are conserved residues.
graminicola (Mg), Alternaria alternata (Aa), Aspergillus oryzae (Ao), Botrytis cinerea (Bc), Corynespora cassiicola (Cc) and Gallus gallus (Gg). Positions of key amino acid residues are marked with a star. Shaded residues are amino acid residues that have been altered and linked with SDHI resistance. Bold amino acids are conserved residues.
Relationship between mutant Sdh variants and SDHI sensitivity
3,3, ,44 show the relationship between the different Sdh variants and the sensitivity ranges measured for the different SDHI fungicides. Mutants carrying B‐S221P are clustered into three different phenotypic groups depending on the parental strain. In comparison with IRE30 mutants, B‐S221P mutants derived from IPO323 have higher levels of resistance to all SDHIs tested, in particular for boscalid with EC50 levels above 12.5 µg/mL. For one IPO323‐derived mutant, M152, carrying S221P with no other mutations detected in the SdhB, SdhC and SdhD genes, the sensitivity to bixafen and isopyrazam was much lower than for IPO323. For all other Sdh variants tested, there was no clear strain‐dependent difference in SDHI sensitivity. Most mutants with multiple mutations in Sdh genes (B‐P155L/B‐H267Y, B‐S221P/C‐R54G and B‐I13V/D‐D129E/D‐V154A) were more sensitive to most SDHIs tested relative to mutants carrying single amino acid substitutions (B‐S221P, B‐H267Y and D‐D129E). Mutants carrying variant B‐H267L were highly resistant to all four SDHIs tested. High levels of boscalid resistance were measured for variants carrying B‐C137R, B‐S221P (only IPO323‐derived mutants), B‐H267F, H267Y, C‐N86K, D‐D129E and D‐D129G. Sensitivity levels to bixafen and isopyrazam were similar for mutants carrying different Sdh variants, with the exception of B‐I269V and B‐S221T mutants that were inhibited more strongly with bixafen than isopyrazam. Interestingly, high levels of resistance to both bixafen and isopyrazam were only measured for mutants carrying B‐H267L or C‐N86K.
µg/mL. For one IPO323‐derived mutant, M152, carrying S221P with no other mutations detected in the SdhB, SdhC and SdhD genes, the sensitivity to bixafen and isopyrazam was much lower than for IPO323. For all other Sdh variants tested, there was no clear strain‐dependent difference in SDHI sensitivity. Most mutants with multiple mutations in Sdh genes (B‐P155L/B‐H267Y, B‐S221P/C‐R54G and B‐I13V/D‐D129E/D‐V154A) were more sensitive to most SDHIs tested relative to mutants carrying single amino acid substitutions (B‐S221P, B‐H267Y and D‐D129E). Mutants carrying variant B‐H267L were highly resistant to all four SDHIs tested. High levels of boscalid resistance were measured for variants carrying B‐C137R, B‐S221P (only IPO323‐derived mutants), B‐H267F, H267Y, C‐N86K, D‐D129E and D‐D129G. Sensitivity levels to bixafen and isopyrazam were similar for mutants carrying different Sdh variants, with the exception of B‐I269V and B‐S221T mutants that were inhibited more strongly with bixafen than isopyrazam. Interestingly, high levels of resistance to both bixafen and isopyrazam were only measured for mutants carrying B‐H267L or C‐N86K.
In planta fitness studies on SDHI‐resistant UV mutants of M.
planta fitness studies on SDHI‐resistant UV mutants of M. graminicola
graminicola
A selection of field strains (BC1, BC4 and IRE30) and Sdh mutants derived from IRE30 were further examined with regard to their in planta pathogenicity and the ability to produce spores (Table
planta pathogenicity and the ability to produce spores (Table 5). After 21 days, the inoculated areas were completely necrotic and covered with pycnidia for the three field isolates. All mutants produced lesions with pycnidia, but some areas of green leaf tissue were still observed within the inoculated areas. The largest areas of necrosis were observed for mutants M74 and M79 (data not shown). In comparison with IRE30, where 36
5). After 21 days, the inoculated areas were completely necrotic and covered with pycnidia for the three field isolates. All mutants produced lesions with pycnidia, but some areas of green leaf tissue were still observed within the inoculated areas. The largest areas of necrosis were observed for mutants M74 and M79 (data not shown). In comparison with IRE30, where 36 ng of pathogen DNA were detected per 50
ng of pathogen DNA were detected per 50 ng of total DNA after 25 days, the level of pathogen DNA was lower for the mutants, with measured values between 5 and 30
ng of total DNA after 25 days, the level of pathogen DNA was lower for the mutants, with measured values between 5 and 30 ng. However, half of the mutants tested produced more pycnidiospores than IRE30 after 21 days. The amount of spores harvested per leaf after 21 days was at least five‐fold higher than the initial amount of spores inoculated for all mutants and strains tested. Although some small differences in symptom expression, fungal growth and spore production were measured for individual mutants, these differences were not linked to a particular Sdh variant (e.g. B‐S221P, B‐H267N, B‐H267Y and D‐D129E).
ng. However, half of the mutants tested produced more pycnidiospores than IRE30 after 21 days. The amount of spores harvested per leaf after 21 days was at least five‐fold higher than the initial amount of spores inoculated for all mutants and strains tested. Although some small differences in symptom expression, fungal growth and spore production were measured for individual mutants, these differences were not linked to a particular Sdh variant (e.g. B‐S221P, B‐H267N, B‐H267Y and D‐D129E).
Table 5
In planta fitness studies on field strains and succinate dehydrogenase inhibitor (SDHI)‐resistant laboratory mutants of Mycosphaerella graminicola.
planta fitness studies on field strains and succinate dehydrogenase inhibitor (SDHI)‐resistant laboratory mutants of Mycosphaerella graminicola.
| Strains/mutants* | SDHI‐resistant Sdh alterations† | Amount of DNA (pg)‡ | Amount of spores§ | |||
|---|---|---|---|---|---|---|
| 7 days | 14 days | 21 days | 25 days | |||
| BC1 | 43 ± ± 43 43 | 2544 ± ± 17 17 | 6 047 047 ± ± 1 1 668 668 | 32 270 270 ± ± 1 1 750 750 | 3.67E + 05 | |
| BC4 | 334 ± ± 50 50 | 1625 ± ± 774 774 | 7 419 419 ± ± 1 1 663 663 | 29 010 010 ± ± 2 2 000 000 | 3.58E + 05 | |
| IRE30 | 184 ± ± 44 44 | 100 ± ± 1.8 1.8 | 17 135 135 ± ± 615 615 | 36 245 245 ± ± 1 1 495 495 | 3.00E + 05 | |
| M77 | B‐S221P | 248 ± ± 49 49 | 1658 ± ± 487 487 | 1 968 968 ± ± 114 114 | 15 330 330 ± ± 360 360 | 3.33E + 05 |
| M99 | B‐S221P | 57 ± ± 12 12 | 148 ± ± 15 15 | 10 749 749 ± ± 791 791 | 17 115 115 ± ± 145 145 | 2.67E + 05 |
| M125 | B‐S221P | 169 ± ± 0.5 0.5 | 100 ± ± 29 29 | 4 556 556 ± ± 941 941 | 5 334 334 ± ± 2 2 168 168 | 2.17E + 05 |
| M73 | B‐H267N | 281 ± ± 7.6 7.6 | 73.4 ± ± 2.3 2.3 | 12 140 140 ± ± 860 860 | 21 045 045 ± ± 2 2 015 015 | 5.67E + 05 |
| M126 | B‐H267N | 143 ± ± 16 16 | 229 ± ± 48 48 | 3 174 174 ± ± 205 205 | 14 630 630 ± ± 440 440 | 6.50E + 05 |
| M74 | B‐H267Y | 290 ± ± 45 45 | 1482 ± ± 74 74 | 12 500 500 ± ± 890 890 | 16 115 115 ± ± 275 275 | 4.17E + 05 |
| M79 | B‐H267Y | 252 ± ± 31 31 | 419 ± ± 104 104 | 8 086 086 ± ± 81 81 | 29 770 770 ± ± 3 3 310 310 | 2.92E + 05 |
| M80 | B‐H267Y | 63 ± ± 6.7 6.7 | 189 ± ± 31 31 | 9 490 490 ± ± 2 2 820 820 | 12 384 384 ± ± 2 2 787 787 | 1.42E + 05 |
| M121 | B‐H267Y | 256 ± ± 39 39 | 572 ± ± 3.7 3.7 | 4 641 641 ± ± 15 15 | 9 479 479 ± ± 1 1 251 251 | 4.42E + 05 |
| M81 | D‐D129E | 159 ± ± 22 22 | 344 ± ± 52 52 | 6 831 831 ± ± 812 812 | 24 370 370 ± ± 0 0 | 1.33E + 05 |
 ng of total DNA extracted from inoculated 2‐cm segments of four leaves. Mean values of two measurements
ng of total DNA extracted from inoculated 2‐cm segments of four leaves. Mean values of two measurements ±
± standard errors are given.
standard errors are given.DISCUSSION
With the exception of B‐C137R, the resistance‐related amino acid substitutions discovered in the mutant library are all positioned around the quinone binding pocket of the Sdh complex of M. graminicola formed by subunits B, C and D. Most (e.g. B‐S221P/T, B‐H267F/L/N/Y, B‐I269V and D‐D129E/G/T) are directly involved in forming the cavity in which SDHI fungicides bind. Docked carboxin (Fig.
graminicola formed by subunits B, C and D. Most (e.g. B‐S221P/T, B‐H267F/L/N/Y, B‐I269V and D‐D129E/G/T) are directly involved in forming the cavity in which SDHI fungicides bind. Docked carboxin (Fig. 2) is positioned in a similar manner as found in the crystal structure of the avian Sdh complex (Huang et
2) is positioned in a similar manner as found in the crystal structure of the avian Sdh complex (Huang et al., 2006). This confirms that the docking procedure provides reasonable binding mode predictions. Based on the binding energies, the model predicts that bixafen and isopyrazam will bind more strongly than boscalid, whereas carboxin will be less effective. The model can also predict the binding efficacies of SDHI fungicides to mutated Sdh variants (Table
al., 2006). This confirms that the docking procedure provides reasonable binding mode predictions. Based on the binding energies, the model predicts that bixafen and isopyrazam will bind more strongly than boscalid, whereas carboxin will be less effective. The model can also predict the binding efficacies of SDHI fungicides to mutated Sdh variants (Table 2). The prediction that B‐H267L will be resistant to most SDHIs tested was in line with fungicide sensitivity testing (Table
2). The prediction that B‐H267L will be resistant to most SDHIs tested was in line with fungicide sensitivity testing (Table 3). Docking studies also demonstrated a different binding for fluopyram with the Sdh complex of M.
3). Docking studies also demonstrated a different binding for fluopyram with the Sdh complex of M. graminicola ((2,2, ,3),3), a finding consistent with the absence of cross‐resistance to fluopyram in highly boscalid‐resistant isolates of A.
graminicola ((2,2, ,3),3), a finding consistent with the absence of cross‐resistance to fluopyram in highly boscalid‐resistant isolates of A. alternata (Avenot and Michailides, 2010), C.
alternata (Avenot and Michailides, 2010), C. cassiicola and Podosphaera xanthii (Ishii et
cassiicola and Podosphaera xanthii (Ishii et al., 2011). However, the model was not always consistent with biological data, in particular for B‐H267Y, where stronger binding was predicted for carboxin and bixafen. In addition, the predicted binding modes for isopyrazam and penthiopyrad were unexpected because the orientation of the amide bond was found to be maintained across carboxamides in docking studies performed at Syngenta (G. Scalliet, personal communication). For isopyrazam, the pyrazole ring was predicted to be oriented towards the bottom of the quinone binding site. The docking simulations used in this study were performed without any bias towards a known binding mode of related fungicides. Further refinement of the model, taking into account residues at longer distances and specific interactions between inhibitors and key residues, will further improve the predictive power.
al., 2011). However, the model was not always consistent with biological data, in particular for B‐H267Y, where stronger binding was predicted for carboxin and bixafen. In addition, the predicted binding modes for isopyrazam and penthiopyrad were unexpected because the orientation of the amide bond was found to be maintained across carboxamides in docking studies performed at Syngenta (G. Scalliet, personal communication). For isopyrazam, the pyrazole ring was predicted to be oriented towards the bottom of the quinone binding site. The docking simulations used in this study were performed without any bias towards a known binding mode of related fungicides. Further refinement of the model, taking into account residues at longer distances and specific interactions between inhibitors and key residues, will further improve the predictive power.
The diversity and high frequency of particular resistant mutants generated by UV mutagenesis demonstrate a potentially high risk for resistance development in M. graminicola to SDHIs. In total, 16 key amino acid substitutions conferring resistance to SDHIs were identified at five different positions in SdhB (137, 221, 265, 267 and 269) and two each in SdhC (85 and 86) and SdhD (127 and 129). In addition, several other amino acid substitutions (B‐I13V, B‐P155L, B‐D166G, C‐R54G, D‐V96A and D‐V154A) were found in combination with these 16, with most contributing to a more sensitive phenotype. Sixty‐four of the 124 mutants isolated and characterized carried B‐H267Y. Because B‐H267Y variants have lower carboxin resistance than B‐H267L variants, only nine of which were isolated, a higher fitness cost for B‐H267L variants seems likely. Shima et
graminicola to SDHIs. In total, 16 key amino acid substitutions conferring resistance to SDHIs were identified at five different positions in SdhB (137, 221, 265, 267 and 269) and two each in SdhC (85 and 86) and SdhD (127 and 129). In addition, several other amino acid substitutions (B‐I13V, B‐P155L, B‐D166G, C‐R54G, D‐V96A and D‐V154A) were found in combination with these 16, with most contributing to a more sensitive phenotype. Sixty‐four of the 124 mutants isolated and characterized carried B‐H267Y. Because B‐H267Y variants have lower carboxin resistance than B‐H267L variants, only nine of which were isolated, a higher fitness cost for B‐H267L variants seems likely. Shima et al. (2009) reported different levels of succinate‐cytochrome c reductase activity from crude mitochondrial fractions of wild‐type and carboxin‐resistant UV mutants of As.
al. (2009) reported different levels of succinate‐cytochrome c reductase activity from crude mitochondrial fractions of wild‐type and carboxin‐resistant UV mutants of As. oryzae, including counterparts of the M.
oryzae, including counterparts of the M. graminicola B‐H267Y, B‐H267L, B‐H276N and D‐D129E variants. The basal SDH activity of all mutants was significantly lower than that of the wild‐type, with the lowest relative activity measured for B‐H267L (18%) and the highest for D‐D129E (56%). In the presence of 100
graminicola B‐H267Y, B‐H267L, B‐H276N and D‐D129E variants. The basal SDH activity of all mutants was significantly lower than that of the wild‐type, with the lowest relative activity measured for B‐H267L (18%) and the highest for D‐D129E (56%). In the presence of 100 µm carboxin, the basal SDH activity of B‐H267L was fully retained, whereas the basal activity of D‐D129E was reduced to 33%. The isolate‐dependent SDHI sensitivity phenotype with B‐S221P mutants cannot be explained by differences in the amino acid sequences of the SdhA, SdhB and SdhC subunits. In comparison with IPO323, only one residue is different in IRE30 (D‐R47P). It is likely that other changes in the mitochondrial electron transport chain are responsible. IRE30 is unique because it carries cytochrome b F129L alleles which, in comparison with G143A alleles, confer intermediate levels of resistance to QoI fungicides (Lucas and Fraaije, 2008). Fisher et
µm carboxin, the basal SDH activity of B‐H267L was fully retained, whereas the basal activity of D‐D129E was reduced to 33%. The isolate‐dependent SDHI sensitivity phenotype with B‐S221P mutants cannot be explained by differences in the amino acid sequences of the SdhA, SdhB and SdhC subunits. In comparison with IPO323, only one residue is different in IRE30 (D‐R47P). It is likely that other changes in the mitochondrial electron transport chain are responsible. IRE30 is unique because it carries cytochrome b F129L alleles which, in comparison with G143A alleles, confer intermediate levels of resistance to QoI fungicides (Lucas and Fraaije, 2008). Fisher et al. (2004) showed that modified cytochrome b quinone outside sites can affect cytochrome c reductase activity using yeast cytochrome b as a model. The combination of SdhB S221P and cytochrome b F129L might have a more deleterious effect on fitness than other combinations of Sdh variants and cytochrome b alleles. Increased in
al. (2004) showed that modified cytochrome b quinone outside sites can affect cytochrome c reductase activity using yeast cytochrome b as a model. The combination of SdhB S221P and cytochrome b F129L might have a more deleterious effect on fitness than other combinations of Sdh variants and cytochrome b alleles. Increased in vitro sensitivity to boscalid has also been reported for QoI reduced‐sensitive field strains of Alternaria solani carrying cytochrome b F129L alleles (Pasche et
vitro sensitivity to boscalid has also been reported for QoI reduced‐sensitive field strains of Alternaria solani carrying cytochrome b F129L alleles (Pasche et al., 2005) and QoI‐resistant laboratory mutants of B.
al., 2005) and QoI‐resistant laboratory mutants of B. cinerea (Markoglou et
cinerea (Markoglou et al., 2006). One IPO323‐derived mutant (M152) was less resistant to carboxin than other mutants carrying B‐S221P, and showed increased sensitivity to bixafen and isopyrazam. For this mutant, additional mutations causing changes in other components of the mitochondrial electron transport chain cannot be excluded.
al., 2006). One IPO323‐derived mutant (M152) was less resistant to carboxin than other mutants carrying B‐S221P, and showed increased sensitivity to bixafen and isopyrazam. For this mutant, additional mutations causing changes in other components of the mitochondrial electron transport chain cannot be excluded.
The field strains were all well inhibited in vitro with the multi‐site inhibitor chlorothalonil, with EC50 recorded between 0.040 and 0.336
vitro with the multi‐site inhibitor chlorothalonil, with EC50 recorded between 0.040 and 0.336 µg/mL. Ranges for the SDHIs were tight, but some strains, G303 and R39‐1, in particular, showed slightly increased EC50 values for one or more SDHIs. Sequencing revealed that strain R39‐1 carried two unique alterations in SdhC, I29V and R54G. These residues are too far (>25
µg/mL. Ranges for the SDHIs were tight, but some strains, G303 and R39‐1, in particular, showed slightly increased EC50 values for one or more SDHIs. Sequencing revealed that strain R39‐1 carried two unique alterations in SdhC, I29V and R54G. These residues are too far (>25 Å) from the quinone binding site to have a direct effect on fungicide binding. However, C‐R54G was also found in combination with B‐S221P in one mutant and increased the sensitivity to boscalid and bixafen, in comparison with most IPO323‐derived mutants carrying B‐S221P alone (Table
Å) from the quinone binding site to have a direct effect on fungicide binding. However, C‐R54G was also found in combination with B‐S221P in one mutant and increased the sensitivity to boscalid and bixafen, in comparison with most IPO323‐derived mutants carrying B‐S221P alone (Table 3). Intensive monitoring since 2003 of M.
3). Intensive monitoring since 2003 of M. graminicola field strains for SDHI sensitivity by agrochemical companies has shown a fully sensitive picture (FRAC SDHI Working Group, unpublished). A wide range of EC50 values were measured for epoxiconazole (<0.0005 to 1.5
graminicola field strains for SDHI sensitivity by agrochemical companies has shown a fully sensitive picture (FRAC SDHI Working Group, unpublished). A wide range of EC50 values were measured for epoxiconazole (<0.0005 to 1.5 µg/mL) and prothioconazole‐desthio (0.0006–0.180
µg/mL) and prothioconazole‐desthio (0.0006–0.180 µg/mL). Prothioconazole‐desthio is approximately 100‐fold more active than prothioconazole (B. A. Fraaije, unpublished), and in
µg/mL). Prothioconazole‐desthio is approximately 100‐fold more active than prothioconazole (B. A. Fraaije, unpublished), and in planta and/or in
planta and/or in vivo metabolism of prothioconazole by the fungus itself has been suggested to contribute to the inhibition of CYP51 (Parker et
vivo metabolism of prothioconazole by the fungus itself has been suggested to contribute to the inhibition of CYP51 (Parker et al., 2011). Increased sensitivity to azoles, both in
al., 2011). Increased sensitivity to azoles, both in vitro and in
vitro and in planta (Fraaije et
planta (Fraaije et al., 2011), has been directly linked to changes in the CYP51 target protein, and novel CYP51 variants are continuously evolving (Brunner et
al., 2011), has been directly linked to changes in the CYP51 target protein, and novel CYP51 variants are continuously evolving (Brunner et al., 2008; Cools and Fraaije, 2008; Cools et
al., 2008; Cools and Fraaije, 2008; Cools et al., 2011; Leroux and Walker, 2011). There is a concern that the activity of the most active azoles might erode to such an extent that field performance will be compromised, as reported for some older azoles (Clark, 2006). The use of mixtures as a resistance management strategy will only work with effective noncross‐resistant fungicides acting simultaneously at key fungal developmental stages. Further monitoring of sensitivity shifts to both SDHI and DMI fungicides is required, and an improved understanding of the underlying resistance mechanisms will lead to the exploitation of the chemical diversity within these fungicide classes.
al., 2011; Leroux and Walker, 2011). There is a concern that the activity of the most active azoles might erode to such an extent that field performance will be compromised, as reported for some older azoles (Clark, 2006). The use of mixtures as a resistance management strategy will only work with effective noncross‐resistant fungicides acting simultaneously at key fungal developmental stages. Further monitoring of sensitivity shifts to both SDHI and DMI fungicides is required, and an improved understanding of the underlying resistance mechanisms will lead to the exploitation of the chemical diversity within these fungicide classes.
Although these laboratory studies cannot provide direct evidence of the potential for field resistance, the knowledge arising from this investigation will enable the prediction of which mutations are most likely to be selected in field populations by different SDHIs, and also aid in the development of DNA‐based diagnostics based on allele‐specific PCR. In addition to the sequence information on Sdh complexes in field strains of M. graminicola, different mutations conferring resistance to SDHIs were identified. The early detection of resistant alleles at low frequencies, coupled with risk evaluation, will allow the implementation of anti‐resistance strategies to prolong the lifetime and cost‐effectiveness of this class of fungicides.
graminicola, different mutations conferring resistance to SDHIs were identified. The early detection of resistant alleles at low frequencies, coupled with risk evaluation, will allow the implementation of anti‐resistance strategies to prolong the lifetime and cost‐effectiveness of this class of fungicides.
EXPERIMENTAL PROCEDURES
Isolation of fungal strains
Field population sampling and M. graminicola isolation from plants were performed as described by Fraaije et
graminicola isolation from plants were performed as described by Fraaije et al. (2007). Conidial suspensions were streaked onto yeast extract peptone dextrose agar (YEPD agar; ForMedium, Norwich, Norfolk, UK) amended with penicillin G sodium and streptomycin sulphate at 100
al. (2007). Conidial suspensions were streaked onto yeast extract peptone dextrose agar (YEPD agar; ForMedium, Norwich, Norfolk, UK) amended with penicillin G sodium and streptomycin sulphate at 100 µg/mL, and incubated for 5 days at 23
µg/mL, and incubated for 5 days at 23 °C. Single colonies subsequently grew in the form of yeast‐like budding cells. Single spore strains were propagated by transferring single colonies to fresh YEPD agar plates. Strains were either used directly in sensitivity assays or stored in 80% glycerol at −80
°C. Single colonies subsequently grew in the form of yeast‐like budding cells. Single spore strains were propagated by transferring single colonies to fresh YEPD agar plates. Strains were either used directly in sensitivity assays or stored in 80% glycerol at −80 °C. All strains used in this study are listed in Table
°C. All strains used in this study are listed in Table 1.
1.
Generation of SDHI‐resistant UV mutants
Ten thousand spores of M. graminicola strains IPO323 or IRE30 were plated out onto YEPD agar amended with carboxin at 50
graminicola strains IPO323 or IRE30 were plated out onto YEPD agar amended with carboxin at 50 µg/mL. Plates were exposed to UV light at 500 or 600
µg/mL. Plates were exposed to UV light at 500 or 600 J/m2 using a Stratagene UV cross‐linker (model 1800; Stratagene Corporation, La Jolla, CA, USA), which resulted in approximately 20% survival. Putative carboxin‐resistant mutants were isolated from growing colonies after 14 days of incubation at 21
J/m2 using a Stratagene UV cross‐linker (model 1800; Stratagene Corporation, La Jolla, CA, USA), which resulted in approximately 20% survival. Putative carboxin‐resistant mutants were isolated from growing colonies after 14 days of incubation at 21 °C in the dark. These mutants were grown for another generation on carboxin‐amended YEPD agar, harvested in sterile distilled water and stored in 80% (v/v) glycerol at −80
°C in the dark. These mutants were grown for another generation on carboxin‐amended YEPD agar, harvested in sterile distilled water and stored in 80% (v/v) glycerol at −80 °C for further use.
°C for further use.
Isolation of fungal genomic DNA, PCR and DNA sequencing
Genomic DNA for PCR was extracted and quantified according to the protocol of Rudd et al. (2010). To amplify genes encoding for SdhB, SdhC and SdhD, specific primers for each subunit (see Table
al. (2010). To amplify genes encoding for SdhB, SdhC and SdhD, specific primers for each subunit (see Table 6) were designed based on the available genome sequence for strain IPO323 downloaded from the Joint Genome Institute (JGI) M.
6) were designed based on the available genome sequence for strain IPO323 downloaded from the Joint Genome Institute (JGI) M. graminicola genome website (URL: http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html). PCR was carried out in an Applied Biosystems (Foster City, CA, USA) Veriti 96‐well thermal cycler in a final volume of 30
graminicola genome website (URL: http://genome.jgi‐psf.org/Mycgr3/Mycgr3.home.html). PCR was carried out in an Applied Biosystems (Foster City, CA, USA) Veriti 96‐well thermal cycler in a final volume of 30 µL containing 2.5 units of Easy‐A high‐fidelity PCR cloning enzyme (Stratagene Corporation), 3
µL containing 2.5 units of Easy‐A high‐fidelity PCR cloning enzyme (Stratagene Corporation), 3 µL of 10 × Easy‐A reaction buffer, 200
µL of 10 × Easy‐A reaction buffer, 200 µm of each deoxynucleoside triphosphate (dNTP), 1.0
µm of each deoxynucleoside triphosphate (dNTP), 1.0 µm of each primer and 30
µm of each primer and 30 ng of fungal template DNA. Amplification conditions consisted of 94
ng of fungal template DNA. Amplification conditions consisted of 94 °C for 2
°C for 2 min, followed by 35 cycles at 94
min, followed by 35 cycles at 94 °C for 20
°C for 20 s, 57
s, 57 °C (SdhB), 66
°C (SdhB), 66 °C (SdhC) or 58
°C (SdhC) or 58 °C (SdhD) for 30
°C (SdhD) for 30 s, and 72
s, and 72 °C for 2
°C for 2 min, with a final DNA extension at 72
min, with a final DNA extension at 72 °C for 8
°C for 8 min. PCR products were separated in ethidium bromide‐stained 1% agarose gels run in 1 × tris(hydroxymethyl)aminomethane (Tris)–borate–ethylenediaminetetraacetic acid (EDTA) buffer and exposed to UV light to visualize DNA fragments. PCR products were cloned into pGEM‐T easy vector (Promega Corporation, Madison, WI, USA) according to the manufacturer's instructions, and subsequently sequenced with M13 sequence primers using a dideoxy chain termination reaction method. Sequences were assembled and further aligned with Vector NTI and ClustalW software.
min. PCR products were separated in ethidium bromide‐stained 1% agarose gels run in 1 × tris(hydroxymethyl)aminomethane (Tris)–borate–ethylenediaminetetraacetic acid (EDTA) buffer and exposed to UV light to visualize DNA fragments. PCR products were cloned into pGEM‐T easy vector (Promega Corporation, Madison, WI, USA) according to the manufacturer's instructions, and subsequently sequenced with M13 sequence primers using a dideoxy chain termination reaction method. Sequences were assembled and further aligned with Vector NTI and ClustalW software.
Table 6
Polymerase chain reaction (PCR) primers to amplify succinate dehydrogenase (Sdh) genes from Mycosphaerella graminicola.
| Primers | Target | Sequence (5′–3′) | Amplicon size (bp) |
|---|---|---|---|
| Mgsdhbf1 | SdhB | ACTCTTCTCACATACCACACA | 1051 |
| Mgsdhbr1 | CTTTCCAATCATCTCGTTCCAT | ||
| Mgsdhcf1 | SdhC | GGCACATCGCGTCTCACG | 736 |
| Mgsdhcr1 | TCCCTCATTTCCCCATCCAC | ||
| Mgsdhdf1 | SdhD | CTCACCCTCACCGTCGCC | 715 |
| Mgsdhdr1 | CCATCTACAACTTCTGCTCAATC |
Phenotyping and genotyping of carboxin‐resistant UV mutants and field strains
Wells of flat‐bottomed microtitre plates (TPP92696, Techno Plastic Products, Trasadingen, Switzerland) were filled with 100 µL of double‐strength Sabouraud dextrose liquid medium (SDLM; Oxoid, Basingstoke, Hampshire, UK) without fungicide or amended with different fungicide concentrations. The following concentrations were used to test field strains: bixafen, boscalid and isopyrazam (0.00023, 0.00092, 0.0037, 0.0146, 0.0586, 0.234, 0.938, 1.875, 3.75, 7.5 and 15
µL of double‐strength Sabouraud dextrose liquid medium (SDLM; Oxoid, Basingstoke, Hampshire, UK) without fungicide or amended with different fungicide concentrations. The following concentrations were used to test field strains: bixafen, boscalid and isopyrazam (0.00023, 0.00092, 0.0037, 0.0146, 0.0586, 0.234, 0.938, 1.875, 3.75, 7.5 and 15 µg/mL), carboxin (0.0977, 0.195, 0.391, 0.781, 1.56, 3.13, 6.25, 12.5, 25, 50 and 100
µg/mL), carboxin (0.0977, 0.195, 0.391, 0.781, 1.56, 3.13, 6.25, 12.5, 25, 50 and 100 µg/mL), chlorothalonil (0.0195, 0.0391, 0.0781, 0.156, 0.313, 0.625, 1.25, 2.5, 5, 10 and 20
µg/mL), chlorothalonil (0.0195, 0.0391, 0.0781, 0.156, 0.313, 0.625, 1.25, 2.5, 5, 10 and 20 µg/mL), epoxiconazole (0.00014, 0.00051, 0.00192, 0.0072, 0.0270, 0.101, 0.379, 1.42, 5.33, 20 and 75
µg/mL), epoxiconazole (0.00014, 0.00051, 0.00192, 0.0072, 0.0270, 0.101, 0.379, 1.42, 5.33, 20 and 75 µg/mL) and prothioconazole‐desthio (Parker et
µg/mL) and prothioconazole‐desthio (Parker et al., 2011), a metabolite of prothioconazole (0.00034, 0.0010, 0.0030, 0.0091, 0.0274, 0.0823, 0.247, 0.741, 2.22, 6.67 and 20
al., 2011), a metabolite of prothioconazole (0.00034, 0.0010, 0.0030, 0.0091, 0.0274, 0.0823, 0.247, 0.741, 2.22, 6.67 and 20 µg/mL). For the mutants, higher concentrations of SDHIs were used: bixafen, boscalid and isopyrazam (0.00076, 0.0031, 0.0122, 0.0488, 0.195, 0.781, 3.13, 12.5 and 25
µg/mL). For the mutants, higher concentrations of SDHIs were used: bixafen, boscalid and isopyrazam (0.00076, 0.0031, 0.0122, 0.0488, 0.195, 0.781, 3.13, 12.5 and 25 µg/mL) and carboxin (0.488, 0.977, 1.95, 3.91, 7.81, 15.6, 31.3, 62.5, 125, 250 and 500
µg/mL) and carboxin (0.488, 0.977, 1.95, 3.91, 7.81, 15.6, 31.3, 62.5, 125, 250 and 500 µg/mL). Before dilution in media, chemicals were dissolved in acetone, with the exception of chlorothalonil, which was dissolved in dimethylsulphoxide (DMSO). One hundred microlitres of spore suspension (2.5 × 104 spores/mL) of M.
µg/mL). Before dilution in media, chemicals were dissolved in acetone, with the exception of chlorothalonil, which was dissolved in dimethylsulphoxide (DMSO). One hundred microlitres of spore suspension (2.5 × 104 spores/mL) of M. graminicola strains or UV mutants were added to each well. Plates were incubated for 4 days at 23
graminicola strains or UV mutants were added to each well. Plates were incubated for 4 days at 23 °C, and growth was measured at 630
°C, and growth was measured at 630 nm using a FLUOstar OPTIMA microplate reader (BMG Labtech, Offenburg, Germany) in well‐scanning mode with a 2 × 2 matrix of scanning points within a 3‐mm diameter. Fungicide sensitivities were determined as the 50% effective concentration to inhibit growth (EC50 in µg/mL) using a dose–response relationship according to OPTIMA software. Based on the phenotyping results, the SdhB, SdhC and SdhD genes of mutants with differences in SDHI sensitivity were sequenced. Sequence analysis identified putative resistance‐conferring mutations. Subsequently, pyrosequencing assays were developed to screen the remaining mutants for the presence of these mutations. The SdhB, SdhC and SdhD genes of mutants testing negative by pyrosequencing for the identified mutations were also sequenced. This was repeated until all mutants were genotyped.
nm using a FLUOstar OPTIMA microplate reader (BMG Labtech, Offenburg, Germany) in well‐scanning mode with a 2 × 2 matrix of scanning points within a 3‐mm diameter. Fungicide sensitivities were determined as the 50% effective concentration to inhibit growth (EC50 in µg/mL) using a dose–response relationship according to OPTIMA software. Based on the phenotyping results, the SdhB, SdhC and SdhD genes of mutants with differences in SDHI sensitivity were sequenced. Sequence analysis identified putative resistance‐conferring mutations. Subsequently, pyrosequencing assays were developed to screen the remaining mutants for the presence of these mutations. The SdhB, SdhC and SdhD genes of mutants testing negative by pyrosequencing for the identified mutations were also sequenced. This was repeated until all mutants were genotyped.
Pyrosequencing assays
Primers (forward primer, reverse primer and sequence primer) were designed with Pyrosequencing Assay Design Software (Version 1.0.6; Biotage, Uppsala, Sweden) targeting different regions of SdhB, SdhC and SdhD in which SDHI resistance‐conferring mutations were identified (Table 7). Polymerase chain reactions (PCRs) were carried out in a Biometra T3 thermocycler (Biotron, Göttingen, Germany) with 0.2 units of Red Hot DNA polymerase (ABgene, Epsom, Surrey, UK) using 20
7). Polymerase chain reactions (PCRs) were carried out in a Biometra T3 thermocycler (Biotron, Göttingen, Germany) with 0.2 units of Red Hot DNA polymerase (ABgene, Epsom, Surrey, UK) using 20 mm (NH4)SO4, 75
mm (NH4)SO4, 75 mm Tris‐HCl (pH 9.0), 0.01% Tween‐20, 1.5
mm Tris‐HCl (pH 9.0), 0.01% Tween‐20, 1.5 mm MgCl2, 150
mm MgCl2, 150 µm of each dNTP, 0.5
µm of each dNTP, 0.5 µm of each primer and 20
µm of each primer and 20 ng template DNA in a final volume of 20
ng template DNA in a final volume of 20 µL. Cycling conditions were as follows: 94
µL. Cycling conditions were as follows: 94 °C for 2
°C for 2 min, followed by 40 cycles at 94
min, followed by 40 cycles at 94 °C for 10
°C for 10 s, 61
s, 61 °C (SdhB codon 221) or 65
°C (SdhB codon 221) or 65 °C (other targets) for 20
°C (other targets) for 20 s and 72
s and 72 °C for 30
°C for 30 s, with a final DNA extension at 72
s, with a final DNA extension at 72 °C for 5
°C for 5 min. Single‐stranded biotinylated PCR products were prepared using the Pyrosequencing Vacuum Prep Tool (Biotage). Three microlitres of Streptavidin Sepharose HP beads (GE Healthcare, Uppsala. Sweden) were added to 40
min. Single‐stranded biotinylated PCR products were prepared using the Pyrosequencing Vacuum Prep Tool (Biotage). Three microlitres of Streptavidin Sepharose HP beads (GE Healthcare, Uppsala. Sweden) were added to 40 µL of binding buffer (10
µL of binding buffer (10 mm Tris‐HCl, pH 7.6, 2 m NaCl, 1
mm Tris‐HCl, pH 7.6, 2 m NaCl, 1 mm EDTA, 0.1% Tween‐20) and mixed with 15
mm EDTA, 0.1% Tween‐20) and mixed with 15 µL of PCR product and 25
µL of PCR product and 25 µL of sterile distilled water for 10
µL of sterile distilled water for 10 min at room temperature using an Orbis plate shaker (Mikura, Lower Beeding, West Sussex, UK). Beads containing the captured DNA product were aspirated onto filters after applying a vacuum, washed with 70% (v/v) ethanol for 5
min at room temperature using an Orbis plate shaker (Mikura, Lower Beeding, West Sussex, UK). Beads containing the captured DNA product were aspirated onto filters after applying a vacuum, washed with 70% (v/v) ethanol for 5 s, rendered single stranded with denaturation solution (0.2 m NaOH) for 10
s, rendered single stranded with denaturation solution (0.2 m NaOH) for 10 s and neutralized with washing buffer (10
s and neutralized with washing buffer (10 mm Tris‐acetate, pH 7.6) for 5
mm Tris‐acetate, pH 7.6) for 5 s. The vacuum was released and beads with biotinylated single‐stranded DNA were transferred into a PSQ 96‐well plate (Biotage) containing 45
s. The vacuum was released and beads with biotinylated single‐stranded DNA were transferred into a PSQ 96‐well plate (Biotage) containing 45 µL of annealing buffer (20
µL of annealing buffer (20 mm Tris‐acetate, 2
mm Tris‐acetate, 2 mm magnesium acetate, pH 7.6) and 0.5
mm magnesium acetate, pH 7.6) and 0.5 mm sequence primer. Pyrosequencing reactions were performed and analysed on a PyroMark Q96ID according to the manufacturer's instructions using a PyroMark Gold Q96 QSA Reagent Kit (Biotage), and the nucleotide dispensation orders are shown in Table
mm sequence primer. Pyrosequencing reactions were performed and analysed on a PyroMark Q96ID according to the manufacturer's instructions using a PyroMark Gold Q96 QSA Reagent Kit (Biotage), and the nucleotide dispensation orders are shown in Table 7.
7.
Table 7
Pyrosequencing assay to detect mutations in SdhB and SdhD.
| Target | Amino acid substitutions | Primer sets* |
|---|---|---|
| SdhB | S221P, S221T | F: CATTCTCTGCGCATGCTG* |
| R: CGTTGATCCATCGGTATGACT | ||
| S: CCGAGTTCCACCAGT | ||
| SdhB | R265P, H267F, H267L, H267N, H267Y, I269V | F: CGCAAGGACGCACTCAACA* |
| R: GGGCAGGTCCTTGAGCAATT | ||
| S: GTCCTTGAGCAATTCAG | ||
| SdhD | I127V, D129E, D129G, D129T | F: CCCACCCGTATCATTCTGACA* |
| R: GAGCTTGCGGGTTTTGGG | ||
| S: CGCCATGAGGGGAAG |
In silico studies on the binding of SDHIs to the Sdh complex of M.
silico studies on the binding of SDHIs to the Sdh complex of M. graminicola
graminicola
For docking studies of fungicides in the M. graminicola Sdh complex, the crystallographic coordinates of the avian Sdh complex were taken from the Protein Data Bank (PDB). The Sdh complex of chicken (PDB code 2FBW) is similar to that of M.
graminicola Sdh complex, the crystallographic coordinates of the avian Sdh complex were taken from the Protein Data Bank (PDB). The Sdh complex of chicken (PDB code 2FBW) is similar to that of M. graminicola (70% sequence identity) and also contains a bound carboxin molecule. By replacing all residues within a 14‐Å radius of the bound carboxin with the corresponding residues of the M.
graminicola (70% sequence identity) and also contains a bound carboxin molecule. By replacing all residues within a 14‐Å radius of the bound carboxin with the corresponding residues of the M. graminicola Sdh complex using PyMOL, a structural model of the fungal quinone binding site was prepared. All fungicides tested were also prepared in PyMOL. The Molegro Virtual Docker (MVD; Molegro ApS, Aarhus, Denmark) was used for docking simulations. For docking simulations, the structural model and all fungicides were first subjected to energy minimization in MVD. For each resistance‐related mutation in one of the subunits, the corresponding mutant protein was built and optimized. Fungicides were docked within a sphere (radius, 12
graminicola Sdh complex using PyMOL, a structural model of the fungal quinone binding site was prepared. All fungicides tested were also prepared in PyMOL. The Molegro Virtual Docker (MVD; Molegro ApS, Aarhus, Denmark) was used for docking simulations. For docking simulations, the structural model and all fungicides were first subjected to energy minimization in MVD. For each resistance‐related mutation in one of the subunits, the corresponding mutant protein was built and optimized. Fungicides were docked within a sphere (radius, 12 Å) that encompassed the quinone binding site. For docking, the default settings were used, except for the grid resolution which was set to 0.2, and a total of 25 runs was performed with a maximum of 500 iterations. The best superimposed positions with lowest MolDock energy scores were used in this study. For the visualization of the docked fungicides, PyMOL was used.
Å) that encompassed the quinone binding site. For docking, the default settings were used, except for the grid resolution which was set to 0.2, and a total of 25 runs was performed with a maximum of 500 iterations. The best superimposed positions with lowest MolDock energy scores were used in this study. For the visualization of the docked fungicides, PyMOL was used.
In planta fitness studies on Sdh mutants and field strains of M.
planta fitness studies on Sdh mutants and field strains of M. graminicola
graminicola
In planta pathogenicity tests were carried out on 2–3‐week‐old wheat seedlings of cultivar Riband according to Keon et
planta pathogenicity tests were carried out on 2–3‐week‐old wheat seedlings of cultivar Riband according to Keon et al. (2007). In total, 18 leaves per strain were inoculated with spore suspensions [2.5 × 105 spores/mL in 0.1% (v/v) Tween‐20] using a mini ‘mist’ sprayer which covered up to a 4‐cm length of the leaf (approximately 100
al. (2007). In total, 18 leaves per strain were inoculated with spore suspensions [2.5 × 105 spores/mL in 0.1% (v/v) Tween‐20] using a mini ‘mist’ sprayer which covered up to a 4‐cm length of the leaf (approximately 100 µL of inoculum per leaf segment). After 72
µL of inoculum per leaf segment). After 72 h of incubation at 100% relative humidity in polystyrene boxes, inoculated plants were incubated at 16
h of incubation at 100% relative humidity in polystyrene boxes, inoculated plants were incubated at 16 °C with a 16‐h light period at 88% relative humidity for up to 25 days to allow symptoms to develop. Visual assessments, recording the extent of pycnidia formation and chlorosis, were conducted 21 days after inoculation. DNA was also extracted from four 2‐cm inoculated leaf segments at several time points (7, 14, 21 and 25 days after inoculation) and the amount of pathogen DNA was quantified in 50‐ng DNA samples according to Fraaije et
°C with a 16‐h light period at 88% relative humidity for up to 25 days to allow symptoms to develop. Visual assessments, recording the extent of pycnidia formation and chlorosis, were conducted 21 days after inoculation. DNA was also extracted from four 2‐cm inoculated leaf segments at several time points (7, 14, 21 and 25 days after inoculation) and the amount of pathogen DNA was quantified in 50‐ng DNA samples according to Fraaije et al. (2005) using the CY5 probe to quantify the presence of the cytochrome b gene of M.
al. (2005) using the CY5 probe to quantify the presence of the cytochrome b gene of M. graminicola. Spores were also counted from three inoculated leaves after 21 days. Three 2‐cm leaf segments were added to 2‐mL tubes and incubated overnight in the dark at room temperature in the presence of 300
graminicola. Spores were also counted from three inoculated leaves after 21 days. Three 2‐cm leaf segments were added to 2‐mL tubes and incubated overnight in the dark at room temperature in the presence of 300 µL of 0.2% Tween‐20 solution. Spores were harvested after two rounds of spinning and shaking at 4.0
µL of 0.2% Tween‐20 solution. Spores were harvested after two rounds of spinning and shaking at 4.0 m/s in a FastPrep machine (Savants Instruments, Holbrook, NY, USA). The amount of spores collected was determined microscopically using a haemocytometer.
m/s in a FastPrep machine (Savants Instruments, Holbrook, NY, USA). The amount of spores collected was determined microscopically using a haemocytometer.
ACKNOWLEDGEMENTS
Part of this work was supported by the Department of Environment, Food and Rural Affairs (DEFRA) through the Sustainable Arable LINK programme, project LK09133, in collaboration with Home‐Grown Cereals Authority (HGCA), BASF, Bayer CropScience, DuPont, Syngenta, Velcourt, Scottish Agricultural College (SAC) and Agricultural Development Advisory Service (ADAS). Rothamsted Research receives grant‐aided support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK. Thanks are also due to Gert Kema, Andreas Mehl, Mike Shaw, Gerd Stammler and Suvi Viljanen‐Rollinson for supplying M. graminicola strains.
graminicola strains.
REFERENCES
- Avenot, H.F. and Michailides, T.J. (2010) Progress in understanding molecular mechanisms and evolution of resistance to succinate inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop Prot. 29, 643–651. [Google Scholar]
- Avenot., H. , Sellam, A. , Karaoglanidis, G. and Michailides, T. (2008) Characterization of mutations in the iron–sulphur subunit of succinate dehydrogenase correlating with boscalid resistance in Alternaria alternata from California pistachio. Phytopathology, 98, 736–742. [Abstract] [Google Scholar]
- Avenot, H. , Sellam, A. and Michailides, T. (2009) Characterization of mutations in the membrane‐anchored subunits AaSDHC and AaSDHD of succinate dehydrogenase from Alternaria alternata isolates conferring resistance to the fungicide boscalid. Plant Pathol. 58, 1134–1143. [Google Scholar]
- Bearchell, S.J. , Fraaije, B.A. , Shaw, M.W. and Fitt, B.D.L. (2005) Wheat archive links long‐term fungal population dynamics to air pollution. Proc. Natl. Acad. Sci. USA, 102, 5438–5442. [Europe PMC free article] [Abstract] [Google Scholar]
- Berry, P.M , Kindred, D.R. and Paveley, N.D. (2008) Quantifying the effects of fungicides and disease resistance on the greenhouse gas emissions associated with wheat production. Plant Pathol. 57, 1000–1008. [Google Scholar]
- Brunner, P.C. , Stefenato, F.L. and McDonald, B.A. (2008) Evolution of the CYP51 gene in Mycosphaerella graminicola: evidence for intragenic recombination and selective replacement. Mol. Plant Pathol. 9, 305–316. [Europe PMC free article] [Abstract] [Google Scholar]
- Clark, W.S. (2006) Septoria tritici and azole performance. In: Fungicide resistance: are we winning the battle but losing the war? Aspects Appl. Biol. 78, 127–132. [Google Scholar]
- Cools, H.J. and Fraaije, B.A. (2008) Mechanisms of resistance to azole fungicides in Mycosphaerella graminicola . Pest Manag. Sci. 64, 681–684. [Abstract] [Google Scholar]
- Cools, H.J. , Fraaije, B.A. , Parker, J.E. , Kelly, D.E. , Lucas, J.A. and Kelly, S.L. (2011) Impact of recently emerged sterol 14α‐demethylase (CYP51) variants of Mycosphaerella graminicola on azole fungicide sensitivity. Appl. Environ. Microbiol. 77, 3830–3837. [Europe PMC free article] [Abstract] [Google Scholar]
- Fisher, N. , Brown, A.C. , Sexton, G. , Cook, A. , Windass, J. and Meunier, B. (2004) Modelling the Qo site of crop pathogens in Saccharomyces cerevisiae cytochrome b . Eur. J. Biochem. 271, 2264–2271. [Abstract] [Google Scholar]
- Fraaije, B.A. , Cools, H.J. , Fountaine, J. , Lovell, D.J. , Motteram, J. , West, J.S. and Lucas, J.A. (2005) The role of ascospores in further spread of QoI‐resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola . Phytopathology, 95, 933–941. [Abstract] [Google Scholar]
- Fraaije, B.A. , Cools, H.J. , Kim, S.‐H. , Motteram, J. , Clark, W.S. and Lucas, J.A. (2007) A novel substitution I381V in the sterol 14α‐demethylase (CYP51) of Mycosphaerella graminicola is differentially selected by azole fungicides. Mol. Plant Pathol. 8, 245–254. [Abstract] [Google Scholar]
- Fraaije, B.A. , Burnett, F.J. , Clark, W.S. , Paveley, N. , Norman, K. and Lucas, J.A. (2011) Understanding Evolution and Selection of Azole Resistance Mechanisms in UK Populations of Mycosphaerella graminicola. London: Agriculture & Horticulture Development Board. HGCA project report 475. [Google Scholar]
- Griffin, M.J. and Fischer, N. (1985) Laboratory studies on benzimidazole resistance in Septoria tritici . EPPO Bull. 15, 505–511. [Google Scholar]
- Hardwick, N.V. , Jones, D.R. and Royle, D.J. (2001) Factors affecting diseases of winter wheat in England and Wales, 1989–98. Plant Pathol. 50, 453–462. [Google Scholar]
- Huang, L.‐S. , Sun, G. , Cobessi, D. , Wang, A.C. , Shen, J.T. , Tung, E.Y. , Anderson, V.E. and Berry, E.A. (2006) 3‐Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J. Biol. Chem. 281, 5965–5972. [Europe PMC free article] [Abstract] [Google Scholar]
- Ishii, H. , Miyamoto, T. , Ushio, S. and Kakishima, M. (2011) Lack of cross‐resistance to a novel succinate dehydrogenase inhibitor, fluopyram, in highly boscalid‐resistant isolates of Corynespora cassiicola and Podosphaera xanthii . Pest Manag. Sci. 67, 474–482. [Abstract] [Google Scholar]
- Ito, Y. , Muraguchi, H. , Seshime, Y. , Oita, S. and Yanagi, S.O. (2004) Flutolanil and carboxin resistance in Coprinus cinereus conferred by a mutation in the cytochrome b 560 subunit of succinate dehydrogenase complex (complex II). Mol. Gen. Genomics, 272, 328–335. [Abstract] [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J. , Baker, J.M. , Beale, M.H. , Hammond‐Kosack, K. and Rudd, J.J. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant–Microbe Interact. 20, 179–193. [Abstract] [Google Scholar]
- Leroux, P. and Walker, A.S. (2011) Multiple mechanisms account for resistance to sterol 14α‐demethylation inhibitors in field isolates of Mycosphaerella graminicola . Pest Manag. Sci. 67, 44–59. [Abstract] [Google Scholar]
- Leroux, P. , Albertini, C. , Gautier, A. , Gredt, M. and Walker, A.S. (2007) Mutations in the CYP51 gene correlated with changes in sensitivity to sterol 14 alpha‐demethylation inhibitors in field isolates of Mycosphaerella graminicola . Pest Manag. Sci. 63, 688–698. [Abstract] [Google Scholar]
- Leroux, P. , Gredt, M. , Leroch, M. and Walker, A.S. (2010) Exploring mechanisms of resistance to respiratory inhibitors in field strains of Botrytis cinerea, the causal agent of grey mold. Appl. Environ. Microbiol. 76, 6615–6630. [Europe PMC free article] [Abstract] [Google Scholar]
- Lucas, J.A. and Fraaije, B.A. (2008) QoI resistance in Mycosphaerella graminicola: what have we learned so far? In: Modern Fungicides and Antifungal Compounds V (Dehne H.W., Deising H.B., Gisi U., Kuck K.‐H., Russell P.E. and Lyr H., eds), pp. 71–77. Braunschweig: Deutsche Phytomedizinische Gesellschaft. [Google Scholar]
- Markoglou, A.N. , Malandrakis, A.A. , Vitoratos, A.G. and Ziogas, B.N. (2006) Characterization of laboratory mutants of Botrytis cinerea resistant to QoI fungicides. Eur. J. Plant Pathol. 115, 149–162. [Google Scholar]
- Matsson, M. , Ackrell, B.A. , Cochran, B. and Hederstedt, L. (1998) Carboxin resistance in Paracoccus denitrificans conferred by a mutation in the membrane‐anchor domain of succinate: quinine reductase. Arch. Microbiol. 170, 27–37. [Abstract] [Google Scholar]
- Miyamoto, T. , Ishii, H. , Stammler, G. , Koch, A. , Ogawara, T. , Tomita, Y. , Fountaine, J.M. , Ushio, S. , Seko, T. and Kobori, S. (2010) Distribution and molecular characterization of Corynespora cassiicola isolates resistant to boscalid. Plant Pathol. 59, 873–881. [Google Scholar]
- Parker, J.E. , Warrilow, A.G.S. , Cools, H.J. , Martel, C.M. , Nes, W.D. , Fraaije, B.A. , Lucas, J.A. , Kelly, D.E. and Kelly, S.L. (2011) Mechanism of binding of prothioconazole to Mycosphaerella graminicola CYP51 differs from that of other azole antifungals. Appl. Environ. Microbiol. 77, 1460–1465. [Europe PMC free article] [Abstract] [Google Scholar]
- Pasche, J.S. , Piche, L.M. and Gudmestead, N.C. (2005) Effect of the F129L mutation in Alternaria solani on fungicides affecting mitochondrial respiration. Plant Dis. 89, 269–278. [Abstract] [Google Scholar]
- Rudd, J. , Antoniw, J. , Marshall, R. , Motteram, J. , Fraaije, B. and Hammond‐Kosack, K. (2010) Identification and characterisation of Mycosphaerella graminicola secreted or surface‐associated proteins with variable intragenic coding repeats. Fungal Genet. Biol. 47, 19–32. [Abstract] [Google Scholar]
- Shima, Y. , Ito, Y. , Kaneko, S. , Hatabayashi, H. , Watanabe, Y. , Adachi, Y. and Yabe, K. (2009) Identification of three mutant loci conferring carboxin‐resistance and development of a novel transformation system in Aspergillus oryzae . Fungal Genet. Biol. 46, 67–76. [Abstract] [Google Scholar]
- Skinner, W. , Bailey, A. , Renwick, A. , Keon, J. , Gurr, S. and Hargreaves, J. (1998) A single amino‐acid substitution in the iron–sulphur protein subunit of succinate dehydrogenase determines resistance to carboxin in Mycosphaerella graminicola (Septoria tritici). Curr. Genet. 34, 393–398. [Abstract] [Google Scholar]
- Stammler, G. , Brix, H.D. , Nave, B. , Gold, R. and Schoefl, U. (2008) Studies on biological performance of boscalid and its mode of action In:Modern Fungicides and Antifungal Compounds V (Dehne H.W., Deising H.B., Gisi U., Kuck K.‐H., Russell P.E. and Lyr H., eds), pp. 45–51. Braunschweig: Deutsche Phytomedizinische Gesellschaft. [Google Scholar]
- White, G.A. , Thorn, G.D. and Georgopoulos, S.G. (1978) Oxathiin carboxamides highly active against carboxin‐resistant succinic dehydrogenase complexes from carboxin‐selected mutants of Ustilago maydis and Aspergillus nidulans . Pestic. Biochem. Physiol. 9, 165–182. [Google Scholar]
Articles from Molecular Plant Pathology are provided here courtesy of Wiley
Full text links
Read article at publisher's site: https://doi.org/10.1111/j.1364-3703.2011.00746.x
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc6638775?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Fungal Disease Tolerance with a Focus on Wheat: A Review.
J Fungi (Basel), 10(7):482, 13 Jul 2024
Cited by: 1 article | PMID: 39057367 | PMCID: PMC11277790
Review Free full text in Europe PMC
Impact of Exposure to Pyraclostrobin and to a Pyraclostrobin/Boscalid Mixture on the Mitochondrial Function of Human Hepatocytes.
Molecules, 28(20):7013, 10 Oct 2023
Cited by: 1 article | PMID: 37894492 | PMCID: PMC10609024
Biology, taxonomy, genetics, and management of Zymoseptoria tritici: the causal agent of wheat leaf blotch.
Mycology, 14(4):292-315, 08 Aug 2023
Cited by: 1 article | PMID: 38187886 | PMCID: PMC10769150
Review Free full text in Europe PMC
Double Mutations in Succinate Dehydrogenase Are Involved in SDHI Resistance in Corynespora cassiicola.
Microorganisms, 10(1):132, 09 Jan 2022
Cited by: 1 article | PMID: 35056581 | PMCID: PMC8779870
Effects of Pesticides on Longevity and Bioenergetics in Invertebrates-The Impact of Polyphenolic Metabolites.
Int J Mol Sci, 22(24):13478, 15 Dec 2021
Cited by: 4 articles | PMID: 34948274 | PMCID: PMC8707434
Go to all (58) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - JF916683
Protein structures in PDBe
-
(1 citation)
PDBe - 2FBWView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen Mycosphaerella graminicola.
PLoS One, 7(4):e35429, 19 Apr 2012
Cited by: 61 articles | PMID: 22536383 | PMCID: PMC3334918
Emergence of succinate dehydrogenase inhibitor resistance of Pyrenophora teres in Europe.
Pest Manag Sci, 72(10):1977-1988, 03 Mar 2016
Cited by: 19 articles | PMID: 26823120
Detection of Zymoseptoria tritici SDHI-insensitive field isolates carrying the SdhC-H152R and SdhD-R47W substitutions.
Pest Manag Sci, 72(12):2203-2207, 08 Apr 2016
Cited by: 21 articles | PMID: 26941011
A review of current knowledge of resistance aspects for the next-generation succinate dehydrogenase inhibitor fungicides.
Phytopathology, 103(9):880-887, 01 Sep 2013
Cited by: 95 articles | PMID: 23593940
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (1)
Grant ID: BBS/E/C/00005193
 1
1





