Abstract
Free full text

WDFY4 is required for cross-presentation in response to viral and tumor antigens
Abstract
During the process of cross presentation, viral or tumor-derived antigens are presented to CD8+ T cells by the Batf3-dependent CD8α+/XCR1+ classical dendritic cell (cDC1). We designed a functional CRISPR screen for novel regulators of cross presentation, and identified the BEACH-domain containing protein WDFY4 as essential for cross-presentation of cell-associated antigens by cDC1. WDFY4 was not, however, required for MHC class II presentation or for cross-presentation by monocyte-derived DCs. In contrast to Batf3−/− mice, Wdfy4−/− mice have normal lymphoid and non-lymphoid cDC1 populations that produce IL-12 and protect against Toxoplasma gondii infection. However similar to Batf3−/− mice, Wdfy4−/− mice fail to prime virus- specific CD8+ T cells in vivo or induce tumor rejection, revealing a critical role for cross-presentation in anti-viral and anti-tumor immunity.
One Sentence Summary:
WDFY4 is required for cross-presentation in vivo, and is necessary for anti-viral and anti-tumor immunity.
Presentation of antigens as peptides bound to proteins of the major histocompatibility complex (MHC) is the principal mechanism by which innate cells promote antigen-specific T cell immunity (1). Classical dendritic cells (cDC) are particularly efficient antigen presenting cells and comprise two major functionally distinct subsets, the cDC1 and cDC2 (2–4). The cDC1 lineage (2,5) is the most efficient at priming cytotoxic CD8+ T cells to exogenously-derived antigens, a process termed cross-presentation (6–10). This specialization was observed in Batf3−/− mice that specifically lack cDC1 development and cannot mount cytotoxic T cell responses to viruses and tumors (10–24). However, since these studies have only analyzed these responses in the context of mice lacking cDC1, the role of cross-presentation versus other cDC1-specific effector functions, such as IL-12 mediated protection against Toxoplasma gondii (25), has remained incompletely understood.
Cross-presentation has been studied using different cell types and various forms of antigen, and not all findings have been confirmed in vivo (26). DCs generated from monocytes (moDCs) or whole bone marrow cultured in vitro with GM-CSF with or without IL-4 (27–29) are heterogeneous, resembling both macrophages and DCs (30), and use a cross-presentation program divergent from the cDC1 in vivo (26,31,32). Studies of moDCs have produced two major models of cross-presentation; one that involves transport of exogenous antigen to the cytosolic proteasome before peptide loading in the endoplasmic reticulum (ER) (1,7,33–35), and another where peptide loading occurs directly in phagosomes by fusion with vesicles containing the peptide-loading-complex (36,37). The latter pathway may be regulated by the SNARE family member Sec22b, although two recent studies of Sec22b deficient mice arrived at different conclusions as to the role of Sec22b in T cell priming to cell-associated antigens in vivo (38,39). These differences highlight the need for systematic investigation into the mechanisms of cross-presentation in vivo (39,40).
We established a screen for novel cellular components required for cross-presentation, and optimized in vitro conditions to replicate this process in cDC1. The efficiency and cell-type specificity of cross-presentation can vary, depending on whether the antigen is soluble or associated with cells or pathogens (32). Bacterial-associated antigen in the form of heat-killed Listeria monocytogenes expressing ovalbumin (HKLM-OVA) is efficiently cross-presented by cDC1 to OT-I T cells, but not presented by cDC2 (Fig. 1A). In contrast, soluble OVA is crosspresented by both cDC1 and cDC2 lineages, with 3–10 fold lower efficiency in cDC2 (Fig. 1B). Presentation of SIINFEKL peptide to OT-I cells is equally efficient in cDC1 and cDC2, as expected (Fig. 1C). Previous studies have suggested that the majority of antigens undergo translocation to the cytosol during cross-presentation in vivo (1,7,35). We found that cell-associated antigens, which are presented only by cDC1 and not cDC2, are Tap1-dependent, suggesting presentation through the cytosolic pathway (Fig. 1D). In contrast, soluble antigens were presented by both Tap1−/− cDC1 and cDC2, with only slight differences in efficiency compared to WT cDCs (Fig. 1E). For these reasons, we concluded that use of cell-associated antigens in a screen would best emphasize cDC1-specific processing functions.
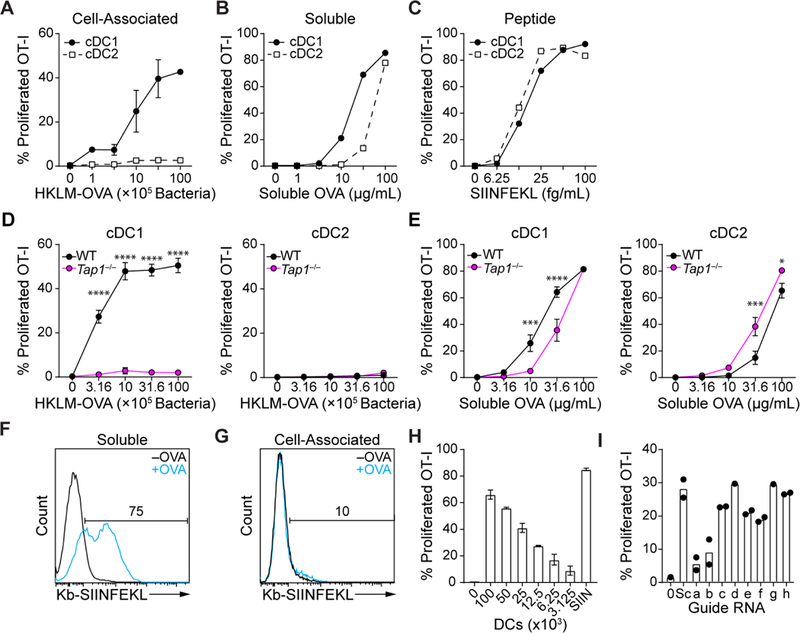
(A-C) Increasing concentrations of HKLM-OVA (A), soluble ovalbumin (B) or SIINFEKL peptide were cultured with sort-purified cDCland cDC2 for three days with CFSE-labeled OT-I T cells and assayed for proliferation (CFSE−CD44+). (D, E) WT or Tap1−/− sort-purified cDCl and cDC2 were cultured for three days with varying concentrations of HKLM-OVA (D) or soluble OVA (E) and CFSE-labeled OT-I T cells and assayed for proliferation (CFSE−CD44+). (F-G) Sorted cDCl were cultured with 100 μg/mL soluble ovalbumin (F) or 106 splenocytes osmotically loaded with OVA (G), cultured for 48 hours and stained with 25-D1.16 and analyzed by flow cytometry. (H) CFSE-labeled OT-I cells were cultured with the indicated number of whole Flt3L-generated DCs and 107 HKLM-OVA or 25 fg/mL SIINFEKL peptide (SIIN) and proliferation measured as in (A). (I) c-Kithi bone marrow progenitors from Cas9 transgenic mice were infected with retroviruses expressing various sgRNAs (Supplementary Table 2), cultured with Flt3L for seven days, and infected cDCs tested for cross-presentation to CFSE-labeled OT-I T cells as in (H), Sc=Scramble. Activated T cells gated as CFSE−CD44+. For all figures data indicate mean ±SEM. For all figures, statistical analysis was performed using 2-way ANOVA with Tukey’s multiple comparisons test. *P<0.05; ***P<0.001; ****P<0.0001
Screening could be done using either biochemical detection of peptide:MHC complexes (p:MHC) or using a T cell response. The antibody 25-D1.16 can directly measure SIINFEKL:Kb complexes on the cell surface (41). 25-D1.16 detected a robust signal from soluble OVA processed by cDC1 (Fig. 1F), but no signal was detected using an immunogenic dose of cell-associated antigen (Fig. 1G). T cells can respond to only a few hundred p:MHC (42,43), implying that the detection limit for 25-D1.16 is greater than that for T cells. Thus, we decided to use T cell proliferation as the readout and determined that 104 cDCs can produce reliable and specific signal of OT-I proliferation (Fig. 1H). We considered gene candidates based on expression in cDCs, relative cDC1 specificity and gene ontology (Table S1). We expressed single guide RNAs (sgRNAs) (44) for candidates (Table S2) by retrovirus under the U6 promoter and infected DC progenitors from Cas9 transgenic mice (45) (fig. S1A). Cells were cultured in Flt3L for 7d, sorted to purify infected cDCs, and tested for cross-presentation (Fig. 1I, fig. S1B,C).
Cross-presentation was substantially impaired by two independent sgRNAs for WD Repeat- and FYVE Domain-Containing Protein 4 (Wdfy4), a member of the BEACH (Beige and Chediak-Higashi) domain containing family of proteins (46) (Fig. 2A, fig S1C). WDFY4 is highly expressed in mouse and human cDC1 (fig S2), with 80% species similarity (47). Wdfy4 is one of 9 BEACH-domain containing proteins (46) and has three closely related family members. However, CRISPR targeting using sgRNAs for Wdfyl, Wdfy2, Wdfy3 did not impair cross-presentation, in contrast to Wdfy4 (Fig. 2B). Thus, Wdfy4 appears to be unique within this gene family for supporting cross-presentation by cDC1.
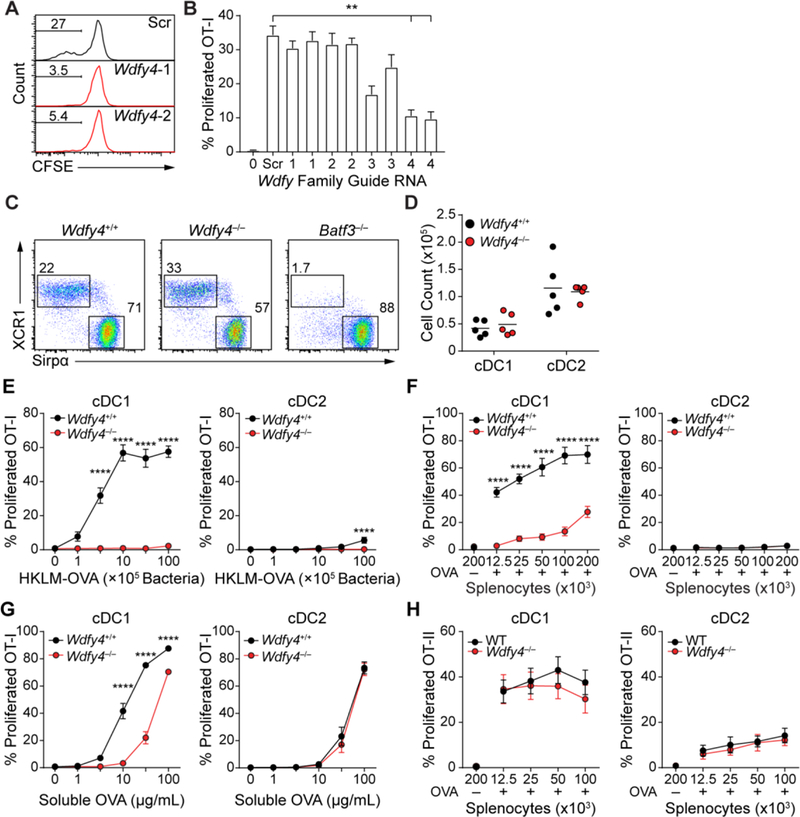
(A) Cross-presentation was measured for Cas9-transgenic cDCl expressing two sgRNA (1 and 2; middle and bottom) for Wdfy4 or a scramble control (top) that were generated as described in (Fig. 1I). T cell proliferation shown by percentages of CFSE− OT-I cells. (B) Cross-presentation by cDCl expressing sgRNAs for Wdfy1, Wdfy2, Wdfy3 and Wdfy4 was measured as described in (Fig. 1I). Scr=Scramble. Activated T cells gated as CFSE−CD44+. Data indicate mean ± SEM of 3 independent experiments (C) cDC1 and cDC2 development was assessed by flow cytometry in WT, Wdfy4−/− and Batf3−/− mice, plots were pre-gated as B220−CD11c+MHCII+ and then gated as cDC1 (XCR1+Sirpα−) or cDC2 (XCR1−Sirpα+). (D) absolute cell numbers of cDC1 and cDC2 in WT and Wdfy4−/− mice. Each dot indicates one mouse, bar indicates mean. (E) FACS sorted cDC1 and cDC2 from spleens of WT and Wdfy4−/− mice were assayed for presentation to OT-I (CFSE−CD44+) in response to the indicated concentrations of HKLM-OVA. (F) FACS sorted cDC1 and cDC2 from spleens of bone marrow chimeric mice with WT or Wdfy4−/− bone marrow were assayed for presentation to OT-I (CFSE−CD44+) in response to indicated concentrations of OVA-loaded irradiated splenocytes from MHCI TKO mice. A negative control of splenocytes osmotically pulsed in the absence of OVA, OVA-, was included (G) FACS sorted cDC1 and cDC2 from spleens of WT and Wdfy4−/− mice were assayed for presentation to OT-I (CFSE−CD44+) in response to indicated concentrations of soluble OVA. (H) FACS sorted cDC1 and cDC2 from spleens of WT and Wdfy4−/− mice were assayed for presentation to OT-II (CFSE−CD44+) in response to indicated concentrations of OVA-loaded irradiated splenocytes from MHCII KO mice. A negative control of splenocytes osmotically pulsed in the absence of OVA, OVA-, was included. For all figures, data indicate mean ±SEM for three independent experiments, *P<0.05; **P<0.01; ****P<0.0001 using 2-way ANOVA with Tukey’s multiple comparisons test.
To evaluate the in vivo function of Wdfy4, we obtained mice with exon 4 deleted by CRISPR/Cas9 genome editing, leading to translational termination due to a reading frame shift when exon 3 splices to exon 5 (fig. S3). Wdfy4−/− mice are viable, born in normal Mendelian ratios and have normal development of hematopoietic lineages, including cDCs (Fig. 2C and andD,D, fig. S4), which express Irf8 and have normal turnover kinetics (fig. S4H and I), and T cells (fig. S5). In particular, cDC1 develop in Wdfy4−/− mice, unlike Batf3−/− mice, and express CD24, XCR1 and CD103 normally (Fig. 2 C and andD,D, fig. S4B and F). However, cDC1 from Wdfy4−/− mice show a striking defect in cross-presentation of both cell-associated and bacterial-associated antigen in vitro (Fig. 2 E and andF,F, fig. S6A) and show reduced efficiency for soluble OVA presentation compared with WT cDC1 (Fig. 2G). Notably, Wdfy4−/− cDC1 cross-present soluble OVA with the efficiency of cDC2, which are not influenced by the loss of Wdfy4 (Fig. 2G). However, Wdfy4−/− cDC1 can directly present antigen introduced into the cytoplasm by osmotic shock or virus, a process that is equally efficient in cDC1 and cDC2 (fig. S6B to D), suggesting that Wdfy4−/− cDC1 have the capacity to present endogenous antigens on MHCI.
MoDCs can cross-present both soluble and cell-associated antigens in vitro (27,48,49), but use a distinct transcriptional program from cDC1 (31). We find that moDCs derived from WT and Wdfy4−/− mice cross-present antigens with the same efficiency, both for cell-associated (fig. S6E) and soluble OVA (fig. S6F), suggesting that moDCs use a Wdfy4-independent pathway for cross-presentation. The defect in cross-presentation by Wdfy4−/− cDC1 is specific, since MHC class II antigen processing was unchanged in Wdfy4−/− mice for both cell-associated and soluble antigens (Fig. 2H; fig. S6G). MHC class II antigen processing by B cells is also normal in Wdfy4−/− mice (fig. S7A), which are able to generate germinal center B cells and T follicular helper (TFH) cells in response to immunization with sheep red blood cells (fig. S7B to E).
cDCs from Wdfy4−/− mice expressed normal levels of MHCI at steady state and after activation (fig. S8A and B), upregulated costimulatory molecules CD80/86 and expressed cytokines normally (fig S8C to F). Loss of Wdfy4 also did not influence gene expression in cDC1s at steady state or after activation in tumor-bearing mice (fig. S8G and H). Despite their inability to cross-present, Wdfy4−/− cDC1 are capable of taking up and degrading soluble antigens normally (fig. S9A and B) and phagocytosing labeled HKLM-OVA, as seen both microscopically (fig. S9C) and by quantification of this phagocytosis as measured by FACS (fig. S9D).
To explore the mechanism of action of WDFY4, we analyzed various cellular compartments of wild-type and Wdfy4−/− cDC1 by confocal microscopy and found minimal differences in distribution of MHCI stores, ER, early endosomes, lysosomes, late endosomes, or the peptide-loading complex at steady state (fig. S10) or Rab43 (a molecule previously described to be involved in cross-presentation (32)), p62 (autophagic vesicles), Rab7 (late endosomes), or Lamp1 (lysosomes) after antigen phagocytosis (fig. S11). Electron microscopy of WDFY4-deficient ex vivo cDC1 showed the presence of large and numerous lipid bodies throughout the cytoplasm that were not present in wild-type cells (fig. S12, fig. S13A and C). However, these lipid bodies were not present in Flt3L-derived cDC1 from Wdfy4−/− mice (S13B and C), which still have a defect in cross-presentation of cell-associated antigen (fig. S13D), suggesting that the lipid bodies are not necessary to cause the defect in cross-presentation in Wdfy4−/− cDC1.
To determine interacting partners of WDFY4, we generated four individually FLAG-tagged sub-regions of WDFY4 spanning the entire protein (Fig. 3A). We stably transduced these fragments into the murine DC line JAWSII (50), and performed affinity purification-mass spectrometry (AP-MS) to isolate WDFY4 binding partners. We found 143 candidates enriched by different regions of the WDFY4 protein, with the largest number binding to the FL4 fragment of WDFY4 that contains the PH, BEACH, WD40, and FYVE domains (Fig. 3A, Table S3). We performed gene ontology analysis to determine the biological processes most likely influenced by WDFY4 (51). The fragments FL1 and FL2 of WDFY4 associated with proteins involved in “protein complex assembly,” and therefore may be involved in forming multimeric protein structures or scaffolding vesicular machinery (Fig. 3B, Table S4). FL3 and FL4 associated with proteins involved in “protein localization,” “vesicle transport,” and “cytoskeletal organization,” suggesting a role for WDFY4 in proper subcellular vesicular targeting (Fig. 3C, Table S5). Notably, FL4 associated with components critical to the formation, function, and trafficking of endocytic vesicles, including clathrin (Cltc, Clta) (52), subunits of the AP-2 clathrin adaptor complex (Ap2a1, Ap2a2, Ap2b1) (52), modulators of cytoskeleton dynamics (Iqgap1, Actn4) (53,54), and several members of the vacuolar-type (H+) ATPase (Atp6v0a3, Atp6va1, Atp6v1f) (55) (Fig. 3D, Table S3 and S6). FL4 also selectively associated with Hsp90ab1, a member of the HSP90 chaperone family involved in endosome-to-cytosol translocation of antigen during cross-presentation (56–59) (Figure 3D to toF).F). While heat-shock proteins such as Hspa8 and Hsp1a1 can appear as artefacts in AP-MS data due to their function as chaperones (60), Hsp90ab1 is rarely detected in this manner, and therefore its association may represent a functional interaction with WDFY4.
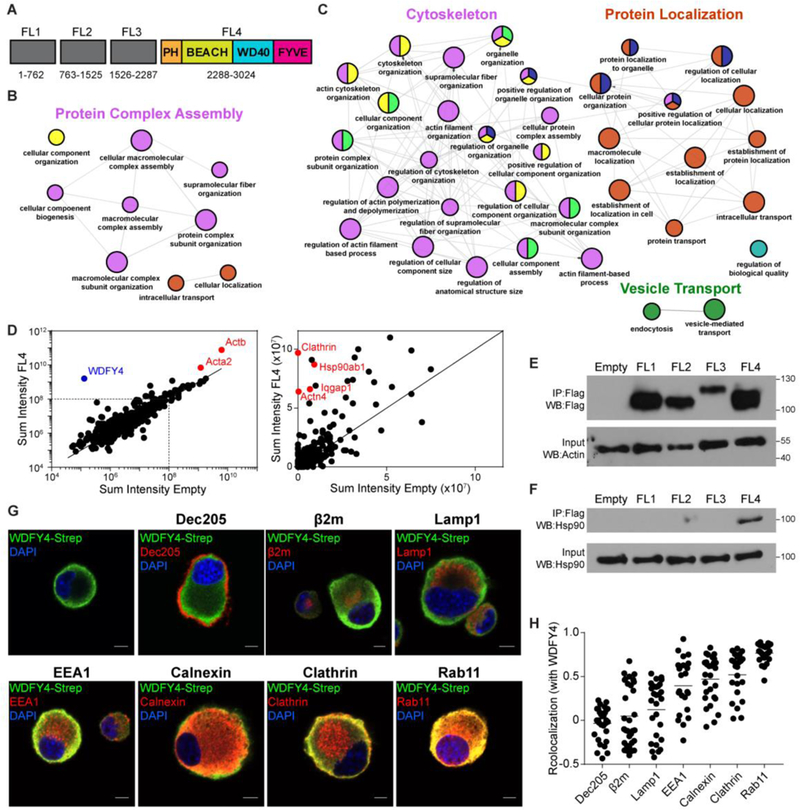
(A) Diagram of truncated fragments of WDFY4 protein, showing predicted domains within FL4 fragment. Numbers indicate amino-acid locations of fragments. (B) ClueGO visualization of gene ontology terms enriched after immunoprecipitation of fragments from (A) in the mouse DC line JAWSII, expressing either FL1 or FL2 fragments. Small circles P<.001, large circles P<.0001. Colors indicate Gene Ontology (GO) term groups. (C) ClueGO visualization of gene ontology terms enriched after immunoprecipitation of fragments from (A) in the mouse DC line JAWSII expressing either FL3 or FL4 fragments. Small circles P<3×10−5, large circles P<3×10−6. Colors indicate GO term groups. (D) Scatterplot of representative data for sum intensity of proteins found after mass spectrometry between FL4 expressing and empty-vector expressing JAWSII cells. (E) Western blot of Flag immunoprecipitates from HEK293 cells transfected with empty-vector or Flag-tagged WDFY4 fragments 1 to 4 (top), and input control for β-actin (bottom) (F) Western blot for endogenous Hsp90 in Flag immunoprecipitates from HEK293 transfected with empty-vector or Flag-tagged WDFY4 fragments 1 to 4 (top) and input control for endogenous Hsp90 (bottom) (G) Confocal microscopy of JAWSII cells overexpressing full length Twin-Strep-tagged WDFY4, stained for anti-Strep (green), various cellular markers (red), and DAPI (blue). Scale bars indicate 5μm. (H) Quantification of co-localization between WDFY4 and intracellular markers from images in (G), each dot represents one cell, bar indicates the mean.
We then sought to determine which vesicles WDFY4 may be acting on by determining its intracellular location. We visualized full-length Twin-Strep-tagged (61) WDFY4 in JAWSII cells by confocal microscopy and found that it localized to the periphery of the cytosol near the plasma membrane (Fig 3G). WDFY4 was poorly co-localized with the cell surface receptor DEC-205, intracellular MHCI stores and lysosomes, but demonstrated moderate co-localization with early endosomes and the ER. cDC1 have been previously shown to have well defined and extensive ER structures which may extend throughout the cytosol near vesicular compartments (62), and lead to co-localization with components of the endosomal pathway. WDFY4 shows highest correlation with endosomal markers clathrin and Rab11 (Fig 3G and andH),H), suggesting that it localizes to an endosomal compartment near the plasma membrane. Taken together, these data suggest that WDFY4 functions in trafficking between the cell surface and endosomes and thus may regulate multimeric protein assembly required for proper formation and localization of endocytic vesicles.
We then examined the role of WDFY4 in cross-presentation of cell-associated antigens in vivo. CFSE-labeled OT-I T cells showed strong in vivo proliferation induced by immunization with OVA-loaded splenocytes when transferred into Wdfy4+/− mice, but not Wdfy4−/− mice (Fig. 4A and andB),B), confirming an in vivo defect in cross-presentation. IL-12 produced by cDC1 in response to soluble tachyzoite antigen (STAg) is required for innate immune protection against T. gondii, as illustrated by the susceptibility of Batf3−/− mice to lethal infection by this pathogen (25). In contrast, Wdfy4−/− mice are resistant to T. gondii infection, similar to Wdfy4+/− mice (Fig. 4C). These results indicate that cross-presentation is not required for innate protection against T. gondii, and that Wdfy4−/− cDC1 are not globally impaired in function.
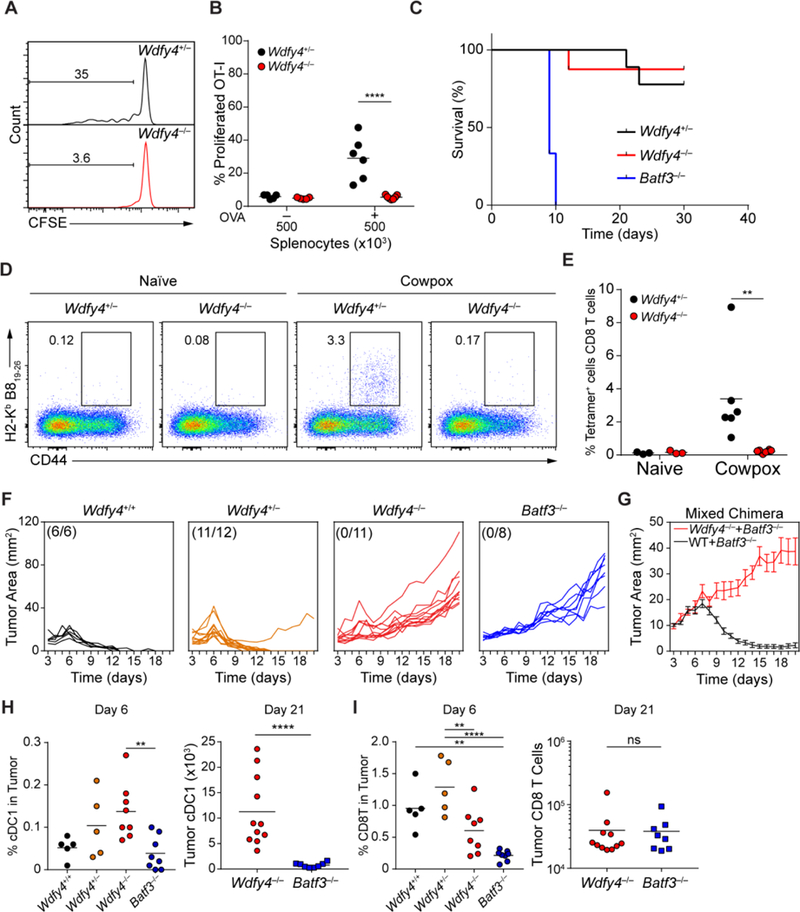
(A-B) Representative flow cytometry analysis of in vivo cross-presentation to 500K irradiated splenocytes loaded with OVA injected i.v. into mice of the indicated genotypes one day after injection of 500K CFSE-labeled OT-I cells. Mice were harvested three days after antigen injection, quantified in (B). Data pre-gated on OT-I cells and shown as percentage of CFSE− cells (A) or CFSE−CD44+ cells (B). Data are pooled from three independent experiments; each point represents one mouse. (C) Survival of mice of the indicated genotypes to injection of 200 Pru.luc T. gondii tachyzoites over 30 days. WDFY4+/− n=9, WDFY4−/− n=8, Batf3−/− n=3. (D-E) Representative flow cytometry plots of CD8 T cells (pre-gate CD4−CD3+CD8+) in lungs of naïve or cowpox infected mice, quantified in (E). Each dot represents one mouse, bar indicates mean. (F) Mice of the indicated genotypes were injected with 1×106 fibrosarcoma cells s.c. and tumors were measured daily starting at day three after injection. (G) Mixed bone marrow chimeras with bone marrow of indicated genotypes were injected into lethally irradiated CD45.1+ WT B6 mice. Eight weeks later, mice were injected with 1×106 fibrosarcoma cells s.c. and tumors were measured daily starting at day three after injection. Data show mean ± SEM of 9 mice per group. (H) Quantification of cDC1 in tumors at either day 6 or day 21 after injection taken from mice of indicated genotypes. Gated as B220−CD11c+MHCII+CD103+CD11b−. Each dot indicates one mouse, bar indicates mean. (I) Quantification of CD8 T cells in tumors at either day 6 or day 21 after injection taken from mice of indicated genotypes. Gated as CD45+TCRβ+CD8α+CD4−. Each dot indicates one mouse, bar indicates mean. For all figures, ** P<.01; **** P<0.0001 using 2-way ANOVA with Tukey’s multiple comparisons test.
We also evaluated CD8+ T cell responses of Wdfy4−/− mice to cowpox virus infection, a model in which effective CD8+ T cell priming is thought to be mediated primarily by Batf3-dependent cells through cross-presentation (73). Batf3−/− mice that lack cDC1 (10–13) have a defect in priming antigen-specific CD8+ T cells to several viruses (10–13), but these studies only indirectly show that this is due to a lack of cross-presentation, since a loss of alternative functions of cDC1 could conceivably be the cause. However, we now show that Wdfy4−/− mice, that retain cDC1 cells which cannot cross-present, also have a severely impaired antigen-specific CD8+ T cell responses to cowpox virus (Fig. 4D and andE,E, fig. S14A to D). This defect in cross-presentation is not restricted to cowpox virus, since Wdfy4−/− mice also show a defect in priming CD8+ T cells to infection by West Nile virus (fig. S14E). However, Wdfy4−/− mice show normal priming of CD4+ T cells to West Nile, indicating that WDFY4 functions for in vivo cross-presentation to CD8+ T cells, but not for priming of CD4+ T cells (fig. S14F).
Studies with Batf3−/− mice suggested that cDC1 were required for tumor rejection (10). To examine the role of cross-presentation directly, we evaluated growth of the highly immunogenic 1969 regressor fibrosarcoma (15) in WT, Wdfy4+/−, Wdfy4−/− and Batf3−/− mice (Fig. 4F). Tumors were readily rejected by WT mice, but not by Batf3−/− mice (Fig. 4F) as expected (15). However, tumors were also rejected by heterozygous Wdfy4+/− mice, but grew uncontrolled in Wdfy4−/− mice similar to Batf3−/− mice (Fig. 4F and S15A). These results with germline-deficient Wdfy4−/− mice indicate an in vivo requirement for WDFY4 in tumor rejection, but do not pinpoint its function to cDC1. To test whether the in vivo defect in Wdfy4−/− mice is cDC1-intrinsic, we generated mixed bone-marrow (BM) chimeras using mixtures of either WT: Batf3−/− or Wdfy4−/− : Batf3−/− BM (Fig. 4G). WT: Batf3−/− chimeras rejected tumors normally, but Wdfy4−/− : Batf3−/− chimeras, in which cDC1 develop only from the Wdfy4−/− BM, failed to control tumor growth (Fig. 4G). These results indicate that the defect in tumor rejection results from loss of Wdfy4 expression in cDC1. Notably, in Wdfy4−/− mice, cDC1 do infiltrate into tumors as they expand (Fig. 4H, fig. S15B), yet they induce less recruitment of CD8+ T cells to tumors, similar to the lack of CD8+ T cells in tumors in Batf3 −/− mice (Fig. 4I).
WDFY4 is one of nine mammalian BEACH-domain containing proteins (BDCP) that typically also contain a PH-like domain and WD repeats (46). BDCP function as protein scaffolds that regulate intracellular vesicle fission and fusion events, and several are associated with human diseases (46). For example, mutations in Lyst cause the Chediak-Higashi syndrome, a primary immunodeficiency disorder characterized by defective neutrophil phagolysosome formation and cytotoxic T cell degranulation (63,64). Mutations in Lrba result in immune dysregulation in regulatory T cells due to improper trafficking of CTLA4 from endosomes to lysosomes by the clathrin adaptor AP-1 (65,66). WDFY3, the closest WDFY4 homolog, regulates recruitment of polyubiquitinated protein aggregates to autophagosomes by interactions with p62, Atg5, Atg12, Atg16L, LC3 and TRAF6 (67–70). Although cross-presentation of cell-associated antigens does not involve autophagy (71), WDFY4 conceivably may regulate vesicular trafficking pathways, a concept supported by its localization to submembrane endosomes and its interaction with endocytic and cytoskeletal machinery. These WDFY4-dependent trafficking pathways may be required for translocation of dead-cell antigen ligated by the cDC1-specific receptor CLEC9A (72) to specific compartments to promote cross-presentation (73). Further investigation of the mechanisms of WDFY4 may elucidate novel components of the cross-presentation pathway and thus offer therapeutic targets for human disease.
Acknowledgements:
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility. We would like to thank E. Tonc for blinded microscopy analysis. We gratefully thank D. Oakley and J. Fitzpatrick for assistance with confocal microscopy which was performed at Washington University Center for Cellular Imaging (WUCCI). supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CDI-CORE-2015–505), the Foundation for Barnes-Jewish Hospital (3770) and the National Institute for Diabetes and Digestive and Kidney Diseases (P30 DK020579). We thank R. Tomaino and the Taplin mass spectrometry core at Harvard University for performing mass spectrometry experiments.
Funding: This work was supported by NIH grant T32 AI007163–40 (D.J.T.); NCI grants T32 CA 009621 (J.T.D.), 5R01 CA 190700 (R.D.S.), P30 CA 091842 (W.E.G.), 5R01 CA 193318 (N.M.); NIAID grant U19-Al109948 (W.Y.); NIH grant F30DK108498 (V.D.); NSF grant DGE-1143954 (P.B.); NIH grant U19-Al109725 (H.W.V.); Howard Hughes Medical Institute (K.M.M.). WDFY4-deficient mice used in this study were generated by the JAX KOMP program (U42OD011185).
Footnotes
Competing interests: H.W.V. is a founder of Casma Therapeutics, Cambridge, MA.
Data and materials availability: Microarray data has been deposited in the NCBI gene expression omnibus at accession number GSE118652, Proteomics data has been deposited in the Proteomics Identifications (PRIDE) archive px-submission #298619 (accession # PENDING), all other data is available in manuscript or supplementary materials. WDFY4 knockout mice were generated by and are available through the KOMP2 program at The Jackson Laboratory (029334).
Reference List
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aat5030
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/362/6415/694.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Suppression of melanoma by mice lacking MHC-II: Mechanisms and implications for cancer immunotherapy.
J Exp Med, 221(12):e20240797, 29 Oct 2024
Cited by: 0 articles | PMID: 39470607 | PMCID: PMC11528124
BEACH domain proteins function as cargo-sorting adaptors in secretory and endocytic pathways.
J Cell Biol, 223(12):e202408173, 08 Nov 2024
Cited by: 0 articles | PMID: 39514288 | PMCID: PMC11554844
Immunotherapy response induces divergent tertiary lymphoid structure morphologies in hepatocellular carcinoma.
Nat Immunol, 25(11):2110-2123, 25 Oct 2024
Cited by: 1 article | PMID: 39455893
In vivo dendritic cell reprogramming for cancer immunotherapy.
Science, 386(6719):eadn9083, 18 Oct 2024
Cited by: 0 articles | PMID: 39236156 | PMCID: PMC7616765
Optimization of the Irf8 +32-kb enhancer disrupts dendritic cell lineage segregation.
Nat Immunol, 25(11):2043-2056, 07 Oct 2024
Cited by: 0 articles | PMID: 39375550
Go to all (139) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE118652
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
An Important Role for CD4+ T Cells in Adaptive Immunity to Toxoplasma gondii in Mice Lacking the Transcription Factor Batf3.
mSphere, 5(4):e00634-20, 15 Jul 2020
Cited by: 6 articles | PMID: 32669460 | PMCID: PMC7364223
CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3.
Blood, 121(9):1574-1583, 07 Jan 2013
Cited by: 77 articles | PMID: 23297132
CD8α(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites.
Immunity, 35(2):249-259, 01 Aug 2011
Cited by: 276 articles | PMID: 21867928 | PMCID: PMC3171793
Dendritic Cells and CD8 T Cell Immunity in Tumor Microenvironment.
Front Immunol, 9:3059, 20 Dec 2018
Cited by: 245 articles | PMID: 30619378 | PMCID: PMC6306491
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NCI NIH HHS (4)
Grant ID: R01 CA190700
Grant ID: T32 CA009621
Grant ID: R01 CA193318
Grant ID: P30 CA091842
NIAID NIH HHS (3)
Grant ID: T32 AI007163
Grant ID: U19 AI109948
Grant ID: U19 AI109725
NIAMS NIH HHS (1)
Grant ID: P30 AR073752
NIDDK NIH HHS (2)
Grant ID: F30 DK108498
Grant ID: P30 DK020579
NIH HHS (1)
Grant ID: U42 OD011185
National Cancer Institute (4)
Grant ID: 5R01CA190700
Grant ID: 5R01CA193318
Grant ID: P30CA091842
Grant ID: T32CA009621
National Institute of Allergy and Infectious Diseases (1)
Grant ID: U19-AI109948





