Abstract
Free full text

Burkitt-like lymphoma with 11q aberration: a germinal center-derived lymphoma genetically unrelated to Burkitt lymphoma
Abstract
Burkitt-like lymphoma with 11q aberration is characterized by pathological features and gene expression profile resembling those of Burkitt lymphoma but lacks the MYC rearrangement and carries an 11q-arm aberration with proximal gains and telomeric losses. Whether this lymphoma is a distinct category or a particular variant of other recognized entities is controversial. To improve the understanding of Burkitt-like lymphoma with 11q aberration we performed an analysis of copy number alterations and targeted sequencing of a large panel of B-cell lymphoma-related genes in 11 cases. Most patients had localized nodal disease and a favorable outcome after therapy. Histologically, they were high grade B-cell lymphoma, not otherwise specified (8 cases), diffuse large B-cell lymphoma (2 cases) and only one was considered as atypical Burkitt lymphoma. All cases had a germinal center B-cell signature and phenotype with frequent LMO2 expression. The patients with Burkitt-like lymphoma with 11q aberration had frequent gains of 12q12-q21.1 and losses of 6q12.1-q21, and lacked common Burkitt lymphoma or diffuse large B-cell lymphoma alterations. Potential driver mutations were found in 27 genes, particularly involving BTG2, DDX3X, ETS1, EP300, and GNA13. However, ID3, TCF3, or CCND3 mutations were absent in all cases. These results suggest that Burkitt-like lymphoma with 11q aberration is a germinal center-derived lymphoma closer to high-grade B-cell lymphoma or diffuse large B-cell lymphoma than to Burkitt lymphoma.
Introduction
Our knowledge of lymphomas in children and young adults has increased dramatically in the last years with the identification of several subtypes that predominantly occur in this age subgroup.1–4 One of these recently recognized categories is Burkitt-like lymphoma with 11q aberration (BLL-11q) which has morphological, phenotyp ic, and gene expression profiles resembling those of Burkitt lymphoma (BL), but lacks MYC rearrangements according to standard methods of detection, such as fluorescence in situ hybridization (FISH). Alternatively, these tumors carry an 11q-arm aberration characterized by proximal gains and telomeric losses.4 In comparison with BL, BLL-11q seems to have more complex karyotypes, a certain degree of cytological pleomorphism, sporadically a follicular pattern and a high incidence of nodal presentation.4,5 Very similar cases have also been reported in the post-transplant setting,6 although its incidence in other immunocompromised conditions, such as human immunodeficiency infection, is still unclear.7,8
BLL-11q has been incorporated in the revised World Health Organization (WHO) classification as a provisional category1 because its precise taxonomy as a particular variant of BL, diffuse large B-cell lymphoma (DLBCL) or a distinct form of high-grade B-cell lymphoma (HGBCL) is still controversial.1,4–6,9–11 The clarification of the biological nature of this uncommon subtype of lymphoma is clinically relevant because of increasing interest in defining the most appropriate management strategies for specific subtypes of lymphomas in pediatric and young adult patients.12 Recent DNA copy number alteration and next-generation sequencing studies have provided a comprehensive catalog of genomic aberrations in BL and DLBCL which clearly distinguish these entities.13–17 In this study we performed an integrated analysis of genomic and mutational alterations with a complete annotation of clinical and pathological features of BLL-11q with the goal of obtaining insights to refine the understanding of the pathogenesis of these tumors and improve their diagnosis.
Methods
Sample selection and DNA/RNA extraction
To identify BLL-11q cases we initially reevaluated the presence of MYC translocations in 95 cases diagnosed as BL, atypical BL or HGBCL, not otherwise specified (NOS), in our Hematopathology Unit between 2000-2016. Three consultation cases from centers belonging to the Sociedad Española de Hematología y Oncología Pediátricas (SEHOP) were also analyzed. Cases were reviewed by three pathologists (BG-F, EC, ESJ). DNA and RNA were extracted using standard protocols (Qiagen, Hilden, Germany). This study was approved by the Institutional Review Board of the Hospital Clinic of Barcelona. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
Immunohistochemistry and fluorescence in situ hybridization
Immunohistochemical analysis using a panel of antibodies detecting common B-and T-cell markers as well as LMO2 and MYC was performed and interpreted as previously reported (Online Supplementary Table S1).18,19
MYC breaks and MYC/IGH fusions were analyzed by FISH using the XL MYC BA Probe (Metasystems, Altlussheim, Germany) and LSI IGH/MYC/CEP 8 Tri-Color Dual Fusion Probe Kit (Vysis-Abbott, Abbott Park, IL, USA) respectively. The 11q alteration was studied with a custom FISH probe using BAC clones (Invitrogen Inc.) for proximal gains (RP11-414G21-spectrum green) and terminal losses (RP11-629A20-spectrum red) combined with CEP11-spectrum aqua (Vysis-Abbott Inc.). The FISH constellation in a normal case is characterized by two signals per probe, while the pattern corresponding to the 11q gain/loss or gain/amplification/loss aberration would be two blue, three up to five green signals and one red signal. The probe was tested in an independent series of eight non-Hodgkin B-cell lymphomas and four MYC-negative HGBCL with lack of the 11q alteration by array analysis and all showed the normal pattern described above.
Copy number analysis
DNA was hybridized on Oncoscan FFPE or SNP array platform (ThermoFisher Scientific, Waltham, MA, USA) and analyzed as described previously (Online Supplementary Methods).2 Published copy number data on MYC-positive BL20 and DLBCL13 were reanalyzed for comparison.
Sequencing approaches
The mutational status of 96 B-cell lymphoma-related genes (Online Supplementary Table S2) was examined by target next-generation sequencing in ten BLL-11q cases and four MYC-negative 11q-negative cases using a NGS SureSelect XT Target Enrichment System Capture strategy (Agilent Technologies, Santa Clara, CA, USA) before sequencing in a MiSeq instrument (Illumina, San Diego, CA, USA) (Online Supplementary Methods). Additionally, analysis of hotspots of mutation in ID3, TCF3 and CCND3 genes, ETS1 exon 1 (transcript NM_005238) and verification of variants in specific cases was performed by Sanger sequencing using primers described in Online Supplementary Table S3.
Gene expression analysis
Cell of origin was determined by Lymph2Cx assay (Nanostring, Seattle, WA, USA) as previously published.21 Gene expression levels of MYC and ETS1 were investigated by real-time quantitative polymerase chain reaction (Online Supplementary Methods) using Taqman assays described in Online Supplementary Table S4.
Statistical methods
The χ2 method was used for categorical variables and Student t-tests for continuous variables. Non-parametric tests were applied when necessary. The P-values for multiple comparisons were adjusted using the Benjamini–Hochberg correction. Survival curves were estimated with the Kaplan-Meier method. Statistical analyses were carried out with SPSS v22 and R software v3.1.3.
Results
Identification of cases with Burkitt-like lymphoma with 11q aberration
To identify BLL-11q cases we reevaluated the presence of MYC translocations in 95 cases diagnosed as having BL, atypical BL or HGBCL, NOS. We confirmed the presence of MYC rearrangements in 78 cases (82.1%), of which 67 (70.5%) were classified as BL. Since the 11q aberration has been found mainly in children and young adults (<40 years old),4 we analyzed the 60 patients under 40 years and the 35 older patients separately (Online Supplementary Figure S1).
In the younger cohort (n=60), the 46 (76.7%) cases with MYC translocations were classified as having BL. To find BLL-11q cases, we initially used the Oncoscan platform in the remaining 14 MYC-negative patients and detected the presence of the 11q gain/loss alteration in eight of them. Additionally, we found a copy number pattern consistent with the presence of 11q alteration in three recent consultation cases from SEHOP (Online Supplementary Figures S1 and S2). Then, among those BLL-11q cases we were able to verify the presence of the 11q aberration by FISH in all ten evaluable cases (Online Supplementary Figure S3 and Supplementary Table S5). The morphological, clinical, and genetic features and consensus diagnosis of the 11 BLL-11q identified in our files are summarized in Table 1. The six cases negative for the MYC rearrangement and 11q aberrations by Oncoscan were re-classified as DLBCL (3 cases) or HGBCL, NOS (3 cases). The DLBCL had predominant centroblastic morphology, germinal center phenotype, very high proliferative index and a focal “starry sky” pattern (see Online Supplementary Results). The absence of 11q alterations was also verified using the 11q FISH probe in four of these MYC/11q-negative cases with available material (Online Supplementary Figure S1A).
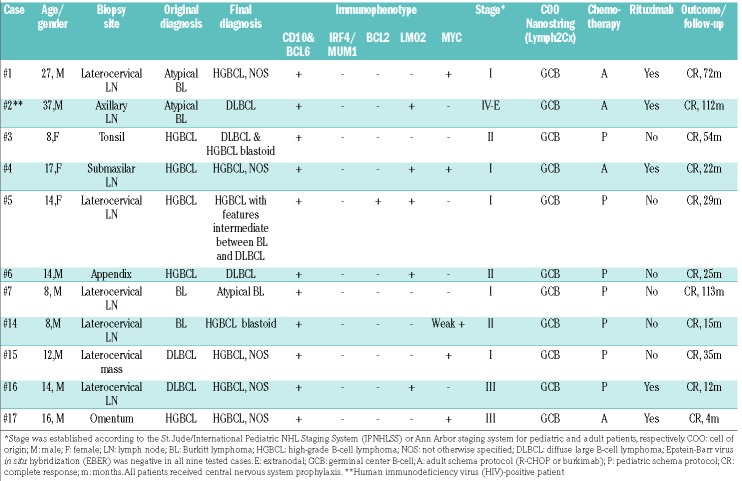
Table 1.
Pathological and clinical features of 11 cases of Burkitt-like lymphoma with 11q aberration.
In the 35 older patients (≥40 years old), a MYC translocation was found in 32 cases; one was classified as DLBCL, 21 as BL, and ten were HGBCL with double-or triple-hit aberrations (BCL2 and/or BCL6 translocations). Only three cases were negative for MYC translocations and were classified as HGBCL, NOS (Online Supplementary Figure S1B and Online Supplementary Results). We screened these cases with the 11q FISH probe and all three were negative for the 11q aberration.
Clinical and morphological results of cases of Burkitt-like lymphoma with 11q aberration
The 11 patients with BLL-11q had a mean age of 15 years (range, 8-37 years); eight were male (Table 1). Eight tumors were reclassified morphologically as HGBCL, NOS, two as DLBCL and only one case was considered atypical BL. None of the cases was considered as typical BL (Figure 1). Six cases showed a “starry sky” pattern and two had a nodular growth pattern with the presence of a disrupted follicular dendritic cell meshwork (Figure 1C). Ki67 was very high in all the samples, as in BL. All cases had a germinal center (GC) phenotype and GC B-cell (GCB) signature by Nanostring Lymph2Cx assay. MUM1/IRF4 was negative in all 11 cases. One case expressed BCL2 (Figure 1D). LMO2, a germinal center marker that is usually seen in GCB-DLBCL but not in BL18 was expressed in five cases (Figure 1A, B). Interestingly, using a 40% cutoff,19 five cases were positive for MYC expression. However, only one case showed diffuse and intense positivity while the other four cases had either only positivity in around 50% of the cells or the intensity was not that expected in typical BL. Additionally, MYC RNA levels were significantly lower in BLL-11q than in MYC-positive BL (relative expression 0.07 vs. 0.36, P=0.019) (Online Supplementary Figure S4A). Epstein-Barr virus hybridization was negative in the nine cases tested.
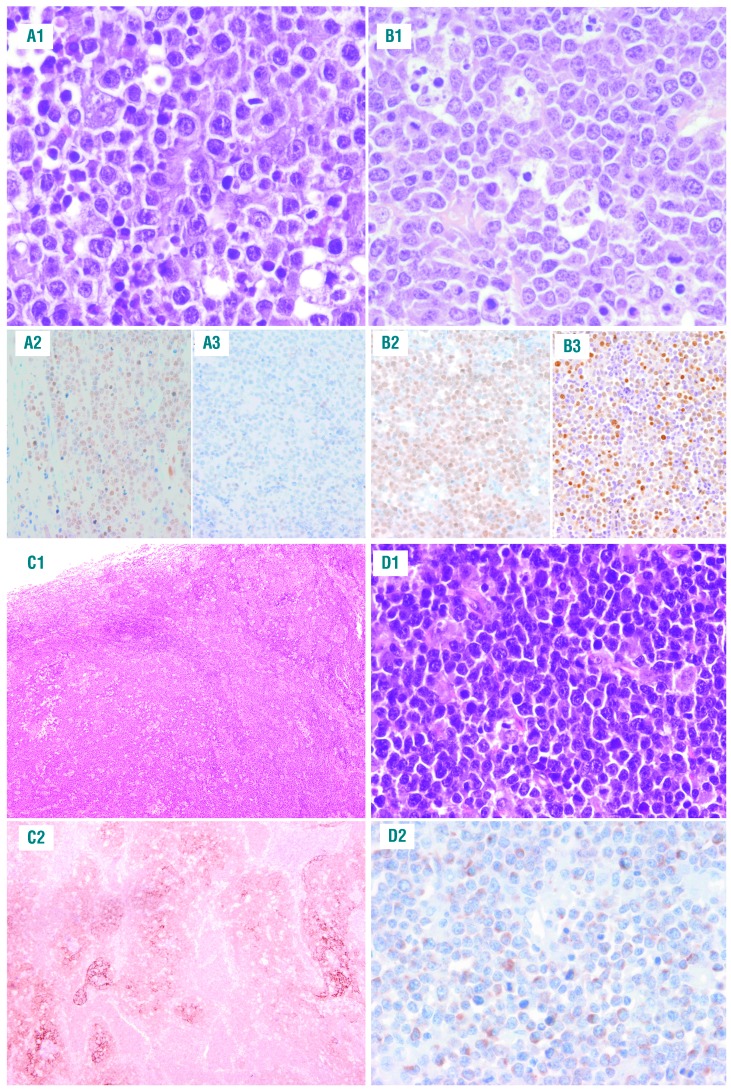
Morphological features of cases of Burkitt-like lymphoma with 11q aberration. (A1-A3) Case #2 shows the typical morphology of diffuse large B-cell lymphomas with large and irregular cells resembling centroblasts. This case was positive for (A2) LMO2 and negative for (A3) MYC. (B1-B3) Case #4 corresponds to a tumor with high-grade B-cell lymphoma (HGBCL) morphology. It is composed mostly of medium-sized cells with mild heterogeneity. Note the “starry sky” pattern. This case was positive for (B2) MYC and (B3) LMO2 expression. (C1-C2; case #7) Lymph node with nodular architecture and a “starry sky” pattern with large follicles and a disrupted follicular cell meshwork highlighted with (C2) CD21. (D1-D2; case #5) This is a case with HGBCL features with expression of (D2) BCL2 in the neoplastic cells.
Clinically, BLL-11q frequently had a nodal localized presentation (8/11) in the head and neck region. Two cases had an extranodal presentation, one in the context of an acute appendicitis and the other debuted as an omental mass. Eight patients (73%) had stage I-II, and one patient presented in an advanced stage (IV-E) with widespread disease in the context of chronic human immunodeficiency virus infection. All cases were treated with chemotherapy, including rituximab in five. All patients were alive with no disease after a median followup of 29 months (Table 1).
Copy number analysis
The copy number analysis of all the 11 BLL-11q cases showed a total of 78 alterations (mean 7.1; range, 2-15) (Online Supplementary Tables S5 and S6). Seven cases had the typical 11q gain/loss pattern (Figure 2A-B, Online Supplementary Figure S2), two cases had only an 11q terminal deletion, one case showed a complex 11q alteration with two gains and two losses, and finally one case had an 11q23.3-q25 copy number neutral loss of heterozygosity in addition to gain (Online Supplementary Figure S2). Two minimal regions of gain were identified (chr11:103326831-111737912/11q22.3-q23.1 and chr11:114767237-116764582/11q23.3.hg19) whereas the minimal region of loss was in chr11:128214400-132020453/11q24.3-q25 (including the ETS1 and FLI1 genes). No cases with homozygous deletions of these two targets were observed in our series. The breakpoint region between gain and loss was not conserved and spanned from chr11:118352769 to chr11:121062860. Amplifications in the 11q arm were observed in four cases, with a minimal region chr11:118347020-120155799/11q23.3, including the USP2 gene (Online Supplementary Figure S5). The most recurrent copy number aberrations other than 11q were 12q12-q21.1 gains and 6q12.1-q21 losses (Figure 2A).
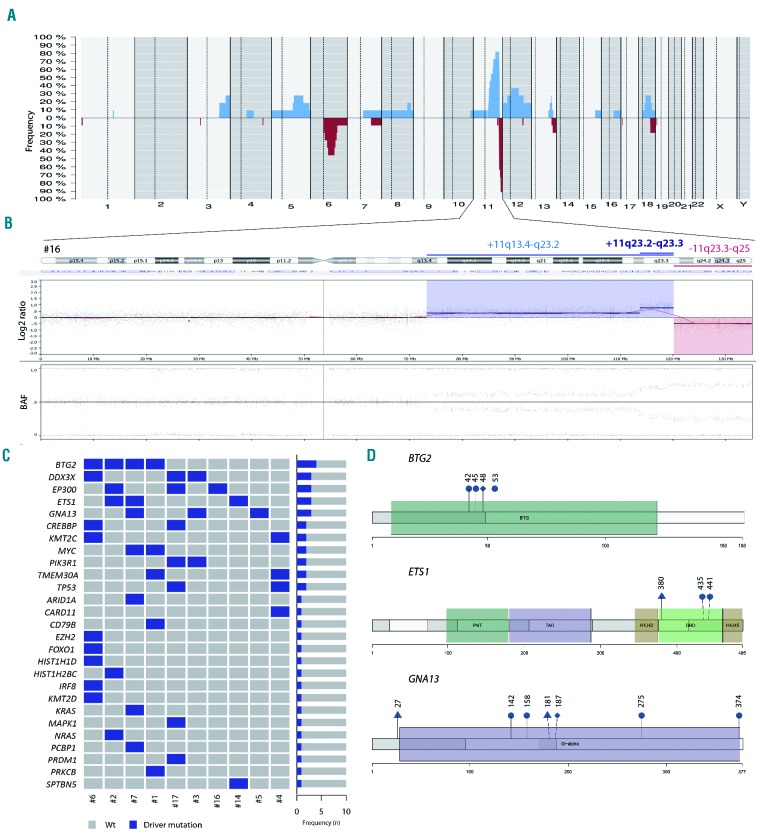
Genetic features of cases of Burkitt-like lymphoma with 11q aberration. (A) Global copy number profile of the 11 cases of Burkitt-like lymphoma (BLL) with 11q aberration. The horizontal axis indicates chromosomes from 1 to Y and p to q. The vertical axis indicates the frequency of the genomic aberration among the cases analyzed. Gains are depicted in blue, losses are depicted in red. (B) Individual copy number profile of case #16 showing a prototypical, gain, loss and amplification in the 11q region. Each probe is aligned from chromosome 1 to Y and p to q arm. (C) Mutational overview of ten cases of BLL with 11q aberration. The heat map shows the case-specific pattern of driver mutations found by next-generation sequencing. Each column represents a case and each row represents a gene. The right bar graph illustrates the mutation frequency of each gene. (D) A diagram of the relative positions of driver mutations is shown for BTG2, ETS1 and GNA13 genes. Domains BTG2: BTG family domain. Domains ETS1: PNT: pointed domain; TAD: transactivation domain; H-1/2: inhibitory a-helices 1/2; DBD: DNA binding domain; H4-5: a-helix 4/5. Domains GNA13: G-alpha: G protein a subunit. Circles indicate missense mutations, triangles indicate truncating mutations and rhombi indicate splicing mutations.
BLL-11q cases displayed similar levels of complexity as MYC-positive BL (7.1 vs. 6 alterations/case),20 but significantly lower than those of GCB-DLBCL (7.1 vs. 19, alterations/case P<0.0001).13 The BLL-11q genomic profile differed from that of BL and DLBCL (Online Supplementary Figure S6). BLL-11q had frequent gains of 5q21.3-q32 and losses of 6q12.1-q21 and lacked the 1q gains seen in MYC-positive BL. BLL-11q also lacked alterations typically seen in GCB-DLBCL such as gains of 2p16.1 and 7p and losses of 1p36.32.
The six tumors negative for both MYC and 11q-aberrations in patients younger than 40 years had similar levels of genomic complexity as those observed in BLL-11q (11.8 vs. 7.1 alterations/case) (Online Supplementary Figure S7A). The unique significant aberration that distinguished the two groups was the presence/absence of the 11q aberration.
The review of the literature regarding other lymphoid neoplasms confirmed that the 11q alteration observed in BLL-11q is mainly absent in other lymphoma entities with the exception of transformed follicular lymphoma (16%) (Online Supplementary Results).22
Next-generation sequencing and gene expression analysis
Target next-generation sequencing showed a total of 49 potential driver mutations affecting 27 different genes (mean=4.9 mutations per case) (Figure 2C, D, Online Supplementary Figures S8 and S9; Online Supplementary Table S7). Interestingly, all cases lacked the typical BL mutations in ID3, TCF3, or CCND3 genes, and their mutational profile was more similar to that of other GC-derived lymphomas with recurrent mutations affecting BTG2 (4 cases), DDX3X, ETS1, EP300, and GNA13 (3 cases each) (Online Supplementary Table S8). Five cases had mutations in epigenetic modifier genes such as EP300, CREBBP, KMT2C, EZH2, ARID1A, KMT2D, HIST1H1D and HIST1H2BC. Two cases had concomitant TMEM30A deleterious mutations associated with a 6q14.1 deletion as seen in DLBCL but not in BL (Figure 2C).14–16
BTG2 mutations, found in four cases, comprised three missense mutations and one deletion in a splicing site. BTG2 is a tumor suppressor gene with an important role in G1/S transition through inhibition of CCND1 in a pRb-dependent mechanism.23 These BTG2-inactivating mutations could release CCND1 inhibition and accelerate G1/S transition. GNA13 mutations were found in three cases including four missense and two nonsense mutations, and one missense mutation in a splicing site. Two MYC missense mutations occurred in the central domain of the protein, but did not affect threonine phosphorylation sites (Online Supplementary Table S7).24 ETS1 mutations have been previously described in BLL-11q and activated B-cell DLBCL13,17 but not in conventional BL (Online Supplementary Table S8).14,15 We detected three coding mutations located on the winged helix-turn-helix DNA-binding domain but the previously described exon 1 mutations (NM_005238) were absent in this series. ETS1 RNA expression was lower in BLL-11q than in MYC-positive BL (relative expression 6.6 vs. 19.3, P<0.001) and was also lower in ETS1-mutated than wild-type BLL-11q (relative expression 1.9 vs. 8.6, P=0.03) (Online Supplementary Figure S4B).
The mutational profile of four MYC-negative/11q alteration-negative cases with material available was analyzed using the same approach. No mutations in BTG2, EP300 or ETS1 genes were observed. Moreover, three out of four did not harbor any BL-related mutation on ID3, TCF3 and CCND3 whereas the fourth case had a mutational profile commonly seen in BL with MYC, DDX3X, SMARC4, CCND3 and TP53 mutations (Online Supplementary Figure S7B).
Discussion
BLL-11q was initially recognized as a particular subset of HGBCL that had an expression profile and some pathological characteristics similar to those of BL but lacked MYC-translocations and alternatively shared a common pattern of gains at 11q23 associated with losses at 11q24-qter. The particular features of these cases raise some uncertainty on their precise categorization as a variant of BL or a tumor related to other HGBCL.1,4–6,9–11 On the other hand, the limited number of cases reported and the different methodologies used for their recognition do not provide a clear view of their incidence and clinic-pathological characteristics.
In this study we searched our files for cases that could be reclassified as BLL-11q among 95 tumors previously classified as BL, atypical BL, or HGBCL, NOS and found eight (8%) cases with the chromosomal aberration. These cases, together with three additional cases received on consultation, were investigated for copy number alterations and mutational profiles and compared to the genomic aberrations recently identified in BL, DLBCL, and HGBCL.13–17 The complexity of BLL-11q was similar to that of MYC-positive BL,20 but significantly lower than that of GCB-DLBCL.13 The BLL-11q genomic profile differed from that of BL and DLBCL (Online Supplementary Figure S6). BLL-11q had frequent gains of 5q21.3-q32 and losses of 6q12.1-q21 and lacked the 1q gains seen in MYC-positive BL. BLL-11q also lacked alterations typically seen in GCB-DLBCL such as gains of 2p16.1 and 7p and losses of 1p36.32. Additionally, we identified a mutational profile in BLL-11q which was different from that of MYC-positive BL since all cases lacked the typical BL mutations in ID3, TCF3, or CCND3 genes and had mutations in BTG2, DDX3X, and ETS1 not seen in BL. In addition, BLL-11q had mutations in epigenetic modifier genes such as EP300, CREBBP, KMT2C, EZH2, ARID1A, KMT2D, HIST1H1D and HIST1H2BC, which are common in DLBCL, particularly of the GCB subtype. Other genes frequently mutated in GCB-DLBCL but not in BL were GNA13 and TMEM30A associated with 6q14.1.14–16
We also compared our results with those of two recent studies on HGBCL (including double-and triple-hit lymphomas).25,26 These cases also have recurrent mutations in histone modifier genes such as KMT2D, CREBBP and EZH2 (Online Supplementary Table S8). Intriguingly, HGBCL, NOS, mainly with MYC-translocations, shared mutations in genes frequently mutated in both BL and GC-DLBCL.25,26 All these observations suggest that BLL-11q is a neoplasm closer to other GC-derived lymphomas rather than BL in which the 11q aberration together with other mutations may play a relevant role in their pathogenesis. While this manuscript was under revision, Wagener et al. published a mutational study of 15 BLL-11q cases. Similar to our findings, no mutations in ID3/TCF3 were identified and those cases carried frequent mutations in GC-DLBCL-associated genes such as GNA13, FOXO1 and EZH2. Intriguingly, that study did not find mutations in BTG2, KMT2D, KMT2C or CREBBP observed in our study.27 Collectively, these findings indicate that the genomic and mutational profiles of BLL-11q are different from those of BL and more similar to those of other GC-derived lymphomas.
In addition to the genetic differences, our BLL-11q cases differed clinically, morphologically and phenotypically from conventional BL and instead showed features more consistent with HGBCL or DLBCL. As in previous studies, all our patients were younger than 40 years, although occasional cases in older patients have been reported.4,5,27 Contrary to BL, most of our cases of BLL-11q presented with localized lymphadenopathy.4,5,27 These cases have a favorable outcome after therapy, although the optimal clinical management remains to be determined. Morphologically, our cases had a prominent “starry sky” pattern and high proliferation rate (>90%) but did not have the typical cytological features of BL since they were better classified as HGBCL with blastoid or intermediate features between HGBCL (8 cases) and DLBCL (2 cases) and only one had features of atypical BL. As previously reported,4 two of our cases displayed a follicular growth pattern, with an underlying meshwork of follicular dendritic cells, raising the differential diagnosis with other pediatric lymphomas such as large B-cell lymphoma with IRF4 rearrangement.3 However, BLL-11q did not express IRF4/MUM1 and often exhibited a “starry sky” pattern with frequent mitotic figures, features that are not usual in large B-cell lymphoma with IRF4 rearrangement. We also identified different immunohistochemical stains that could help in the differential diagnosis from other lymphomas entities. LMO2, a GC marker that is typically downregulated in BL and other lymphomas with MYC translocation,18 was detected in 46% of our BLL-11q cases. In addition, and contrary to BL, MYC expression with a diffuse and intense pattern was only detected in one of our cases while the other four positive cases either exhibited partial positivity or the intensity was weak contrary to the pattern seen in BL.
Negativity for MYC rearrangement is a crucial element for the recognition of these cases. The recommended technique for detecting MYC translocations in clinical practice is FISH analysis using break-apart probes, with the limitation that a subset of 4% of MYC-positive cases are not detected with this method but picked up using MYC/IGH probes.28 The genetic feature that distinguishes BLL-11q is an alteration of the 11q arm that prototypically is characterized by an 11q23.2-q23.3 gain/amplification and 11q24.1-qter loss. Additionally, isolated cases have been recognized with single 11q24.1-qter terminal loss or 11q23 gain with 11q24-qter copy number neutral loss of heterozygosity.4,11 In our study we identified the presence of these 11q alterations using copy number arrays. We also confirmed the presence of 11q alterations by FISH analysis with a custom probe in all tested cases, suggesting that this approach may be useful in clinical practice to identify such cases (Online Supplementary Table S8). The specificity of this FISH approach was also confirmed by the fact that no false positive cases were observed in the 12 lymphoma cases in which the array showed a normal 11q pattern. Nevertheless, more studies on the clinical value of this probe are needed and, for the time being, confirmation of the finding by copy number array would be desirable. The specific 11q alteration observed in BLL-11q should be distinguished from other 11q aberrations such as 11q gains of the 11q24 region that include ETS1 and FLI1, detected in DLBCL,29 or the 11q25 losses missing ETS1 and FLI1 described in some post-transplant lymphoproliferative disorders.30,31 On the other hand, although the 11q23 gain/11q24-qter loss of BLL-11q is mainly absent in other lymphoma entities, its detection should not be considered as a unique feature to diagnose BLL-11q cases since some transformed follicular lymphomas may carry a similar 11q aberration pattern.22
In summary, BLL-11q is a GC-derived lymphoma with genomic and mutational profiles closer to those of HGBCL or GCB-DLBCL rather than BL in which the 11q aberration, together with other mutations, may play a relevant role in pathogenesis. These observations support a reconsideration of the “Burkitt-like” term for these tumors. Although, the most appropriate name is not easy to propose and requires broader discussion and consensus, we think that the term “aggressive B-cell lymphoma with 11q aberration” captures their pathological features. To identify these cases we suggest performing copy number arrays or FISH with the 11q probe in cases with BL, DLBCL, and HGBCL morphology, a GC phenotype and very high proliferative index (>90%), without MYC rearrangements, in young patients. The recognition of these tumors is clinically relevant because they have a favorable outcome after therapy, although further studies are needed to determine the optimal clinical management.
Acknowledgments
We thank the centers of the Sociedad Española de Hematología y Oncología Pediátricas that submitted cases for consultation and Noelia Garcia, Silvia Martín, and Helena Suarez for their excellent technical assistance. We are indebted to the IDIBAPS Genomics Core Facility and to HCB-IDIBAPS Biobank-Tumor Bank and Biobanc de l’HospitaI Infantil Sant Joan de Déu, both integrated in the National Network Biobanks of ISCIII for the sample and data procurement. We thank Prof. Reiner Siebert from the University of Ulm for sharing the 11q
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/9/1822
Funding
This work was supported by Asociación Española Contra el Cáncer (AECC CICPFI6025SALA), Fondo de Investigaciones Sanitarias Instituto de Salud Carlos III (Miguel Servet program CP13/00159 and PI15/00580, to IS), Spanish Ministerio de Economía y Competitividad, Grant SAF2015-64885-R (EC), Generalitat de Catalunya Suport Grups de Recerca (2017-SGR-1107 I.S. and 2017-SGR-1142 to EC), and the European Regional Development Fund “Una manera de fer Europa”. JER-Z was supported by a fellowship from the Generalitat de Catalunya AGAUR FI-DGR 2017 (2017 FI_B01004). EC is an Academia Researcher of the "Institució Catalana de Recerca i Estudis Avançats" (ICREA) of the Generalitat de Catalunya. This work was developed at the Centro Esther Koplowitz, Barcelona, Spain. The group is supported by Acció Instrumental d’Incorporació de Científics i Tecnòlegs PERIS 2016 (SLT002/16/00336) from the Generalitat de Catalunya.
The copy-number data reported in this article have been deposited at the GEO database under accession number GSE116527. Sequencing data have been deposited at the European Nucleotide Archive (ENA, accession number ERP110085).
References
Articles from Haematologica are provided here courtesy of Ferrata Storti Foundation
Full text links
Read article at publisher's site: https://doi.org/10.3324/haematol.2018.207928
Read article for free, from open access legal sources, via Unpaywall:
https://haematologica.org/article/download/9049/64661
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
MYC-rearranged mature B-cell lymphomas in children and young adults are molecularly Burkitt Lymphoma.
Blood Cancer J, 14(1):171, 07 Oct 2024
Cited by: 0 articles | PMID: 39375391 | PMCID: PMC11458770
High-grade B-cell lymphoma with 11q aberration in the HIV setting: a clinicopathological study of 10 cases and literature review.
Infect Agent Cancer, 19(1):42, 11 Sep 2024
Cited by: 0 articles | PMID: 39261841 | PMCID: PMC11391791
What is new in the 5th edition of the World Health Organization classification of mature B and T/NK cell tumors and stromal neoplasms?
J Hematop, 17(2):71-89, 29 Apr 2024
Cited by: 0 articles | PMID: 38683440
Review
Genetic alterations in mature B- and T-cell lymphomas - a practical guide to WHO-HAEM5.
Med Genet, 36(1):59-73, 06 Mar 2024
Cited by: 0 articles | PMID: 38835967 | PMCID: PMC11006337
ETS1 Function in Leukemia and Lymphoma.
Adv Exp Med Biol, 1459:359-378, 01 Jan 2024
Cited by: 0 articles | PMID: 39017852
Review
Go to all (42) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE116527
Nucleotide Sequences
- (1 citation) ENA - ERP110085
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The mutational landscape of Burkitt-like lymphoma with 11q aberration is distinct from that of Burkitt lymphoma.
Blood, 133(9):962-966, 19 Dec 2018
Cited by: 33 articles | PMID: 30567752 | PMCID: PMC6396176
A Diagnostic Approach to the Identification of Burkitt-like Lymphoma With 11q Aberration in Aggressive B-Cell Lymphomas.
Am J Surg Pathol, 45(3):356-364, 01 Mar 2021
Cited by: 18 articles | PMID: 33136583
A comprehensive flow-cytometry-based immunophenotypic characterization of Burkitt-like lymphoma with 11q aberration.
Mod Pathol, 31(5):732-743, 12 Jan 2018
Cited by: 21 articles | PMID: 29327714
Emerging entities: high-grade/large B-cell lymphoma with 11q aberration, large B-cell lymphoma with IRF4 rearrangement, and new molecular subgroups in large B-cell lymphomas. A report of the 2022 EA4HP/SH lymphoma workshop.
Virchows Arch, 483(3):281-298, 09 Aug 2023
Cited by: 5 articles | PMID: 37555980 | PMCID: PMC10541818
Review Free full text in Europe PMC





