Abstract
Free full text

A single‐cell transcriptome atlas of the adult human retina
Abstract
The retina is a specialized neural tissue that senses light and initiates image processing. Although the functional organization of specific retina cells has been well studied, the molecular profile of many cell types remains unclear in humans. To comprehensively profile the human retina, we performed single‐cell RNA sequencing on 20,009 cells from three donors and compiled a reference transcriptome atlas. Using unsupervised clustering analysis, we identified 18 transcriptionally distinct cell populations representing all known neural retinal cells: rod photoreceptors, cone photoreceptors, Müller glia, bipolar cells, amacrine cells, retinal ganglion cells, horizontal cells, astrocytes, and microglia. Our data captured molecular profiles for healthy and putative early degenerating rod photoreceptors, and revealed the loss of MALAT1 expression with longer post‐mortem time, which potentially suggested a novel role of MALAT1 in rod photoreceptor degeneration. We have demonstrated the use of this retina transcriptome atlas to benchmark pluripotent stem cell‐derived cone photoreceptors and an adult Müller glia cell line. This work provides an important reference with unprecedented insights into the transcriptional landscape of human retinal cells, which is fundamental to understanding retinal biology and disease.
Introduction
The eye is a highly specialized sensory organ in the human body. Sight is initiated by the conversion of light into an electrical signal in the photoreceptors of the neurosensory retina. The rod photoreceptors are responsible for light detection at extremely low luminance, while the cone photoreceptors are responsible for color detection and operate at moderate and higher levels. Following preprocessing, by horizontal, bipolar, and amacrine cells, the resultant signal is transferred via ganglion cells to the brain. Neurotransmitter support is provided by Müller glia, retinal astrocytes, and microglial cells. Inherited retinal diseases are becoming the leading cause of blindness in working‐age adults, with loci in over 200 genes associated with retinal diseases (RetNet: https://sph.uth.edu/retnet/), often involving specific retinal cell types. Knowledge of the transcriptome profile of individual retinal cell types in humans is important to understand the cellular diversity in the retina, as well as the study of retinal genes that contribute to disease in individual retinal cell types (Hornan et al, 2007; Farkas et al, 2013; Whitmore et al, 2014; Mustafi et al, 2016; Pinelli et al, 2016).
The transcriptome profiles of whole human retina from adults (Hornan et al, 2007; Farkas et al, 2013; Whitmore et al, 2014; Mustafi et al, 2016; Pinelli et al, 2016) and during fetal development (Kozulin et al, 2009; Hoshino et al, 2017) have been previously described. However, these studies only assayed the averaged transcriptional signatures across all cell types, meaning that information on the cellular heterogeneity in the retina is lost. As such, the transcriptional pathways that underlie the highly specialized function of many human retinal cell types remain unclear, including the rod and cone photoreceptors, Müller glial cells, horizontal cells, and amacrine cells. Recent advances in RNA sequencing and microfluidic platforms have dramatically improved the accessibility of single‐cell transcriptomics, with increased throughput at a lower cost. Critically, single‐cell microfluidics and low‐abundance RNA library chemistries allow accurate profiling of the transcriptome of individual cell types. This has been demonstrated in the mouse, where transcriptome profiles of the mouse retina (Macosko et al, 2015) and retinal bipolar cells (Shekhar et al, 2016) have been described at the single‐cell level using the Drop‐seq method (Macosko et al, 2015). These studies provided a molecular classification of the mouse retina and identified novel markers for specific cell types, as well as novel candidate cell types in the retina. Recently, single‐cell transcriptomics was used to analyze the human retina. Phillips et al (2018) have profiled a total of 139 cells from adult retina using the C1 Fluidigm platform, but the limited number of profiled cells presents challenges in the annotation and accurate identification of individual retinal cell types. Moreover, a flow cytometry approach was used to isolate 65 human fetal cone photoreceptors followed by scRNA‐seq profiling (Welby et al, 2017). During the preparation of this manuscript, Voigt et al (2019) reported scRNA‐seq profiling of 8,217 cells from human retina obtained from a mixed pool of donors that included a healthy patient, a patient with early glaucoma, and one with unknown ocular history.
Herein, we report the generation of a human neural retina transcriptome atlas using 20,009 single cells collected from three healthy donors. Our data provide new insights into the transcriptome profile of major human retinal cell types and establish a high cellular‐resolution reference of the human neural retina, which will have implications for identification of biomarkers and understanding retinal cell biology and diseases.
Results
Preparation of human neural retina samples and generation of single‐cell transcriptome atlas
We obtained post‐mortem human adult eyes approved for research purposes following corneal transplantation. As the transcriptome profile of human retinal pigment epithelial cells has already been reported (Liao et al, 2010; Strunnikova et al, 2010), we focused solely on the neural retina layers. In this study, we extracted the neural retina from 12 donor eyes (Appendix Table S1). We observed consistent cell viability across retinal tissues retrieved within 15 h post‐mortem (Appendix Fig S1A) and found that donor age does not impact negatively on cell viability in the extracted neural retina (Appendix Fig S1B). To minimize potential risk of mRNA degradation due to reduced cell viability, we selected three donor samples retrieved within 15 h post‐mortem and analyzed them with single‐cell RNA sequencing (scRNA‐seq) using the 10X Genomics Chromium platform.
Sequence data from five scRNA‐seq libraries derived from the three neural retinal samples were pooled for processing and analysis. From 23,000 cells, we obtained an average of 40,232 reads per cell and 1,665 UMIs (unique transcripts) per cell. Following quality control and filtering using the Seurat package (Butler et al, 2018), our final dataset contained 20,009 cells, which were taken forward for further analysis.
The scRNA‐seq data were initially analyzed using an unsupervised graph clustering approach implemented in Seurat (version 2.2.1) to classify individual cells into cell populations according to similarities in their transcriptome profiles. Overall, the cells were classified into 18 transcriptionally distinct clusters (Appendix Fig S2). We first assessed the variation between donor samples (Appendix Table S2). Interestingly, although many of the identified clusters are well represented in all three donor retinal samples, we also observed several donor‐specific clusters corresponding to rod photoreceptors (Appendix Fig S3A). In contrast, we observed minimal variation between two different libraries prepared from the same donor sample, supporting the quality of the scRNA‐seq datasets in this study (Appendix Fig S3B). The average expression for all detected genes in each cluster is listed in Dataset EV1.
Identification of major cell types in the human retina using scRNA‐seq
Based on known markers (Blackshaw et al, 2001; Imanishi et al, 2002; Corbo et al, 2007; Soto et al, 2008; Klimova et al, 2015; Macosko et al, 2015; Shekhar et al, 2016; Vecino et al, 2016), we were able to assign cell identities to 16 of the 18 clusters (Fig 1A–D), corresponding to rod photoreceptors (PDE6A, CNGA1, RHO), cone photoreceptors (ARR3, GNGT2, GUCA1C), Müller glia (RLBP1/CRALBP), retinal astrocytes (GFAP), microglia (HLA‐DPA1, HLA‐DPB1, HLA‐DRA), bipolar cells (VSX2, OTX2), retinal ganglion cells (NEFL, GAP43, SNCG), amacrine cells (GAD1, CALB1, CHAT), and horizontal cells (ONECUT1, ONECUT2). The expression of selected marker genes is displayed in t‐SNE plots (Fig 1D). Two clusters (C5 and C14) express markers from multiple retinal cell types (Appendix Fig S4); thus, we were unable to assign cell identities to these two clusters and they were excluded from further analysis. Interestingly, our data demonstrated multiple transcriptionally distinct clusters within the rod photoreceptors (six clusters) and bipolar cells (three clusters). In contrast, only one cluster was detected for cone photoreceptors, Müller glia, retinal ganglion cells, horizontal cells, amacrine cells, retinal astrocytes, and microglia, respectively. Correlation analysis confirmed the similarity between clusters within the same cell type (Fig 1E). As expected, we observed high correlations between the expression levels of transcripts within photoreceptor cell types (rod and cones), as well as glial cells (retinal astrocytes and Müller glia) and other retinal neurons (bipolar cells, retinal ganglion cells, amacrine cells, and horizontal cells). The composition of cell populations across our three donors shows that the majority of the cells in human neural retina were rod photoreceptors (~74%) followed by bipolar cells (~10%). These results are similar to those reported in mice, where rod photoreceptors and bipolar cells form the majority of cells in the retina (Jeon et al, 1998; Macosko et al, 2015).

- A, B
t‐SNE visualization of 20,009 human retinal cells colored by (A) annotation of 18 transcriptionally distinct clusters (C0‐C17) and (B) their distribution in three donor retina samples.
- C
Feature expression heatmap showing expression patterns of major retinal class markers across 16 retinal cell clusters. The size of each circle depicts the percentage of cells expressing the marker within the cluster. Brown color indicates ≥ 10 nTrans (number of transcripts).
- D
t‐SNE plots showing expression of a set of selected marker genes for major retinal classes.
- E
Correlation matrix for the identified 18 clusters. The upper triangle depicts the z‐value for correlation, and the lower triangle depicts the correlation coefficient for gene expression in clusters.
- F
Heatmap of differentially expressed genes used to classify cell types for each cluster compared to all other clusters for the 18 retinal cell clusters. The rows correspond to the top 10 genes most selectively upregulated in individual clusters (P < 0.01, Benjamini–Hochberg correction), and the columns show individual cells ordered by cluster (C0‐C17).
To identify genes whose expression was specific to a given cell type, we performed differential gene expression analysis to identify marker genes for each cluster (Fig 1F). We subsequently extracted membrane‐related proteins from gene ontology annotations to identify potential surface markers, which can be used to develop immuno‐based methods to isolate primary culture of individual retinal cell types. Appendix Table S3 lists the identified markers for individual retinal cell types. We also assessed the gene expression of a panel of commonly known markers in amacrine cells and bipolar cells (Figs EV1 and EV2), as well as a panel of markers for subtype identification recently identified in mouse scRNA‐seq studies (Macosko et al, 2015; Shekhar et al, 2016). In particular, the bipolar clusters can be classified as OFF‐bipolar cells (GRIK1 +: C6) and ON‐bipolar cells (ISL1 +: C8, C11). Further analysis showed that C8 represents rod bipolar cells based on the marker PRKCA, while C11 expresses the marker TTR corresponding to a diffuse bipolar subtype DB4 (Fig EV1). In summary, we profiled the transcriptomes of all major cell types in the human retina in the presented dataset. Due to their abundance, for the subsequent analyses we focused on the photoreceptors and glial cells.
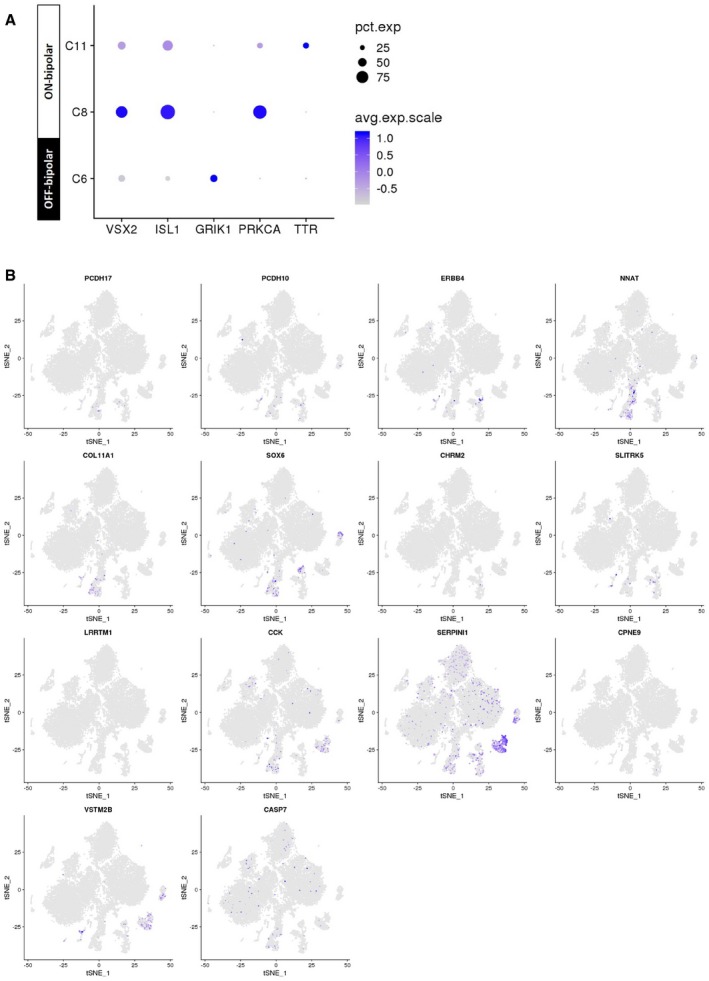
Feature expression heatmap of VSX2 (pan‐bipolar), ISL1 (ON‐bipolar), GRIK1 (OFF‐bipolar), PRKCA (rod bipolar cells), and TTR (DB4 bipolar).
t‐SNE plots showing gene expression for 14 new markers for individual bipolar subtypes identified in a previous mouse scRNA‐seq study (Shekhar et al, 2016).

- A, B
t‐SNE plots showing gene expression in the compiled human retina transcriptome atlas (20,009 cells). (A) 10 commonly used amacrine markers and (B) new markers for amacrine subtypes identified in a previous mouse scRNA‐seq study (Macosko et al, 2015).
Profiling healthy and degenerating human rod photoreceptor subpopulations
We profiled 14,759 rod photoreceptors and showed that they can be classified into six populations with distinct gene expressions (C0, C1, C2, C3, C4, and C7). We assessed these six clusters with a panel of seven known rod or pan‐photoreceptor markers (Fig 2A). Our results suggest differential expression patterns among the seven markers. All seven rod markers are highly abundant, consistent with previous scRNA‐seq studies of mouse and human retina (Macosko et al, 2015; Phillips et al, 2018). The seven markers showed differential expression patterns in the six identified rod photoreceptor clusters. In particular, RHO, GNGT1, and SAG have the highest levels of rod marker detected, followed by NRL, ROM1, GNAT1, and CNGA1. We also noted that ROM1 is expressed in both rod and cone photoreceptors, which is consistent with previous studies (Boon et al, 2008). Importantly, many rod photoreceptor clusters consist of a majority of cells from a single donor (> 90% for C0, C2, and C4 and > 80% for C1 and C7; Fig 2B). It is possible that this observation is due to the systematic biases such as differences in tissue retrieval time, age of donors, or other sample preparation variation. The exception is cluster C3, which is well represented by all three donors.

Feature expression heatmap of a panel of known marker genes for rod photoreceptors across the identified 16 retinal cell clusters. Brown color indicates ≥ 100 nTrans (number of transcripts).
Representation of the three donor retina samples in the six rod photoreceptor clusters.
Violin plot showing high or low expression levels of MALAT1 in rod photoreceptor clusters.
Distribution of rod photoreceptor populations with high MALAT1 expression (MALAT1‐hi) or low MALAT1 expression (MALAT1‐lo) in three donor retina samples.
Fluorescent in situ hybridization analysis of human peripheral retina showing heterogeneous levels of MALAT1 expression in the rod photoreceptors located in the outer nuclear layer (ONL). INL, inner nuclear layer; OPL, outer plexiform layer. Scale bar = 20 μm.
Next, we set out to further define and classify heterogeneity in rod photoreceptors. We observed that MALAT1, a long non‐coding RNA that plays a role in retinal homeostasis and disease (Wan et al, 2017), was robustly expressed in ~66% of the identified rod photoreceptors (9,750 cells), while the rest had little to no expression (5,009 cells; Fig 2C). As such, we utilized MALAT1 expression as a discriminator and investigated differences between rod photoreceptors with high expression (MALAT1‐hi; > 4.68 normalized transcripts per cell) or low expression (MALAT1‐lo; < 4.68 normalized transcripts per cell). MALAT1‐hi and MALAT1‐lo rod photoreceptors were consistently found in each donor and library samples, with MALAT1‐hi accounting for ~66, 90, and 36% of the rods in donors #1, #2, and #3, respectively (Fig 2D). To further validate this finding, we performed RNA in situ hybridization in another three donor retinal samples. We consistently observed the presence of MALAT1‐hi and MALAT1‐lo subpopulations of rod photoreceptors in all retinal samples (Figs 2E and EV3A). Together, these results showed the presence of heterogeneity within rod photoreceptors that can be distinguished by MALAT1 expression.
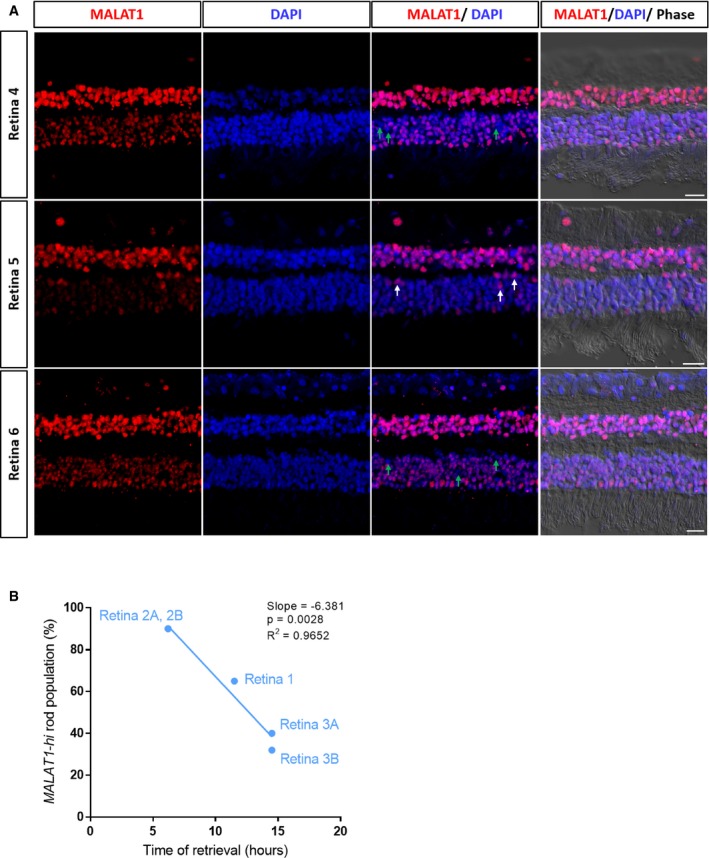
Fluorescent in situ hybridization showing expression of MALAT1 in three donor retina samples (Retina 4–6). Retina 5 from Fig 2E is also displayed here for easier comparison. Green arrows highlight rod photoreceptors with low levels of MALAT1 in Retina 4 and Retina 6, and white arrows highlight rod photoreceptors with high levels of MALAT1 in Retina 5. Scale bars = 20 μm.
Correlation of proportion of MALAT1‐hi rod populations with time of retina retrieval after death for Retina 1–3.
To rule out the possibility that the presence of MALAT1 rod subpopulations was due to donor sample variations, we applied canonical correlation analysis (CCA) to correct for technical and batch artifacts. We found that CCA effectively corrected the donor‐specific effect on rod photoreceptor clusters (Fig 3A and B). The average expression for all detected genes in each cluster is listed in Dataset EV2. Following CCA correction, we identified three rod photoreceptor clusters (CCA0, CCA1, and CCA10), which expressed a panel of seven rod photoreceptor markers and were well represented in all donor samples (Fig 3C). Notably, the majority of cells in CCA0 were low in MALAT1 expression, while CCA1 and CCA10 represented MALAT1‐hi rod subpopulations (Fig 3D and E). This is consistent with our RNA in situ hybridization analysis, where we consistently observed MALAT1‐hi and MALAT1‐lo subpopulations of rod photoreceptors in all retinal samples (Fig EV3A). Collectively, these results provide evidence that MALAT1 heterogeneity in rod photoreceptors is not due to inter‐individual variability.
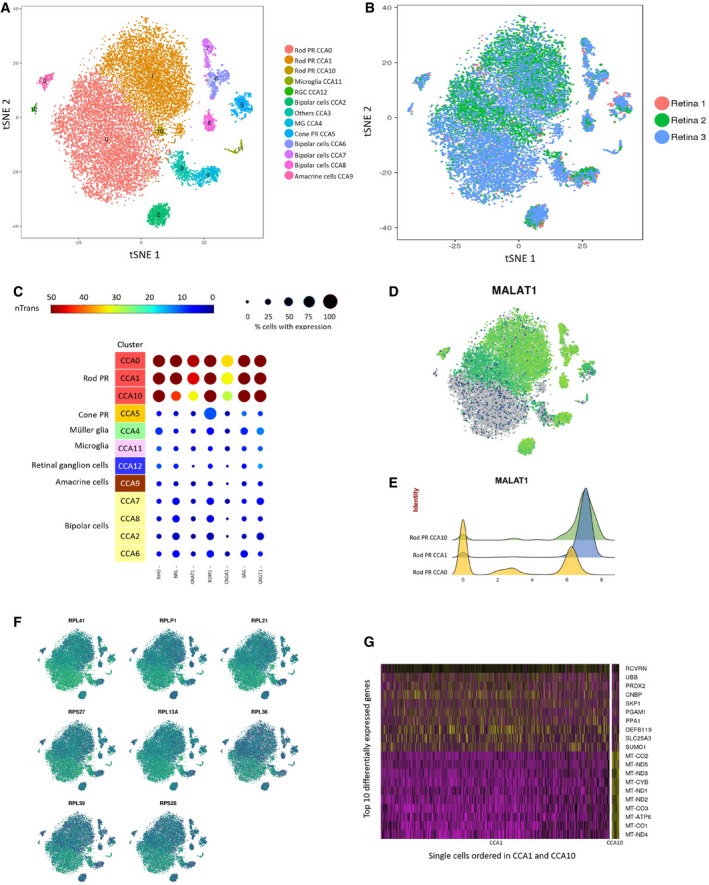
Canonical correlation analysis was used to effectively correct donor‐specific variations in rod photoreceptors.
- A, B
(A) t‐SNE visualization of human retinal cells colored by annotation of 13 transcriptionally distinct clusters (CCA0‐CCA12) and (B) their distribution in three donor retina samples.
- C
Feature expression heatmap showing expression patterns of seven rod photoreceptor markers across 12 retinal cell clusters. The size of each circle depicts the percentage of cells expressing the marker within the cluster. Brown color indicates ≥ 50 nTrans (number of transcripts).
- D
t‐SNE plots showing expression of MALAT1.
- E
Expression pattern of MALAT1 in the rod photoreceptor showing MALAT1‐hi (CCA1, CCA10) and MALAT1‐lo (CCA0) subpopulations. The x‐axis depicts normalized transcript levels.
- F
t‐SNE plots showing expression of major ribosomal genes.
- G
Heatmap of differentially expressed genes between the two MALAT1‐hi clusters CCA1 and CCA10. The rows correspond to top 10 genes most selectively upregulated in individual clusters (P < 0.01, Benjamini–Hochberg correction), and the columns show individual cells ordered in CCA1 and CCA10.
We also considered the possibility that MALAT1‐lo rod subpopulations may represent an artifact of “low‐quality cells” in scRNA‐seq data, due to a low number of sequencing reads or broken cell membrane. In this regard, upregulated levels of mitochondrial‐encoded genes and ribosomal proteins can be used to identify such low‐quality cells in scRNA‐seq data (Ilicic et al, 2016). For our scRNA‐seq dataset, we did not observe upregulation in gene expression for a panel of ribosomal proteins (RPL41, RPLP1, RPL21, RPS27, RPL13A, RPL36, RPL39, and RPS28; Fig 3F). However, the rod cluster CCA10, representing 1.4% of rod photoreceptor cells, showed markedly increased levels of mitochondrial‐encoded genes (MT‐CO2, MT‐ND5, MT‐ND3, MT‐CYB, MT‐ND1, MT‐ND2, MT‐CO3, MT‐ATP6, MT‐CO1, and MT‐ND4; Fig 3G), suggesting that CCA10 represented a low‐quality MALAT1‐hi rod cluster and was excluded from further analysis.
As we utilized post‐mortem retinal samples in this study, we reasoned that MALAT1‐lo subpopulations may potentially reflect the early stages of post‐mortem degeneration in rod photoreceptors. To determine this, we extracted retinal samples from the same donor at different time points of progressive post‐mortem degeneration, with longer time points predicted to have more stressed/dying photoreceptors. Our results showed that there were a high proportion of MALAT1‐hi rod photoreceptors at 7 h post‐mortem (Fig 4, ~95%). However, we observed a marked decrease in MALAT1 expression in rod photoreceptors at 13 h post‐mortem. Similar results were observed for the three retinal samples processed for scRNA‐seq (Fig EV3B). Together, these results demonstrated that MALAT1 is a novel marker for healthy photoreceptors with loss of expression potentially preceding putative cell degeneration. In summary, we showed that scRNA‐seq can be used to profile healthy (CCA1) and degenerating rod photoreceptors (CCA0), which can be distinguished by high or low MALAT1 expression levels, respectively.

Fluorescent in situ hybridization analysis of the same donor human peripheral retina at different time points post‐mortem (7 and 13 h, Retina 7), showing decreases in MALAT1‐hi rod subpopulations in the outer nuclear layer (ONL) at later time point. INL, inner nuclear layer; OPL, outer plexiform layer. Scale bar = 20 μm. White arrows indicated MALAT1‐hi rod photoreceptors.
Transcriptome profile of cone subtypes in the human retina
We detected 564 cone photoreceptor cells in our scRNA‐seq data, which are distinguishable from the other cell types by the expression of the cone marker genes ARR3, CNBG3, GNAT2, GNGT2, GRK7, GUCA1C, PDE6C, PDE6H, OPN1LW, RXRG, and THRB (Fig 5A). All 11 marker genes analyzed show specific expression patterns in the cone cluster (C10). We set out to further assess the composition of the cone cluster. In humans, there are three identified subtypes of cone photoreceptor, which can be distinguished by expression of a sole opsin gene: OPN1SW‐positive S‐cones, OPN1MW‐positive M‐cones, and OPN1LW‐positive L‐cones respond preferentially to shorter, medium, and longer wavelengths responsible for color vision (Roorda & Williams, 1999). Notably, OPN1LW and OPN1MW exhibit ~98% sequence homology and are unable to be distinguished by 3′ sequencing utilized in this study. By quantifying the number of cells that express the opsin genes, our results showed that the majority of the cone cluster are L/M‐cones (73.22%) and S‐cones in much lower proportion (3.19%; Fig 5B), at levels consistent with those estimated by a previous adaptive optics and photobleaching study (Roorda & Williams, 1999). As expected, the identified cone photoreceptors only express either OPN1SW or OPN1LW/MW (Fig 3C).

Feature expression heatmap showing the expression of 11 known cone photoreceptor markers across 16 retinal cell clusters. Brown color indicates ≥ 10 nTrans (number of transcripts).
The proportion of cone photoreceptor subtypes identified in population C10, based on expression of OPN1LW/OPN1MW (L/M‐cones) and OPN1SW (S‐cones).
Scatter plots showing expression of OPN1LW/OPN1MW or OPN1SW in individual cone photoreceptors for population C10. The color depicts expression level for OPN1LW/OPN1MW in individual cells.
Heatmap of top 20 differentially expressed genes between L/M‐cones and S‐cones. The color depicts normalized gene expression (z‐score capped at 2.5).
Expression pattern of glial markers in Muller glia (C9) and retinal astrocytes (C16). The x‐axis depicts normalized transcript levels.
To further study the molecular differences and identify molecular markers for the cone subtypes, we performed differential gene expression analysis to determine genes that can distinguish the cone subtypes. Our results identified a panel of genes that differentially marked S‐cones (e.g., CCDC136, RAMP1, LY75, CADM3, TFPI2, CRHBP, RAB17, UPB1, RRAD, and SLC12A1) and L/M‐cones (e.g., THRB, KIF2A, LBH, PGP, CHRNA3, AHI1, and LIMA1; Fig 3D). We compared this list of cone subtype genes to those identified in scRNA‐seq studies of the macaque and mouse retina, and showed that a number of the cone subtype genes in humans are conserved in macaque and mouse (Macosko et al, 2015; Peng et al, 2019), including S‐cone marker CCDC136 and L/M‐cone marker THRB. Interestingly, CCDC136 is located next to the OPN1SW locus in humans and could possibly be co‐regulated at the transcriptional level. The thyroid hormone receptor THRB is required for the development of M‐cones in mice (Ng et al, 2001) and L/M‐cones in humans as determined by a pluripotent stem cell model (Eldred et al, 2018). Notably, there are two known receptor isoforms for THRB (TRβ1 and TRβ2) and further research to determine the precise roles of THRB isoforms in subtype specification of human cones would be of great interest. Moreover, the transcription factor TBX2 has been implicated in subtype specification of Sws1‐cones in zebrafish and chicken (Alvarez‐Delfin et al, 2009; Enright et al, 2015). In support of these studies, our data showed that TBX2 marks the S‐cones in humans, which is also conserved in macaque (Peng et al, 2019). Together, these results detailed the molecular profiles and identified marker genes that can distinguish the cone subtypes in humans.
Assessment of glial cells in human retina
Next, we focused on two related glial cell types in the human retina, the Müller glia, and the retinal astrocytes. Our scRNA‐seq data have profiled a total of 2,723 Müller glial cells, which classified into a single cluster (C9), and 49 retinal astrocytes, which form a single cluster (C16). Figure 5E shows the expression of a panel of 9 commonly used markers for Müller glia and retinal astrocytes. Our results demonstrated that many of these markers are present in both Müller glia and retinal astrocytes at differential expression levels. VIM, GLUL, and S100A1 marked both Müller glia and retinal astrocytes at high expression levels. GFAP represents a reliable marker for retinal astrocytes, and its expression is consistent with a previous report (Vecino et al, 2016). Notably, Müller glia are low in GFAP expression, indicating they are not in an activated state commonly caused by stresses and reactive gliosis (Fernández‐Sánchez et al, 2015). The S100B is also expressed in retinal astrocytes at varying levels but absent in Müller glia. Conversely, Müller glia can be distinguished from retinal astrocytes by high expression levels of RLBP1, and RGR to a lesser extent. Together, these results provide insights into the differential expression patterns of known glial markers in Müller glia and retinal astrocytes in humans.
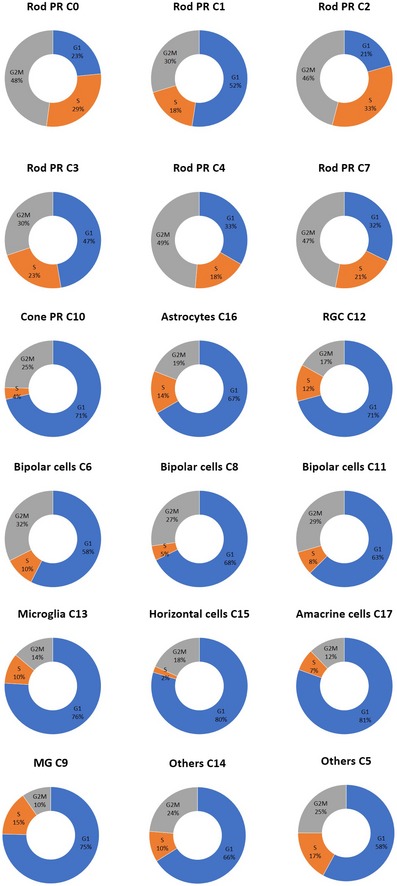
As glial cell proliferation has been linked to a range of pathological conditions including retinal gliosis and retinal injury (Subirada et al, 2018), this provides a means of assessing the health of the profiled retinas. We assigned a cell cycle phase score to each cell using gene expression signatures for the G1, S, G2, and G2/M phases (Kowalczyk et al, 2015; Fig EV4). We determined that most of the Müller glial cells expressed genes indicative of cells in G1 phase (75%), suggesting they are not proliferative. These results demonstrated the absence of hallmarks of gliosis/retinal injury and support the quality of the donor retinas profiled.
Using the human neural retina transcriptome atlas for benchmarking
To demonstrate the use of our dataset as a benchmarking reference, we compared the scRNA‐seq profiles of distinct cell types generated using alternative methods, including fetal human cone photoreceptors, human‐induced pluripotent stem cell‐derived cone photoreceptors (hiPSC‐cone; Welby et al, 2017), and a sample of adult human retina with 139 cells (Phillips et al, 2018). Correlation analysis demonstrated that the adult human retina sample showed highest similarity to rod photoreceptor (0.63; Fig EV5), which is expected as rod photoreceptors represent the majority of the cells in the retina. Interestingly, our results also showed that the transcriptome of hiPSC‐cone after 15 weeks of differentiation exhibited the highest similarity to cone photoreceptors, and low similarities to all other retinal cell types (Figs 6A and EV5). In particular, hiPSC‐cone showed high similarities to fetal cone photoreceptors and adult cone photoreceptors (0.71 and 0.61, respectively), and a much lower similarity to adult rod photoreceptors (0.33). In support of this, principal component analysis demonstrated that the hiPSC‐cone are closer to fetal cone photoreceptors, rather than the adult counterpart (Fig 6B). These results confirmed direct differentiation of hiPSCs to cone photoreceptors with good quality, and the hiPSC‐derived cone photoreceptors are closer to fetal origin compared to their adult counterpart.
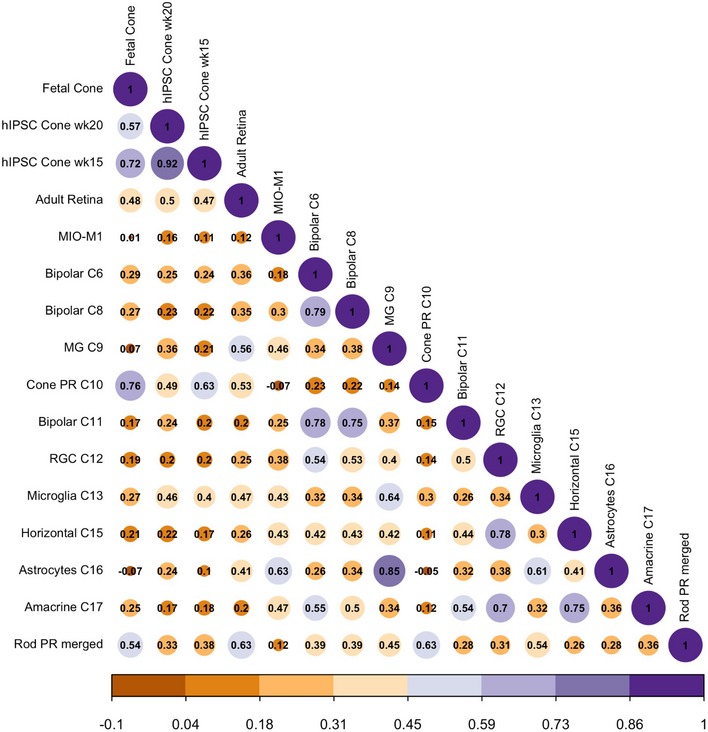
Correlation matrix to benchmark hiPSC‐derived cone photoreceptors (week 15 and week 20; Welby et al, 2017), fetal cone photoreceptors (Welby et al, 2017), adult retina (Phillips et al, 2018), and the human Müller glia cell line MIO‐M1 against all retinal cell types identified in this human neural retina atlas.

Correlation analysis of scRNA‐seq data of hiPSC‐derived cone photoreceptors (week 15) against fetal cone photoreceptors (Welby et al, 2017), as well as adult cone and rod photoreceptors from this human neural retina atlas.
Principal component analysis to assess transcriptome similarity of hiPSC‐derived cone photoreceptors to fetal and adult photoreceptors.
t‐SNE analysis of the human Müller glia cell line MIO‐M1 with the retinal cell types identified in this human neural retina atlas.
Correlation analysis of MIO‐M1 with all major human retinal cell types.
Top ranked differentially expressed genes identified in MIO‐M1 compared to other retinal cell types based on logistic regression score.
In another benchmarking example, we set out to assess the potential differences between in vitro cell lines compared to adult cells in vivo. In this regard, we compared the spontaneously immortalized human Müller glia cell line MIO‐M1 (Limb et al, 2002; Lawrence et al, 2007) to all the retinal cell types identified in our dataset. Using scRNA‐seq, we profiled 7,150 MIO‐M1 cells with 23,987 reads per cell post‐normalization corresponding to 3,421 detected genes. Unsupervised clustering and t‐SNE analysis showed that the MIO‐M1 cells formed one cluster that is transcriptionally distinct from all retina cell types identified in the human neural retina dataset (Fig 6C). Correlation analysis showed that MIO‐M1 displayed similarities to retinal glial cells, with higher similarity to astrocytes compared to Müller glia (0.63 and 0.46, respectively; Fig 6D). In particular, we identified that MIO‐M1 cells express high levels of the thymosin beta 4 gene (TMSB4X), which has been linked to glioma malignancy (Wirsching et al, 2013), and the calcyclin gene (S100A6), which is implicated in macular or cone‐associated diseases (Yoshida et al, 2004; Fig 5E). Together, our results highlight the similarities and differences in MIO‐M1 to adult retinal glial cells in humans.
Discussion
The data presented here describe the generation of a detailed reference transcriptome atlas of the human neural retina at the single‐cell level. The establishment of reference transcriptome maps for individual cell types in the retina provides unprecedented insights into the signals that define retinal cell identity and advance our understanding of the retina. This human neural retina transcriptome data can be used as a benchmark to assess the quality and maturity of pluripotent stem cell‐derived retinal cells, such as retinal ganglion cells (Sluch et al, 2015; Gill et al, 2016; Kobayashi et al, 2018) and photoreceptors (Lakowski et al, 2018). We obtained a mean sequencing depth of 40,232 reads per cell across 23,000 cells, which enabled us to confidently classify the majority of cell types in a complex tissue like the retina. We confirmed that this sequencing depth is sufficient to identify the major cell types. For less transcriptionally distinct cell types, including amacrine and retinal ganglion cells, the ability to resolve subtypes might be improved by increased sample size, greater cell numbers, or ultra‐deep sequencing of those populations. Also, regarding post‐mortem time for the donor retina, we found that at the transcriptome levels there are no obvious variations in all major cell types in neural retina retrieved from 6 to 14 h post‐mortem, with the exception of rod photoreceptors. This potentially suggested that the rod photoreceptors are more sensitive to putative post‐mortem degeneration compared to other retinal cell types. Further studies to optimize methods to preserve donor retinal tissues will help to minimize post‐mortem effects prior to scRNA‐seq processing.
One of the most interesting observations is the presence of heterogeneous subpopulations within known retinal cell types. This highlights the sensitivity of using a scRNA‐seq approach to capture and classify retinal cell types in an unbiased manner. In particular, our results revealed the presence of two rod photoreceptor subpopulations in post‐mortem retina that display differential expression of MALAT1. Notably, the presence of MALAT1‐hi and MALAT1‐lo rod subpopulations was consistently observed in all post‐mortem samples analyzed (n = 7). We further showed that MALAT1‐lo subpopulations represent putative early degenerating rod photoreceptors, a finding that has not previously been reported in humans or any other species. We also noted some heterogeneous MALAT1 expression in other retinal cell types in humans, albeit to a lesser extent compared to rod photoreceptors. Previous studies have demonstrated a role of MALAT1 in regulating the survival of retinal ganglion cells (Li et al, 2017) and in pathogenesis of retinal pigment epithelial cells (Yang et al, 2016). However, the functional role of MALAT1 in photoreceptors remained unclear. Our results demonstrated the loss of MALAT1 expression in rod photoreceptors following longer post‐mortem time with putative degeneration, and suggest MALAT1 as a potential target to enhance rod photoreceptor survival and retinal preservation. Future studies are warranted to investigate the functional role of MALAT1 in photoreceptors, and other retinal cell types in humans. Our transcriptome data also revealed rod photoreceptor clusters specific to particular donor retinas, and we showed that application of the CCA method could effectively correct for these donor/batch variations in rod photoreceptors. Further studies with a larger number of donor samples will allow testing of the feasibility of using scRNA‐seq to comprehensively analyze the retina in different individuals, such as assessment of the effects of aging or degenerative retinal diseases.
Another outcome of this study is the assessment of biomarkers that allow identification of major retinal cell types and subtypes. Our results provide new insights into the cone photoreceptor subtypes in humans. The cone subtypes are traditionally categorized based on expression of different opsins that allowed for cellular detection of light at various wavelengths. While the S‐cones are structurally different from the other two cone subtypes, the L‐cones and M‐cones are structurally similar and difficult to distinguish from each other, except for the opsin they expressed (Viets et al, 2016). We report the first description of the transcriptome profiles of S‐cones in adult humans and highlight novel marker genes that can be used to distinguish them. We also identified the transcriptome and novel marker genes for L/M‐cones; however, given the high sequence homology, particularly at the 3′ end, of OPN1MW and OPN1LW, we could not confidently separate L‐cones and M‐cones. In addition, we show that many of the known Müller glial markers are often expressed in retinal astrocytes, and we also provide a detailed assessment of commonly used retinal glial markers showing the differential expression pattern between Müller glia and retinal astrocytes. Furthermore, we determined that multiple genetic markers, based on binary and/or gradient expression profiles, were required to improve the classification of clustered cell populations. More detailed classification of highly similar cell types may be possible through the combination of single‐cell mRNA and protein measurements using barcoded antibodies, as implemented in the CITE‐seq method (Stoeckius et al, 2017).
Finally, our results highlighted the use of this neural retina transcriptome atlas to benchmark retinal cells derived from stem cells or primary cultures. A major goal of pluripotent stem cell research is to derive cells that are similar to those in adults in vivo, which is important for development of stem cell disease models and regenerative medicine (Hung et al, 2017). Our analysis shows that hiPSC‐derived cone photoreceptors are highly similar to both fetal and adult cones in comparison with all other major retinal cell types. We show that hiPSC‐derived cells are more fetal‐like than adult‐like, which is consistent with other studies (Baxter et al, 2015; Handel et al, 2016). We also benchmark a commonly used Müller glia cell line MIO‐M1 (Limb et al, 2002; Lawrence et al, 2007). Our results showed that while this cell line exhibits similarities to adult retinal glial cells, there are also some differences between MIO‐M1 and adult Müller glia, such as high expression of the glioma‐related gene thymosin beta 4 (TMSB4X) in MIO‐M1. Previous reports have also described differences in gene expression in MIO‐M1 to Müller glia and showed that MIO‐M1 cells express markers for post‐mitotic retinal neurons and neural stem cells (Lawrence et al, 2007; Hollborn et al, 2011). Our results and others highlighted the potential effects of prolonged in vitro culture of primary retinal cells. Collectively, we showed that the human neural retina transcriptome atlas provides an important benchmarking resource to assess the quality of derived retinal cells, which would have implications for stem cell and neuroscience research.
One of the limitations of this study is the finite number of profiled cell types less frequently represented in the retina such as the amacrine cells and the retinal ganglion cells, which are known to be highly complex. The presented dataset is limited in power to accurately identify differences in the transcriptomes of the subtypes in amacrine and retinal ganglion cells. With the identification of surface markers for these retinal cell types in this study, this work lays the foundation for future research using selection and enrichment (Shekhar et al, 2016) of these and other retinal cell types to improve the resolution of the human neural retina transcriptome atlas. Two recent studies have reported the use of surface markers to preselect or enrich for microglia (Masuda et al, 2019) and bipolar cells (Peng et al, 2019) in human tissues prior to scRNA‐seq, which provided a feasible strategy to increase sensitivity to profile cell types less frequently represented. Another limitation is the use of 3′ gene expression profiling, which presents a challenge for distinguishing L‐cones and M‐cones. Given the high sequence homology of OPN1LW and OPN1MW (98%), future studies using full‐length mRNA sequencing of single‐cone photoreceptor cells would provide greater distinction and classification accuracy of the OPN1MW‐ and OPN1LW‐positive cells. Future studies to increase the donor sample size and number of profiled cells with improved capture technologies will further improve the resolution of this human retina transcriptome atlas, allowing more accurate cell type classification and greater statistical power to determine molecular differences between cell populations.
This study describes the transcriptome of human neural retina at a single‐cell level, which identified the transcriptome of all major human retinal cell types. Our findings shed light on the molecular differences between subpopulations within the rod photoreceptors and the cone photoreceptors. The presented dataset provides an important roadmap to define the genetic signals in major cell types in the human retina and can be used as a benchmark to assess the quality of stem cell‐derived cells or primary retinal cells.
Materials and Methods
Human retina collection
Collection of donor samples was approved by the Human Research Ethics Committee of the Royal Victorian Eye and Ear Hospital (HREC13/1151H) and University of Sydney (16/282) and carried out in accordance with the approved guidelines. Informed consent was obtained from all donors, and the experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report. For scRNA‐seq, post‐mortem eye globes were collected by the Lions Eye Donation Service (Royal Victorian Eye and Ear Hospital) for donor cornea transplantation. The remaining eye globes were used for dissection to extract the neural retina. The lens, iris, and vitreous were removed, and the choroid/RPE layers were excluded from the sample collection. Following extraction, the neural retinal samples were dissociated and processed for scRNA‐seq right away. Neural retina samples were dissociated into single cells in dissociation solution (2 mg/ml papain, 120 Units/ml DNase I) for 15 min. The dissociated neural retina was filtered to ensure single‐cell suspension using a 30 μm MACS Smart Strainer (Miltenyi). For the sample from Patient SC, the Dead Cell Removal Kit (Miltenyi) was utilized to remove dead cells prior to scRNA‐seq. However, in our hands we found that the Dead Cell Removal Kit only had a modest improvement in the cell viability (~8% improvement, data not shown). For FISH analysis, post‐mortem eyes were collected by the Lions NSW eye bank and Australian Ocular Biobank.
Single‐cell RNA sequencing (scRNA‐seq)
Single cells from three independent neural retina samples were captured in five batches using the 10X Chromium system (10X Genomics). The cells were partitioned into Gel Bead-In-Emulsions and barcoded cDNA libraries, then prepared using the single‐cell 3′ mRNA kit (V2; 10X Genomics). Single‐cell libraries were sequenced in 100 bp paired‐end configuration using an Illumina Hi‐Seq 2500 at the Australian Genome Research Facility.
Bioinformatics processing
The 10X Genomics cellranger pipeline (version 2.1.0; Zheng et al, 2017) was used to generate fastq files from raw Illumina BCL files (mkfastq). To generate read count matrices from the fastq files, we used cellranger count, which uses the STAR aligner (Dobin et al, 2013), to map high‐quality reads to the transcriptome (GRCh38), and performs UMI counting. To overcome the stringent threshold implemented in cellranger that discards real cells under certain conditions, such as populations of cells with a low RNA content, the –force‐cells parameter was set to 3,000 for the donor 1 library and 5,000 for donor 2 and 3 libraries. Using the barcode rank plots produced by cellranger, these parameters were selected to increase the number of detected cells for further analysis. The cellranger aggregation function (aggr) was used to combine the five libraries and normalize the between‐sample library size differences.
Count data were imported into the Seurat single‐cell analysis software (v2.2.1; https://github.com/satijalab/seurat), and quality control of sequenced libraries was performed to remove outlier cells and genes. Cells with 200–2,500 detected genes and expressing < 10% mitochondrial genes were retained. Genes were retained in the data if they were expressed in ≥ 3 cells. Additional cell–cell normalization was performed using the LogNormalize method, and inherent variation caused by mitochondrial gene expression and the number of unique molecular identifiers (UMIs) per cell was regressed out.
Clustering at a resolution of 0.6 was performed on PCA‐reduced expression data for the top 20 principal components using the graph‐based shared nearest neighbor method (SNN), which calculates the neighborhood overlap (Jaccard index) between every cell and its nearest neighbors. Clustering results were visualized using t‐distributed stochastic neighbor embedding (t‐SNE). Individual samples and sample groups were also visualized using t‐SNE.
Prediction of the cell cycle phase of individual retinal cells was performed in Seurat using cell‐cycle‐specific expression data (Kowalczyk et al, 2015). Briefly, genetic markers associated with G2/M and S phase were used to assign cell scores, and cells expressing neither of the G2/M or S phase markers were classified as being in G1 phase.
Sequencing data for fetal (scRNA‐seq) and hiPSC‐derived cone photoreceptors (bulk RNA‐seq) were obtained from ArrayExpress using the accession numbers E‐MTAB‐6057 and E‐MTAB‐6058 (Welby et al, 2017). Gene expression matrices were generated from the fastq files using the STAR aligner software. scRNA‐seq data from 72 cells were quality‐controlled, filtered, and then normalized with the scran algorithm (Lun et al, 2016) as described (Welby et al, 2017), using the ascend (https://github.com/powellgenomicslab/ascend) package in R, which resulted in 63 high‐quality single‐cell transcriptomes. Bulk RNA‐seq data generated from 6 hiPSC‐derived cone photoreceptor cultures were filtered such that each gene was represented by at least 10 counts in all samples, and normalization was performed in edgeR using the trimmed mean of M method (Robinson & Oshlack, 2010). Preprocessed scRNA‐seq data generated from adult retina (Phillips et al, 2018) were obtained from the Gene Expression Omnibus (GSE98556).
Canonical correlation analysis
Canonical correlation analysis (CCA) was applied to correct donor‐specific effects observed in the rod photoreceptor populations. This was achieved by separating the raw data into five sample‐specific datasets, which were then used as inputs for the RunMultiCCA function in Seurat. For the CCA, we used the most highly variable genes that were shared by all five samples and the recombined data were aligned using the first 20 CC dimensions, selected by biweight midcorrelation (bicor) analysis. Aligned cells were reclustered in Seurat using the first 20 aligned CC dimensions at a resolution of 0.6.
Identification of retinal cell types
Cell types were classified using differential expression analysis, which compared each cluster to all others combined using the Wilcoxon method in Seurat to identify cluster‐specific marker genes. Each retained marker gene was expressed in a minimum of 25% of cells and at a minimum log fold change threshold of 0.25.
In paired cluster analyses, differentially expressed genes were considered significant if the adjusted P‐value was less than 0.01 (Benjamini–Hochberg correction for multiple testing) and the absolute log expression fold change was ≥ 0.5.
Mapping cells between subpopulations in different samples
To compare subpopulations identified in the merged dataset (five samples), we applied scGPS (single‐cell Global Projection between Subpopulations), a machine learning procedure to select optimal gene predictors and to build prediction models that can estimate between‐subpopulation transition scores. The transition scores are the probability of cells from one subpopulation that are in the same class with the cells in the other subpopulation. The scores, therefore, estimate the similarity between any two subpopulations. Here, we compared three main subpopulations from sample Retina 2A with all subpopulations in the sample Retina 2B. The source code of the scGPS method is available with open access (https://github.com/IMB-Computational-Genomics-Lab/scGPS).
Correlation of scRNA‐seq data with retinal cell types
The mean expression levels of cells in each cluster were calculated and used to calculate Pearson's correlations in a pairwise manner with each of the other clusters, and results were deemed significant if the correlation P‐value was less than 0.01.
Pathway analysis
Enrichment analysis for significant differentially expressed genes detected per cluster was performed using Enrichr (Kuleshov et al, 2016). The combined score, computed by taking the log of P‐value from the Fisher exact test and multiplying by the z‐score of the deviation of the expected rank, was used to determine the enrichment ranking for pathways, ontologies, transcription factor network, and protein network analysis.
Fluorescent in situ hybridization
Donor retinas were first dissected from the eye cup. The retinal tissues were subjected to 30% sucrose cryoprotection and were then frozen in −80°C. Sections were cut on a cryostat (Leica CM3050S) and mounted on glass slides (SuperFrost Plus). The retinal samples were fixed in 3.7% (vol/vol) formaldehyde and hybridized with Stellaris RNA FISH Probes (Biosearch Technologies) against MALAT1 labeled with Quasar 570, following the manufacturer's instructions. Briefly, samples were incubated with Quasar 570‐labeled probes at 125 nM in hybridization buffer and hybridized 5 h at 37°C, followed by nuclear counterstain using DAPI. The samples are imaged using a ZEISS confocal laser scanning microscope (ZEISS, LSM700).
Author contributions
HSW and AM coordinated and collected the human donor retinas; GP provided the funding for human donor tissue collection; LF, TN, JSJ, SSCH, RCBW, LZ, TZ, and UG conducted the experiments; SWL, CYL, AS, AAS, RCBW, QN, EW, JCS, TDL, LZ, TZ, and UG processed and/or analyzed the data; P‐YW, AWH, JEP, MCG, and RCBW contributed to experimental design and data analysis; SWL, AWH, JEP, and RCBW wrote the manuscript.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Review Process File
Acknowledgements
This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications) and the ANZ Human Eye Cell Atlas Consortium. The human Müller cell line Moorfields/Institute of Ophthalmology‐Müller 1 (MIO‐M1) was obtained from the UCL Institute of Ophthalmology, London, UK. The authors want to thank David Mackey and Holly Chinnery for critical discussion and helpful feedback for this study, and Prema Finn for technical assistance for the collection of human donor samples. This work was supported by funding from the Ophthalmic Research Institute of Australia (RW, SL), the University of Melbourne De Brettville Trust (RW), and the Kel and Rosie Day Foundation (RW) and GOSHCC (JCS). The Centre for Eye Research Australia receives operational infrastructure support from the Victorian Government.
Data availability
The raw and processed scRNA‐seq files for this analysis are available at ArrayExpress under the accession number E‐MTAB‐7316 (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-7316).
References
- Alvarez‐Delfin K, Morris AC, Snelson CD, Gamse JT, Gupta T, Marlow FL, Mullins MC, Burgess HA, Granato M, Fadool JM (2009) Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc Natl Acad Sci USA 106: 2023–2028 [Europe PMC free article] [Abstract] [Google Scholar]
- Baxter M, Withey S, Harrison S, Segeritz C‐P, Zhang F, Atkinson‐Dell R, Rowe C, Gerrard DT, Sison‐Young R, Jenkins R et al (2015) Phenotypic and functional analyses show stem cell‐derived hepatocyte‐like cells better mimic fetal rather than adult hepatocytes. J Hepatol 62: 581–589 [Europe PMC free article] [Abstract] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL (2001) Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107: 579–589 [Abstract] [Google Scholar]
- Boon CJF, den Hollander AI, Hoyng CB, Cremers FPM, Klevering BJ, Keunen JEE (2008) The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog Retin Eye Res 27: 213–235 [Abstract] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420 [Europe PMC free article] [Abstract] [Google Scholar]
- Corbo JC, Myers CA, Lawrence KA, Jadhav AP, Cepko CL (2007) A typology of photoreceptor gene expression patterns in the mouse. Proc Natl Acad Sci USA 104: 12069–12074 [Europe PMC free article] [Abstract] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [Europe PMC free article] [Abstract] [Google Scholar]
- Eldred KC, Hadyniak SE, Hussey KA, Brenerman B, Zhang P‐W, Chamling X, Sluch VM, Welsbie DS, Hattar S, Taylor J et al (2018) Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362: eaau6348 [Europe PMC free article] [Abstract] [Google Scholar]
- Enright JM, Lawrence KA, Hadzic T, Corbo JC (2015) Transcriptome profiling of developing photoreceptor subtypes reveals candidate genes involved in avian photoreceptor diversification. J Comp Neurol 523: 649–668 [Europe PMC free article] [Abstract] [Google Scholar]
- Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA (2013) Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genom 14: 486 [Europe PMC free article] [Abstract] [Google Scholar]
- Fernández‐Sánchez L, Lax P, Campello L, Pinilla I, Cuenca N (2015) Astrocytes and Müller cell alterations during retinal degeneration in a transgenic rat model of retinitis pigmentosa. Front Cell Neurosci 9: 484 [Europe PMC free article] [Abstract] [Google Scholar]
- Gill KP, Hung SSC, Sharov A, Lo CY, Needham K, Lidgerwood GE, Jackson S, Crombie DE, Nayagam BA, Cook AL et al (2016) Enriched retinal ganglion cells derived from human embryonic stem cells. Sci Rep 6: 30552 [Europe PMC free article] [Abstract] [Google Scholar]
- Handel AE, Chintawar S, Lalic T, Whiteley E, Vowles J, Giustacchini A, Argoud K, Sopp P, Nakanishi M, Bowden R et al (2016) Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell‐derived cortical neurons through single‐cell transcriptomics. Hum Mol Genet 25: 989–1000 [Europe PMC free article] [Abstract] [Google Scholar]
- Hollborn M, Ulbricht E, Rillich K, Dukic‐Stefanovic S, Wurm A, Wagner L, Reichenbach A, Wiedemann P, Limb GA, Bringmann A et al (2011) The human Müller cell line MIO‐M1 expresses opsins. Mol Vis 17: 2738–2750 [Europe PMC free article] [Abstract] [Google Scholar]
- Hornan DM, Peirson SN, Hardcastle AJ, Molday RS, Cheetham ME, Webster AR (2007) Novel retinal and cone photoreceptor transcripts revealed by human macular expression profiling. Invest Ophthalmol Vis Sci 48: 5388–5396 [Abstract] [Google Scholar]
- Hoshino A, Ratnapriya R, Brooks MJ, Chaitankar V, Wilken MS, Zhang C, Starostik MR, Gieser L, La Torre A, Nishio M et al (2017) Molecular anatomy of the developing human retina. Dev Cell 43: 763–779 e4 [Europe PMC free article] [Abstract] [Google Scholar]
- Hung SSC, Khan S, Lo CY, Hewitt AW, Wong RCB (2017) Drug discovery using induced pluripotent stem cell models of neurodegenerative and ocular diseases. Pharmacol Ther 177: 32–43 [Abstract] [Google Scholar]
- Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, Teichmann SA (2016) Classification of low quality cells from single‐cell RNA‐seq data. Genome Biol 17: 29 [Europe PMC free article] [Abstract] [Google Scholar]
- Imanishi Y, Li N, Sokal I, Sowa ME, Lichtarge O, Wensel TG, Saperstein DA, Baehr W, Palczewski K (2002) Characterization of retinal guanylate cyclase‐activating protein 3 (GCAP3) from zebrafish to man. Eur J Neurosci 15: 63–78 [Europe PMC free article] [Abstract] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH (1998) The major cell populations of the mouse retina. J Neurosci 18: 8936–8946 [Europe PMC free article] [Abstract] [Google Scholar]
- Klimova L, Antosova B, Kuzelova A, Strnad H, Kozmik Z (2015) Onecut1 and Onecut2 transcription factors operate downstream of Pax6 to regulate horizontal cell development. Dev Biol 402: 48–60 [Abstract] [Google Scholar]
- Kobayashi W, Onishi A, Tu H‐Y, Takihara Y, Matsumura M, Tsujimoto K, Inatani M, Nakazawa T, Takahashi M (2018) Culture systems of dissociated mouse and human pluripotent stem cell‐derived retinal Ganglion cells purified by two‐step immunopanning. Invest Ophthalmol Vis Sci 59: 776–787 [Abstract] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A (2015) Single‐cell RNA‐seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 25: 1860–1872 [Europe PMC free article] [Abstract] [Google Scholar]
- Kozulin P, Natoli R, O'Brien KMB, Madigan MC, Provis JM (2009) Differential expression of anti‐angiogenic factors and guidance genes in the developing macula. Mol Vis 15: 45–59 [Europe PMC free article] [Abstract] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97 [Europe PMC free article] [Abstract] [Google Scholar]
- Lakowski J, Welby E, Budinger D, Di Marco F, Di Foggia V, Bainbridge JWB, Wallace K, Gamm DM, Ali RR, Sowden JC (2018) Isolation of human photoreceptor precursors via a cell surface marker panel from stem cell‐derived retinal organoids and fetal retinae. Stem Cells 36: 709–722 [Europe PMC free article] [Abstract] [Google Scholar]
- Lawrence JM, Singhal S, Bhatia B, Keegan DJ, Reh TA, Luthert PJ, Khaw PT, Limb GA (2007) MIO‐M1 cells and similar Muller glial cell lines derived from adult human retina exhibit neural stem cell characteristics. Stem Cells 25: 2033–2043 [Abstract] [Google Scholar]
- Li H‐B, You Q‐S, Xu L‐X, Sun L‐X, Abdul Majid AS, Xia X‐B, Ji D (2017) Long non‐coding RNA‐MALAT1 mediates retinal Ganglion cell apoptosis through the PI3K/Akt signaling pathway in rats with glaucoma. Cell Physiol Biochem 43: 2117–2132 [Abstract] [Google Scholar]
- Liao J‐L, Yu J, Huang K, Hu J, Diemer T, Ma Z, Dvash T, Yang X‐J, Travis GH, Williams DS et al (2010) Molecular signature of primary retinal pigment epithelium and stem‐cell‐derived RPE cells. Hum Mol Genet 19: 4229–4238 [Europe PMC free article] [Abstract] [Google Scholar]
- Limb GA, Salt TE, Munro PMG, Moss SE, Khaw PT (2002) In vitro characterization of a spontaneously immortalized human Müller cell line (MIO‐M1). Invest Ophthalmol Vis Sci 43: 864–869 [Abstract] [Google Scholar]
- Lun ATL, Bach K, Marioni JC (2016) Pooling across cells to normalize single‐cell RNA sequencing data with many zero counts. Genome Biol 17: 75 [Europe PMC free article] [Abstract] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM et al (2015) Highly parallel genome‐wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214 [Europe PMC free article] [Abstract] [Google Scholar]
- Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA et al (2019) Spatial and temporal heterogeneity of mouse and human microglia at single‐cell resolution. Nature 566: 388–392 [Abstract] [Google Scholar]
- Mustafi D, Kevany BM, Bai X, Golczak M, Adams MD, Wynshaw‐Boris A, Palczewski K (2016) Transcriptome analysis reveals rod/cone photoreceptor specific signatures across mammalian retinas. Hum Mol Genet 25: 4376–4388 [Europe PMC free article] [Abstract] [Google Scholar]
- Ng L, Hurley JB, Dierks B, Srinivas M, Saltó C, Vennström B, Reh TA, Forrest D (2001) A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet 27: 94–98 [Abstract] [Google Scholar]
- Peng Y‐R, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, Sanes JR (2019) Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176: 1222–1237 e22 [Europe PMC free article] [Abstract] [Google Scholar]
- Phillips MJ, Jiang P, Howden S, Barney P, Min J, York NW, Chu L‐F, Capowski EE, Cash A, Jain S et al (2018) A novel approach to single cell RNA‐sequence analysis facilitates in silico gene reporting of human pluripotent stem cell‐derived retinal cell types. Stem Cells 36: 313–324 [Europe PMC free article] [Abstract] [Google Scholar]
- Pinelli M, Carissimo A, Cutillo L, Lai C‐H, Mutarelli M, Moretti MN, Singh MV, Karali M, Carrella D, Pizzo M et al (2016) An atlas of gene expression and gene co‐regulation in the human retina. Nucleic Acids Res 44: 5773–5784 [Europe PMC free article] [Abstract] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA‐seq data. Genome Biol 11: R25 [Europe PMC free article] [Abstract] [Google Scholar]
- Roorda A, Williams DR (1999) The arrangement of the three cone classes in the living human eye. Nature 397: 520–522 [Abstract] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M et al (2016) Comprehensive classification of retinal bipolar neurons by single‐cell transcriptomics. Cell 166: 1308–1323 e30 [Europe PMC free article] [Abstract] [Google Scholar]
- Sluch VM, Davis C‐HO, Ranganathan V, Kerr JM, Krick K, Martin R, Berlinicke CA, Marsh‐Armstrong N, Diamond JS, Mao H‐Q et al (2015) Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci Rep 5: 16595 [Europe PMC free article] [Abstract] [Google Scholar]
- Soto I, Oglesby E, Buckingham BP, Son JL, Roberson EDO, Steele MR, Inman DM, Vetter ML, Horner PJ, Marsh‐Armstrong N (2008) Retinal Ganglion cells downregulate gene expression and lose their axons within the optic nerve head in a mouse glaucoma model. J Neurosci 28: 548–561 [Europe PMC free article] [Abstract] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck‐Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14: 865 [Europe PMC free article] [Abstract] [Google Scholar]
- Strunnikova NV, Maminishkis A, Barb JJ, Wang F, Zhi C, Sergeev Y, Chen W, Edwards AO, Stambolian D, Abecasis G et al (2010) Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet 19: 2468–2486 [Europe PMC free article] [Abstract] [Google Scholar]
- Subirada PV, Paz MC, Ridano ME, Lorenc VE, Vaglienti MV, Barcelona PF, Luna JD, Sánchez MC (2018) A journey into the retina: Müller glia commanding survival and death. Eur J Neurosci 47: 1429–1443 [Abstract] [Google Scholar]
- Vecino E, Rodriguez FD, Ruzafa N, Pereiro X, Sharma SC (2016) Glia‐neuron interactions in the mammalian retina. Prog Retin Eye Res 51: 1–40 [Abstract] [Google Scholar]
- Viets K, Eldred K, Johnston RJ Jr (2016) Mechanisms of photoreceptor patterning in vertebrates and invertebrates. Trends Genet 32: 638–659 [Europe PMC free article] [Abstract] [Google Scholar]
- Voigt AP, Whitmore SS, Flamme‐Wiese MJ, Riker MJ, Wiley LA, Tucker BA, Stone EM, Mullins RF, Scheetz TE (2019) Molecular characterization of foveal versus peripheral human retina by single‐cell RNA sequencing. Exp Eye Res 184: 234–242 [Europe PMC free article] [Abstract] [Google Scholar]
- Wan P, Su W, Zhuo Y (2017) Precise long non‐coding RNA modulation in visual maintenance and impairment. J Med Genet 54: 450–459 [Europe PMC free article] [Abstract] [Google Scholar]
- Welby E, Lakowski J, Di Foggia V, Budinger D, Gonzalez‐Cordero A, Lun ATL, Epstein M, Patel A, Cuevas E, Kruczek K et al (2017) Isolation and comparative transcriptome analysis of human fetal and iPSC‐derived cone photoreceptor cells. Stem Cell Reports 9: 1898–1915 [Europe PMC free article] [Abstract] [Google Scholar]
- Whitmore SS, Wagner AH, DeLuca AP, Drack AV, Stone EM, Tucker BA, Zeng S, Braun TA, Mullins RF, Scheetz TE (2014) Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA‐Seq. Exp Eye Res 129: 93–106 [Europe PMC free article] [Abstract] [Google Scholar]
- Wirsching H‐G, Krishnan S, Florea A‐M, Frei K, Krayenbühl N, Hasenbach K, Reifenberger G, Weller M, Tabatabai G (2013) Thymosin beta 4 gene silencing decreases stemness and invasiveness in glioblastoma. Brain 137: 433–448 [Abstract] [Google Scholar]
- Yang S, Yao H, Li M, Li H, Wang F (2016) Long non‐coding RNA MALAT1 mediates transforming growth factor Beta1‐induced epithelial‐mesenchymal transition of retinal pigment epithelial cells. PLoS One 11: e0152687 [Europe PMC free article] [Abstract] [Google Scholar]
- Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C et al (2004) Expression profiling of the developing and mature Nrl −/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet 13: 1487–1503 [Abstract] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J et al (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.15252/embj.2018100811
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.15252/embj.2018100811
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.15252/embj.2018100811
Article citations
Single Cell Isolation from Human Diabetic Fibrovascular Membranes for Single-Cell RNA Sequencing.
Bio Protoc, 14(20):e5096, 20 Oct 2024
Cited by: 0 articles | PMID: 39512888 | PMCID: PMC11540046
Ccrk-Mak/Ick signaling is a ciliary transport regulator essential for retinal photoreceptor survival.
Life Sci Alliance, 7(11):e202402880, 18 Sep 2024
Cited by: 0 articles | PMID: 39293864 | PMCID: PMC11412320
Targeting ALDOA to modulate tumorigenesis and energy metabolism in retinoblastoma.
iScience, 27(9):110725, 15 Aug 2024
Cited by: 0 articles | PMID: 39262779 | PMCID: PMC11388021
CilioGenics: an integrated method and database for predicting novel ciliary genes.
Nucleic Acids Res, 52(14):8127-8145, 01 Aug 2024
Cited by: 2 articles | PMID: 38989623 | PMCID: PMC11317154
Single cell dual-omic atlas of the human developing retina.
Nat Commun, 15(1):6792, 09 Aug 2024
Cited by: 1 article | PMID: 39117640 | PMCID: PMC11310509
Go to all (133) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE98556
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Single-cell transcriptome analysis of the Akimba mouse retina reveals cell-type-specific insights into the pathobiology of diabetic retinopathy.
Diabetologia, 63(10):2235-2248, 30 Jul 2020
Cited by: 38 articles | PMID: 32734440
Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks.
Stem Cells, 33(12):3504-3518, 14 Aug 2015
Cited by: 100 articles | PMID: 26235913 | PMCID: PMC4713319
Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration.
Nat Commun, 10(1):4902, 25 Oct 2019
Cited by: 170 articles | PMID: 31653841 | PMCID: PMC6814749
A look into retinal organoids: methods, analytical techniques, and applications.
Cell Mol Life Sci, 78(19-20):6505-6532, 22 Aug 2021
Cited by: 32 articles | PMID: 34420069 | PMCID: PMC8558279
Review Free full text in Europe PMC
Funding
Funders who supported this work.
GOSHCC
Great Ormond Street Hospital Childrens Charity (2)
Grant ID: V4417
Grant ID: V2816
Kel and Rosie Day Foundation
Medical Research Council (1)
Using cell surface antigens for the isolation of human photoreceptor precursor cells for retinal stem cell therapy
Professor Jane Sowden, University College London
Grant ID: MR/M015688/1
National Institute for Health Research (NIHR) (1)
Grant ID: NF-SI-0515-10069
Rosetrees Trust (1)
Grant ID: M257
 4
,
5
,
17
,
‡
4
,
5
,
17
,
‡