Abstract
Free full text

A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury
Abstract
During several months of 2003, a newly identified illness termed severe acute respiratory syndrome (SARS) spread rapidly through the world1,2,3. A new coronavirus (SARS-CoV) was identified as the SARS pathogen4,5,6,7, which triggered severe pneumonia and acute, often lethal, lung failure8. Moreover, among infected individuals influenza such as the Spanish flu9,10 and the emergence of new respiratory disease viruses11,12 have caused high lethality resulting from acute lung failure13. In cell lines, angiotensin-converting enzyme 2 (ACE2) has been identified as a potential SARS-CoV receptor14. The high lethality of SARS-CoV infections, its enormous economic and social impact, fears of renewed outbreaks as well as the potential misuse of such viruses as biologic weapons make it paramount to understand the pathogenesis of SARS-CoV. Here we provide the first genetic proof that ACE2 is a crucial SARS-CoV receptor in vivo. SARS-CoV infections and the Spike protein of the SARS-CoV reduce ACE2 expression. Notably, injection of SARS-CoV Spike into mice worsens acute lung failure in vivo that can be attenuated by blocking the renin-angiotensin pathway. These results provide a molecular explanation why SARS-CoV infections cause severe and often lethal lung failure and suggest a rational therapy for SARS and possibly other respiratory disease viruses.
Supplementary information
The online version of this article (10.1038/nm1267) contains supplementary material, which is available to authorized users.
Main
Recently, ACE2 was identified as a functional SARS coronavirus receptor in cell lines14. But a possible second receptor, CD209L (L-SIGN) has also been identified from in vitro studies15. Thus, it was not known whether ACE2 is indeed crucial for SARS-CoV infections in vivo. To address this question genetically, we infected Ace2 knockout16 and control wild-type mice with SARS-CoV. As reported previously17, SARS-CoV infections of wild-type mice result in viral replication in the lungs and the recovery of large amounts (>107 the tissue culture–infectious dose, which will infect 50% of the cell monolayers (TCID50T) per gram lung tissue) of infectious virus (Fig. 1a). In Ace2 knockout mice, only a very low quantity of infectious SARS-CoV virus could be recovered (<102 TCID50 per gram lung tissue; Fig. 1a), and the copy numbers of SARS-CoV Spike RNA were greatly reduced (Fig. 1b). SARS-CoV infection of mice is associated with the development of mild pathological changes in the lungs (Fig. 1c,d). Moreover, pathologic alterations in lungs were reduced in Ace2 mutant mice compared to wild-type mice (Fig. 1c,d). These data provide the first genetic proof that ACE2 is indeed a crucial in vivo SARS receptor required for effective replication of infectious SARS-CoV.
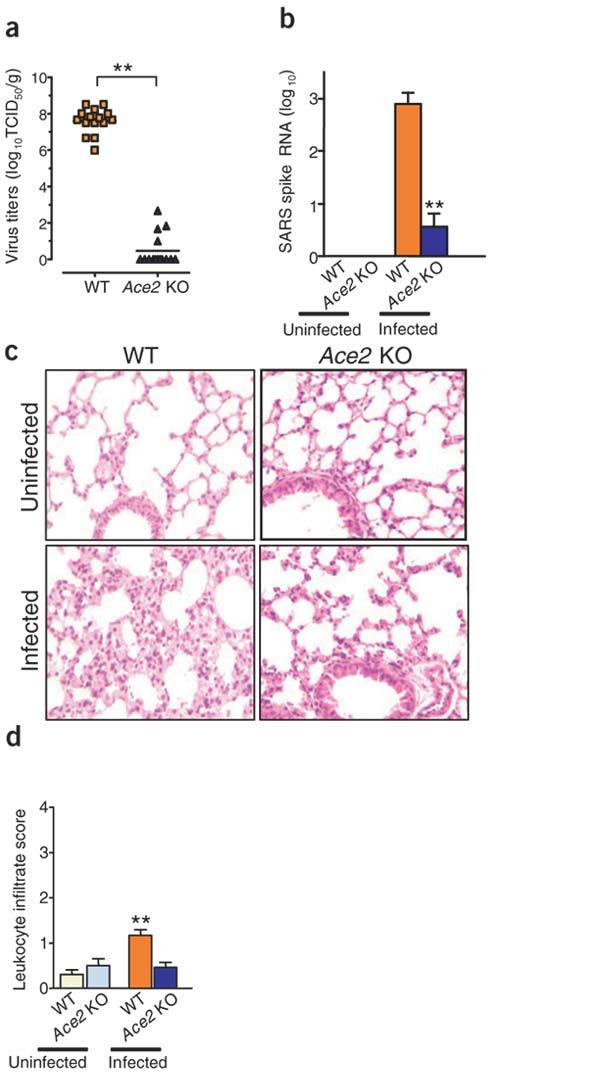
(a,b) SARS-CoV replication (a) and detection of SARS-CoV Spike RNA (b) in wild-type (WT) and Ace2 knockout mice. Viral replication was determined from lung tissue at day 2 of infection. Virus titers (mean log10TCID50 per gram lung tissue) are shown for individual mice. n = 15 per group. SARS-CoV Spike RNA expression was assayed using real-time RT-PCR and normalized to mouse Actb. Data are shown as mean + s.e.m. n = 15 per group. **P < 0.01. (c) Lung histopathology (original magnification ×200) and (d) lung injury scores as defined by leukocyte infiltration of control and SARS-CoV–infected wild-type and Ace2 knockout mice. Lung samples were taken on day 6 after SARS-CoV infection.
We have recently shown that the renin-angiotensin system has a crucial role in severe acute lung injury and that the SARS-CoV receptor ACE2 has a protective role in acute lung failure18 (Fig. 2a). Notably, experimental SARS-CoV infections of wild-type mice in vivo resulted in considerably reduced ACE2 expression in the lungs (Fig. 2b) suggesting that reduced ACE2 expression might have a role in SARS-CoV–mediated severe acute lung pathologies. By contrast, ACE lung expression levels were not overtly changed in SARS-CoV–infected mice (Fig. 2b). We therefore speculated that SARS-CoV might affect lung pathologies through ACE2. To test this idea, we established a defined model system using recombinant SARS-CoV surface-Spike protein, which is the essential ligand for ACE2 binding14. This model system allowed us to avoid possible secondary effects resulting from viral replication or infections in vivo and to directly test whether SARS-CoV Spike protein might adversely affect acute lung injury through modulation of ACE2.
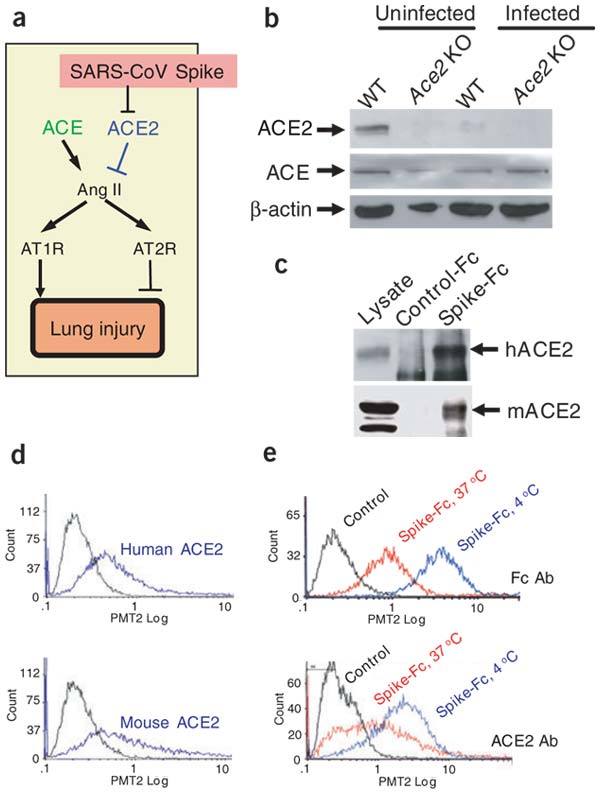
(a) Schematic diagram of the renin-angiotensin system in acute lung failure and proposed SARS-CoV action. (b) Decreased ACE2 protein, but normal ACE levels, in the lungs of SARS-CoV–infected mice. Lung homogenates were prepared from control and SARS-CoV–infected wild-type or Ace2 knockout (KO) mice on day 2 and analyzed by western blot. (c) Binding of recombinant Spike(S-1190)-Fc protein to human ACE2 (hACE2) and mouse ACE2 (mACE2) in pull-down assays. Spike-Fc but not control-Fc protein pulled down hACE2 and mACE2 from total-cell extracts of A549 human alveolar epithelial cells and IMCD mouse kidney epithelial cells, respectively. Total lysates are shown as controls. (d) Binding of Spike-Fc protein to human and mouse ACE2 in cell culture. 293 cells transfected with hACE2 or mACE2 were incubated with Spike-Fc and the binding was detected by FACS (blue lines). Nontransfected 293 cells incubated with Spike-Fc followed by Fc-specific antibodies are shown as controls (black line). (e) Decreased cell-surface expression of ACE2 after binding to Spike-Fc protein at 37 °C compared to 4 °C in Vero E6 cells. ACE2 surface expression was detected at 3 h of incubation with Spike-Fc using an ACE2-specific monoclonal antibody. Similar data were obtained using Fc-specific antibody to directly detect surface-bound Spike-Fc and to avoid masking of the ACE2 epitope. Representative FACS histograms are shown including a background control with an isotype-matched antibody.
We first tested whether recombinant SARS-CoV Spike protein (Supplementary Fig. 1 online) binds to human as well as mouse ACE2 protein using in vitro pull-down assays. Our recombinant Spike-Fc protein indeed pulled down both human and mouse ACE2 (Fig. 2c). SARS-CoV Spike-Fc binding to human and mouse ACE2 was confirmed by FACS binding assays of Spike-Fc to 293 cells overexpressing human or mouse ACE2 (Fig. 2d). Moreover, Spike-Fc bound to endogenous ACE2 in Vero E6 cells (Fig. 2e). Notably, binding of Spike-Fc to endogenous ACE2 in Vero E6 cells resulted in downregulation of ACE2 surface expression (Fig. 2e and Supplementary Fig. 1 online). Spike-Fc also decreased surface levels of human and mouse ACE2 overexpressed in 293 cells (not shown) and triggered syncytia formation of mouse ACE2-transfected but not control CD4-transfected 293 cells (data not shown). Thus, analogous to other virus-receptor interactions19, SARS-CoV Spike protein binding to ACE2 in cell lines or SARS-CoV infections in vivo results in reduced ACE2 protein expression.
Because ACE2 is a crucial SARS-CoV receptor (Fig. 1), SARS-CoV Spike protein binding to ACE2 downmodulates ACE2 expression (Fig. 2), and loss of ACE2 expression results in severe acute respiratory failure18, we tested whether SARS-CoV Spike protein, which is the crucial ACE2 binding protein20,21, could affect the severity of acute lung injury in vivo. Notably, treatment with Spike-Fc protein worsened the lung function in wild-type mice, whereas control-Fc protein showed no apparent effects (Fig. 3a). Moreover, Spike-Fc treatment of acid-challenged wild-type mice augmented the pathological changes in the lung parenchyma (Fig. 3b,c and Supplementary Table 1 online) and increased lung edemas as defined by a wet/dry lung weight ratios (Fig. 3d). We next made a Spike-deletion mutant that only contained the previously mapped ACE2-binding domain (amino acids 318–510)21 fused to human Fc (Fig. 3e and Supplementary Fig. 1 online). This short Spike(S318-510)-Fc protein containing the minimal ACE2 binding site also binds to ACE2 in cell lines using FACS assays and downmodulates the cell-surface expression of ACE2 (Supplementary Fig. 1 online). Treatment with Spike(S318-510)-Fc again worsened acid-induced acute lung injury in wild-type mice (Fig. 3e). Notably, in vivo Spike-Fc protein administration did not affect the severity of lung failure in Ace2 knockout mice (Fig. 3f), indicating that the effect of Spike protein on acute lung injury is ACE2 specific.
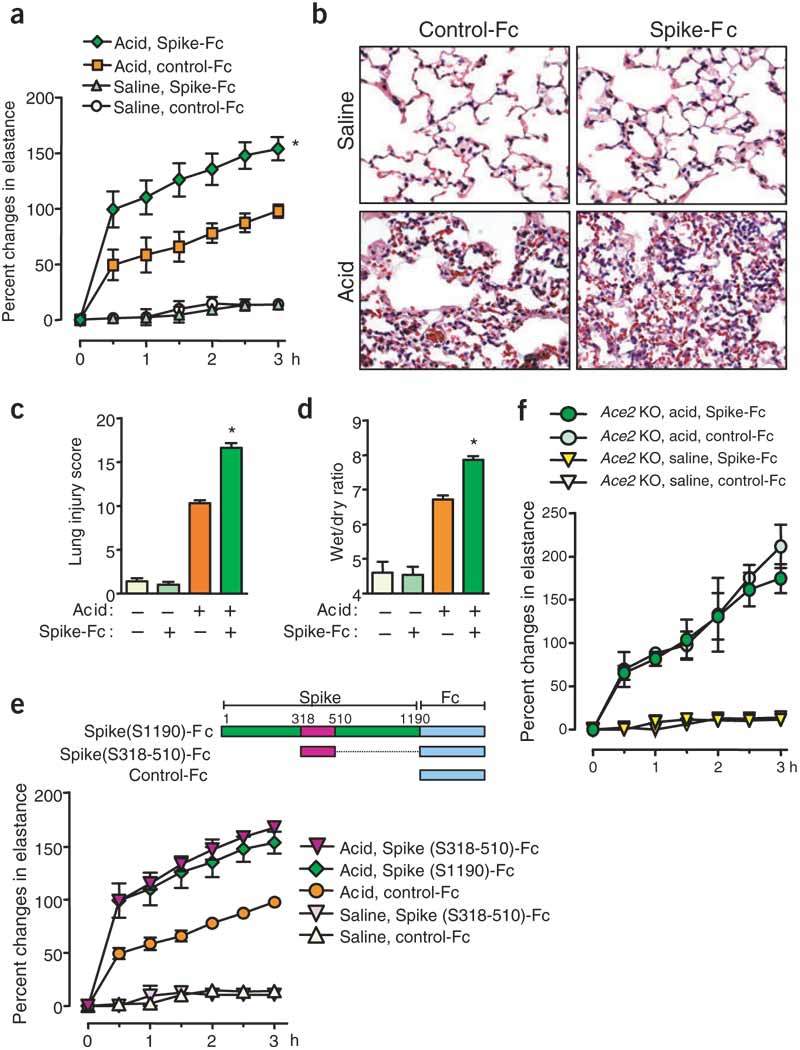
(a) Lung elastance measurements after saline or acid instillation in Spike-Fc protein–(5.5 nmol/kg) or control-Fc– (5.5 nmol/kg) treated wild-type mice. n = 5–7 per group. *P < 0.05 for the whole time course comparing Spike-Fc–treated and control-Fc–treated wild-type mice after acid injury. (b) Lung histopathology. Representative images are shown. Original magnification, ×200. (c) Lung injury score (Supplementary Table 1 online). **P < 0.01 versus control-Fc–treated wild-type. (d) Wet to dry weight ratios of lungs as readout for pulmonary edema in control and Spike-Fc–treated mice in the presence or absence of acid-induced lung injury. *P < 0.05 between control and Spike-Fc–treated mice with acid challenge. (e) Severe acute lung failure by Spike(S318-510)-Fc (5.5 nmol/kg) treatment in acid-challenged mice. The scheme (upper panel) shows the ACE2-binding domain of Spike (S318-510). Lung elastance measurements (lower panel) showed that Spike(S318-510)-Fc induced severe acute lung failure in acid-challenged wild-type mice, comparable to Spike(S1190)-Fc. n = 5–7 per group. P < 0.05 for the whole time course comparing Spike(S318-510)-Fc or Spike(S1190)-Fc–treated and control-Fc–treated wild-type mice after acid injury. (f) Lung elastance measurements after acid instillation in Spike-Fc protein–(S1190; 5.5 nmol/kg) or control-Fc–(5.5 nmol/kg) treated Ace2 knockout (KO) mice. n = 5–7 for each group.
To further clarify whether the intraperitoneally injected Spike-Fc protein directly affected lung pathology in mice, we examined the localization of the injected Spike-Fc protein in lung. Spike-Fc was detected in lung homogenates by western blot using human Fc–specific antibody (Fig. 4a), whereas injected control-Fc was not detected. In addition, using immunohistochemistry, we found that Spike-Fc protein localized to bronchial epithelial cells, inflammatory exudates and alveolar pneumocytes (Fig. 4b). Notably, Spike-Fc primarily localized to severe lesions (Fig. 4b). This localization of Spike-Fc protein is similar to Spike antigen staining in SARS-CoV–infected mice22. Spike-Fc treatment resulted in downregulation of ACE2 protein expression in lungs of acid-treated wild-type mice in vivo (Fig. 4c), consistent with ACE2 protein downregulation in SARS-CoV–infected mice (Fig. 2a) and Spike-Fc protein–treated cells in vitro (Fig. 2e). These results show that the SARS-CoV Spike protein can directly affect the development of severe acute lung failure through ACE2.
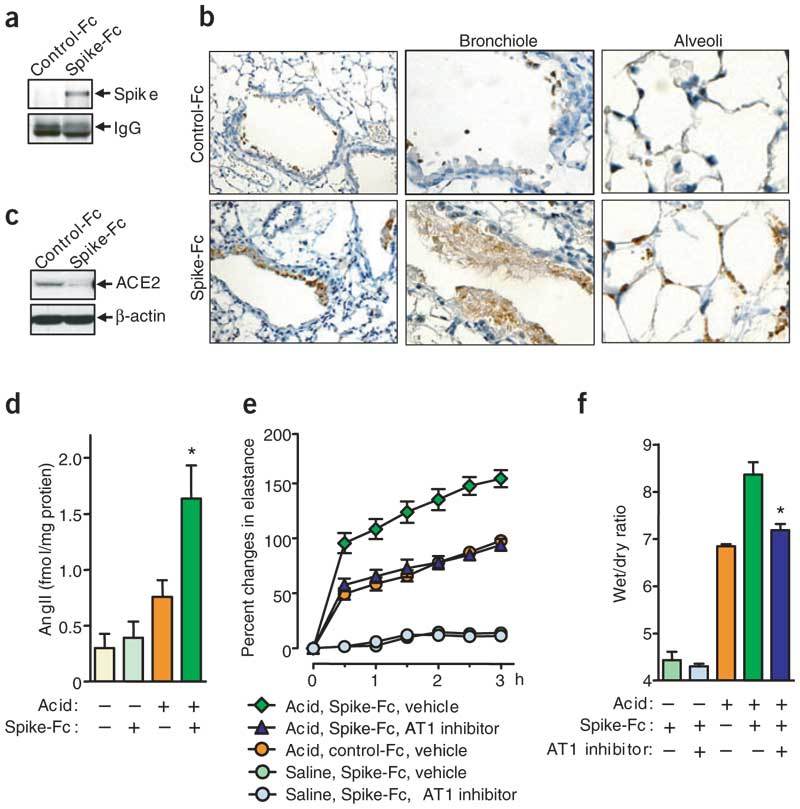
(a,b) Localization of intraperitoneally injected Spike(S-1190)-Fc in lung tissue. (a) Spike-Fc was detected by pull-down assay with Protein G Sepharose and western blot with human Fc–specific antibody. Mouse IgG is shown as loading control. (b) Lung immunohistochemistry to detect Spike(S-1190)-Fc or control-Fc protein using a human Fc–specific antibody. Spike(S-1190)-Fc localizes to bronchial epithelial cells (left; original magnification, ×100), inflammatory exudates cells (middle; original magnification, ×200), and alveolar pneumocytes (right; original magnification, ×200). (c) Decreased ACE2 protein expression in the lungs of Spike(S-1190)-Fc– treated mice. Lung homogenates were prepared from control-Fc– and Spike(S-1190)-Fc–treated wild-type mice and analyzed by western blot with ACE2-specific antibody. (d) AngII peptide levels in lungs of Spike(S1190)-Fc protein– or control–Fc-treated wild-type mice after saline or acid aspiration. AngII levels were determined at 3 h by enzyme immunoassay. Bars, mean ± s.e.m. *P < 0.05 comparing Spike(S1190)-Fc– and control-Fc–treated wild-type mice after acid injury. (e) Lung elastance measurements in acid plus Spike(S1190)-Fc–challenged wild-type mice treated with the AT1R inhibitor losartan (15 mg/kg). n = 4–6 per group. P < 0.05 comparing losartan-treated Spike(S1190)-Fc–challenged mice with vehicle-treated Spike(S1190)-Fc–challenged mice. (f) Wet to dry weight ratios of lungs of acid and Spike(S1190)-challenged mice in the presence or absence of losartan (15 mg/kg). n = 4–6 mice per group. *P < 0.05, comparing losartan-treated wild-type with vehicle-treated wild-type mice at 3 h after acid injury.
ACE2 functions as a carboxypeptidase, cleaving a single residue from angiotensin I (AngI), generating Ang1–9 (refs. 23,24), and a single residue from angiotensin II (AngII) to generate Ang1–7 (ref. 23). The ACE2 homologue ACE, by contrast, cleaves the decapeptide AngI into the octapeptide AngII25. Thus, ACE2 counterbalances the function of ACE and negatively regulates AngII production (Fig. 2a). To test whether Spike-Fc injections indeed affect the function of the renin-angiotensin system, we analyzed AngII levels in the lungs of acid- and Spike-Fc–treated mice. Acid aspiration increased AngII levels in the lungs of wild-type mice. Notably, we observed a further, significant increase in AngII levels in the lung tissue of mice treated with Spike-Fc (Fig. 4d). To confirm whether Spike-Fc promotes lung disease pathogenesis through increased AngII production and functional alterations of the renin-angiotensin system, we blocked the AngII receptor type 1 (AT1R)26 with a specific inhibitor. The AT1R is the crucial receptor that mediates AngII-induced vascular permeability and severe acute lung injury18. Inhibition of the AT1R indeed attenuated acute severe lung injury in Spike-Fc–treated mice (Fig. 4e). Inhibition of the AT1R also attenuated pulmonary edema (Fig. 4f). Taken together, our data show that SARS-CoV Spike can exaggerate acute lung failure through deregulation of the renin-angiotensin system. Moreover, SARS-CoV Spike–mediated lung failure can be rescued by inhibition of AT1R.
It has been estimated that the Spanish flu virus that killed more than 20 million people at the beginning of the twentieth century was lethal to around 0.5% of infected people9,10, whereas the lethality of SARS-CoV infections reached 10% even with modern intensive care treatment1,2,3. Given this very high lethality of SARS and the enormous economic and social impact of the worldwide SARS outbreak, elucidation of the disease pathogenesis is crucial for future treatment in case of renewed outbreaks. Moreover, a recent outbreak of avian influenza A (H5N1) in humans resulted in up to 70% of lethality from acute respiratory failure12. Before the discovery of SARS-CoV, two coronaviruses (HCoV-229E and HCoV-OC43) were known to infect humans, but they caused only self-limiting upper respiratory tract infections (30% of the common colds) and had never been reported to cause severe illness27. The molecular determinants that may account for the dramatic differences in pathogenesis between these human coronaviruses and SARS-CoV were unknown.
Our data provide a molecular explanation for the severe lung failure and lethality associated with SARS: we hypothesize that infections with SARS-CoV result in ACE2 downregulation through binding of SARS-CoV Spike protein to ACE2. Given that ACE2 is a key negative regulatory factor for severity of lung edema and acute lung failure, SARS-CoV Spike protein–mediated ACE2 downregulation then contributes to the severity of lung pathologies. This scenario would explain how this family member of the 'relatively harmless' coronaviruses has turned into a lethal virus. Notably, recent data in a small cohort of individuals with SARS suggested that an insertion deletion ACE polymorphism that affects ACE function correlates with disease severity28, implying that our findings are indeed relevant for humans.
Our data provide a molecular link between SARS pathogenesis and the role of the renin-angiotensin system in lung failure. Recombinant ACE2 protein therefore could not only be a treatment to block spreading of SARS-CoV, but modulation of the renin-angiotensin system could also be used to protect individuals with SARS, and possibly individuals infected with other viruses such as strains of avian influenza A strains, from developing acute severe lung failure and acute respiratory distress syndrome.
Methods
For details of Methods, see Supplementary Methods online.
In vivo SARS infections.
The SARS-CoV (Beijing strain, PUMC01 isolate) used in this study was provided by Z. Wang and Y. Liu (Chinese Academy of Medical Sciences, Chinese National Human Genome Center). All mouse studies were approved by the Ministry of Health Science and Technology division of the People's Republic of China. We intranasally inoculated mice with 100 μl virus (105.23 TCID50). At day 2, mice were killed, and the lungs were removed for further analyses. We assessed lung injury scores as described previously30.
SARS-CoV Spike protein binding experiments.
We cloned the coding sequence of SARS-CoV Spike protein (amino acids 1–1,190 from Urbani strain) or a Spike sequence that only contains the previously mapped21 ACE2-binding domain (amino acids 318–510) to generate a fusion protein with the Fc portion of human IgG1. We purified Spike-Fc protein from transfected CHO cells using affinity chromatography. For in vitro binding assays, we used cell lysates from A549 human alveolar epithelial cells or IMCD mouse kidney epithelial cells, and we pulled down Spike-Fc or control human IgG-Fc protein using Protein G Sepharose followed by western blot. For flow cytometry, we detached Vero E6 cells using a 2 mM mixture of EDTA and PBS and incubated them with Spike-Fc or control human IgG-Fc protein at 4 °C or 37 °C for 3 h. We then incubated cells with ACE2-specific antibodies or a FITC-conjugated human IgG–specific antibody.
Recombinant Spike-Fc in vivo challenge in mice.
We used the mouse model of acid aspiration–induced acute lung injury18 for all Spike-Fc in vivo experiments. Mice received Spike(S1190)-Fc, Spike(S318-510)-Fc or control-Fc (5.5 nmol/kg each) intraperitoneally three times (at 30 min before and at 1 and 2 h after) during acid treatment. For AT1 inhibition of Spike-Fc–mediated acute lung injury, we treated the Spike(S1190)-Fc–challenged mice with the AT1 inhibitor losartan (15 mg/kg). Lung injury scores were assessed as described previously30.
Statistical analyses.
All data are shown as mean ± s.e.m. Measurements at single time points were analyzed by unpaired t-test (virus titers, SARS-CoV Spike-RNA levels), ANOVA with two tailed t-test (percent changes in elastance, wet/dry ratio AngII levels) or Kruskal-Wallis test (histological scores). Time courses were analyzed by repeated measurements (mixed model) ANOVA with Bonferroni post-t tests. All statistical tests were calculated using the GraphPad Prism 4.00 (GraphPad Software) and a JMP (SAS Institute) programs. P < 0.05 was considered to indicate statistical significance.
Accession number.
The Genbank accession number for the SARS coronavirus (Beijing strain, PUMC01 isolate) is AY350750.
Note: Supplementary information is available on the Nature Medicine website.
Acknowledgements
We thank C. Richardson and all members of our laboratory for discussions. We also thank B. Seed for discussion and providing the systems to generate recombinant Spike proteins. We thank Q. Zhu, L. Ruan and L. Zhang for sharing unpublished data of SARS coronavirus infection on mice and protocols of virus infections. This work is supported by the Institute for Molecular Biotechnology of the Austrian Academy of Sciences and the Jubilaeumsfonds of the Austrian National Bank. K.K. is supported by a Marie Curie Fellowship from the EU. C.J. is supported by Beijing Committee of Science and Technology grant H030230010930, National Natural Science Foundation of China innovation group grant 30421003 and SARS donation from Joincare Corporation. A.S. is supported in part by the Canadian Institutes of Health Research and the Canada Foundation for Innovation.
Competing interests
The Institute for Molecular Biotechnology of the Austrian Academy of Sciences has applied for a patent on modulating the renin-angiotension system for treatment of lung edema.
Footnotes
Keiji Kuba, Yumiko Imai and Shuan Rao: These authors contributed equally to this work.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nm1267
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/nm1267.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102438859
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nm1267
Article citations
Psycho-Neuro-EndocrinE-Immunology Therapy of Cancer, Autoimmunity, Geriatric Disorders, Covid-19, and Hypertension.
Methods Mol Biol, 2868:111-132, 01 Jan 2025
Cited by: 0 articles | PMID: 39546228
Combination of the chemokine receptor type 2 (CCR2) antagonist DMX-200 and candesartan for COVID-19: a randomised controlled trial.
BMJ Open, 14(10):e081790, 22 Oct 2024
Cited by: 0 articles | PMID: 39438096 | PMCID: PMC11499840
Unveiling the Interplay-Vitamin D and ACE-2 Molecular Interactions in Mitigating Complications and Deaths from SARS-CoV-2.
Biology (Basel), 13(10):831, 16 Oct 2024
Cited by: 0 articles | PMID: 39452140 | PMCID: PMC11504239
Review Free full text in Europe PMC
Transport of Neutral Amino Acids in the Jejunum of Pigs with Special Consideration of L-Methionine.
Nutrients, 16(19):3418, 09 Oct 2024
Cited by: 0 articles | PMID: 39408384 | PMCID: PMC11478682
Clinical management and complications of acute appendicitis in 3 children with SARS-CoV-2 infection: Case report.
Medicine (Baltimore), 103(43):e40105, 01 Oct 2024
Cited by: 0 articles | PMID: 39470524 | PMCID: PMC11521089
Go to all (2,180) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - AY350750
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice.
Exp Physiol, 93(5):543-548, 10 Apr 2008
Cited by: 201 articles | PMID: 18448662 | PMCID: PMC7197898
Review Free full text in Europe PMC
Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2.
J Virol, 78(19):10628-10635, 01 Oct 2004
Cited by: 179 articles | PMID: 15367630 | PMCID: PMC516384
Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2.
EMBO J, 24(8):1634-1643, 24 Mar 2005
Cited by: 648 articles | PMID: 15791205 | PMCID: PMC1142572
Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus.
Cell Mol Life Sci, 61(21):2738-2743, 01 Nov 2004
Cited by: 155 articles | PMID: 15549175 | PMCID: PMC7079798
Review Free full text in Europe PMC

 2 and
2 and