Abstract
Objectives
The aim was to understand persistence of the virus in body fluids the and immune response of an infected host to severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), an agent of coronavirus disease 2019 (COVID-19).Methods
We determined the kinetics of viral load in several body fluids through real time reverse transcription polymerase chain reaction, serum antibodies of IgA, IgG and IgM by enzyme-linked immunosorbent assay and neutralizing antibodies by microneutralization assay in 35 COVID-19 cases from two hospitals in Guangdong, China.Results
We found higher viral loads and prolonged shedding of virus RNA in severe cases of COVID-19 in nasopharyngeal (1.3 × 106 vs 6.4 × 104, p < 0.05; 7∼8 weeks) and throat (6.9 × 106 vs 2.9 × 105, p < 0.05; 4∼5 weeks), but similar in sputum samples (5.5 × 106 vs 0.9 × 106, p < 0.05; 4∼5 weeks). Viraemia was rarely detected (2.8%, n = 1/35). We detected early seroconversion of IgA and IgG at the first week after illness onset (day 5, 5.7%, n = 2/35). Neutralizing antibodies were produced in the second week, and observed in all 35 included cases after the third week illness onset. The levels of neutralizing antibodies correlated with IgG (rs = 0.85, p < 0.05; kappa = 0.85) and IgA (rs = 0.64, p < 0.05; kappa = 0.61) in severe, but not mild cases (IgG, rs = 0.42, kappa = 0.33; IgA, rs = 0.32, kappa = 0.22). No correlation with IgM in either severe (rs = 0.17, kappa = 0.06) or mild cases (rs = 0.27, kappa = 0.15) was found.Discussion
We revealed a prolonged shedding of virus RNA in the upper respiratory tract, and evaluated the consistency of production of IgG, IgA, IgM and neutralizing antibodies in COVID-19 cases.Free full text

The kinetics of viral load and antibodies to SARS-CoV-2
Abstract
Objectives
The aim was to understand persistence of the virus in body fluids the and immune response of an infected host to severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), an agent of coronavirus disease 2019 (COVID-19).
Methods
We determined the kinetics of viral load in several body fluids through real time reverse transcription polymerase chain reaction, serum antibodies of IgA, IgG and IgM by enzyme-linked immunosorbent assay and neutralizing antibodies by microneutralization assay in 35 COVID-19 cases from two hospitals in Guangdong, China.
Results
We found higher viral loads and prolonged shedding of virus RNA in severe cases of COVID-19 in nasopharyngeal (1.3 × 106 vs 6.4 × 104, p < 0.05; 7~8 weeks) and throat (6.9 × 106 vs 2.9 × 105, p < 0.05; 4~5 weeks), but similar in sputum samples (5.5 × 106 vs 0.9 × 106, p < 0.05; 4~5 weeks). Viraemia was rarely detected (2.8%, n = 1/35). We detected early seroconversion of IgA and IgG at the first week after illness onset (day 5, 5.7%, n = 2/35). Neutralizing antibodies were produced in the second week, and observed in all 35 included cases after the third week illness onset. The levels of neutralizing antibodies correlated with IgG (rs = 0.85, p < 0.05; kappa = 0.85) and IgA (rs = 0.64, p < 0.05; kappa = 0.61) in severe, but not mild cases (IgG, rs = 0.42, kappa = 0.33; IgA, rs = 0.32, kappa = 0.22). No correlation with IgM in either severe (rs = 0.17, kappa = 0.06) or mild cases (rs = 0.27, kappa = 0.15) was found.
Discussion
We revealed a prolonged shedding of virus RNA in the upper respiratory tract, and evaluated the consistency of production of IgG, IgA, IgM and neutralizing antibodies in COVID-19 cases.
Introduction
Data on the persistence of the SARS-CoV-2 virus and immune response of the infected host might guide efforts for the prevention and control of its expansion, as well as treatment of COVID-19 cases. Therefore, we studied the kinetics of viral load in a variety body fluids, IgA, IgG, IgM and neutralizing antibodies against SARS-CoV-2 by monitoring mild and severe COVID-19 patients from designated hospitals in Guangdong, China. Healthy close contacts were used as the control group.
Material and methods
Case information
From 23 January to 27 February 2020, 38 hospitalized COVID-19 cases were consecutively recruited (29 mild and 9 severe cases) with advanced planning to collect clinical samples in two designated hospitals for COVID-19, the Guangdong Seconded Provincial General Hospital and the First Hospital of Foshan in Guangdong, China. These were all the patients hospitalized in the two hospitals during that time. All the cases were laboratory-confirmed COVID-19 cases. One mild and two severe cases were transferred to other hospitals after hospitalization in these two hospitals and were excluded from this study. Finally, 35 COVID-19 cases (28 mild and seven severe cases) were included in this study. Twenty-one healthy individuals who were close contacts of these COVID-19 cases were selected as controls. The demographic and the dates of illness onset were collected accordingly. This study was approved by institutional review board in Guangdong Provincial Centre for Disease Control and prevention. All the COVID-19 cases included in this study were asked for their consent.
Case definitions
The case definition was defined as a patient who had a travel history to endemic areas or direct contact with patients from those areas who had fever or respiratory symptoms within 14 days before illness onset, and laboratory-confirmed respiratory specimens that tested positive for the SARS-CoV-2 (please see supplementary material).
Specimen collection and storage
Specimen collection from COVID-19 cases was conducted in consultation with a healthcare provider, and followed the guideline for clinical diagnosis and treatment (please see supplementary material). Throat swabs, nasopharyngeal swabs, sputum samples, faeces specimens and serum samples were collected prospectively from cases every 3 days from hospitalization until the date of discharge from hospital (Table S1).
Nucleic acid extraction and rRT-PCR
The clinical specimens were treated as previously reported [1]. Total RNA was extracted using a prefilled viral total NA kit-Flex (Fisher Scientific, Labserv, Cat. no. KFRPF-805296) following the manufacturer's instructions. A commercial rRT-PCR kit targeting the ORF1ab and N genes was used to detect SARS-CoV-2 RNA (DaAn Gene, Guangzhou, China. Cat.No.DA0931) (http://en.daangene.com) (supplementary material).
Detection of IgA, IgG and IgM against SARS-CoV-2
IgA, IgG and IgM against SARS-CoV-2 were detected in plasma samples using enzyme-linked immunosorbent assay (ELISA) kits (RayBiotech, GA, USA) (Cat. no., IEQ-CoVS1RBD-IgG, IE-CoVS1RBD-IgA and IE-CoVS1RBD-IgM), according to the manufacturer's instructions.
Microneutralization assay
The microneutralization assay and virus culture are detailed in the supplementary material [2,3].
Statistical analysis
Differences of virus load were determined by Student's t test. Consistency among antibodies was evaluated using the Spearman association test and kappa test. Statistical analyses and graphical presentations were conducted with GraphPad Prism version 8.3 (GraphPad Software, Inc., CA, USA). p < 0.05 was considered statistically significant (please see supplementary material).
Results
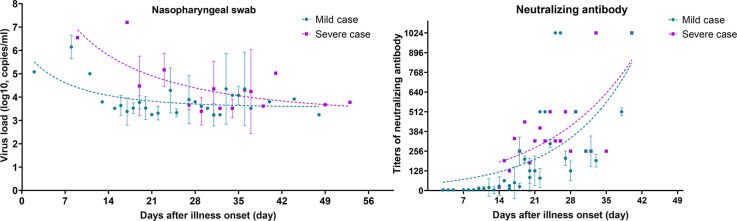
We recruited 35 COVID-19 cases in this study, including 28 mild and seven severe cases. In total, 618 clinical specimens were collected, including 105 nasopharyngeal swabs and 84 throats swabs, and 160 faeces, 60 sputum and 209 serum samples. We found that the viral loads in severe cases was higher than those in mild cases in nasopharyngeal (1.3 × 106 vs 6.4 × 104, p < 0.05; 7~8 weeks) and throat samples (6.9 × 106 vs 2.9 × 105, p < 0.05), but similar in sputum samples (5.5 × 106 vs 0.9 × 106, p < 0.05) (Fig. S1). In both groups, the viral load in upper respiratory tract samples (nasopharyngeal, throat swabs) reached the peak level at the first week after illness onset and declined quickly within the following 2~4 weeks (Figs. 1 A,B). Prolonged shedding of virus RNA could be detected in nasopharyngeal swabs of each mild and severe case at 6~8 weeks after illness onset, while the viral load was extremely low and close to the detection limit of real time RT-PCR (1.7~6.0 × 103 copies/mL). In sputum (Fig. 1C) and faeces samples (only measured in mild cases, Fig. 1D), the viral load was somehow higher than that in upper respiratory tract samples, and reached to 5.6 × 107 copies/mL in peak level at the first week after illness onset, but declined to 1.6 × 103 copies/mL within 2~3 weeks. Viraemia was only detected in one of seven severe cases, and sustained for 5~6 days (Fig. 1E).
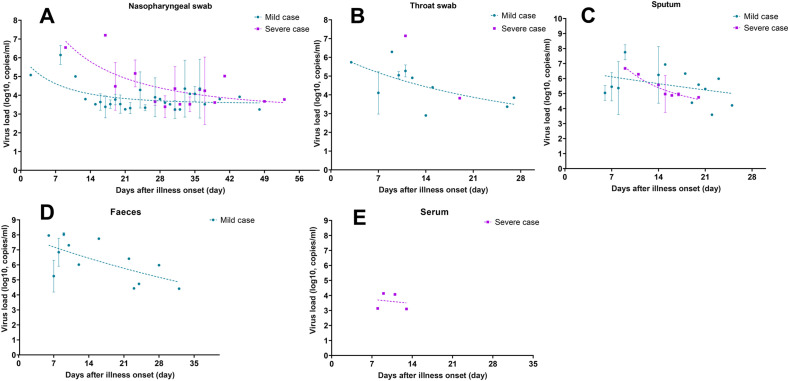
Kinetics of viral load in 35 COVID-19 cases. The copies of SARS like coronavirus 2 (SARS-CoV-2) RNA in nasopharyngeal swab (A), throat swab (B), sputum (C), faeces (D) and serum specimens (E) were estimated by means of real-time reverse-transcriptase polymerase chain reaction. The dashed curve draws with exponential growth model and indicates the dynamic tendency of viral load in variety body fluids. Each dot of viral load present as mean and standard deviation. Purple symbols denote severe cases, and cyan symbols denote mild cases.
IgA in serum samples was detected in 1 of 35 cases at the first week after the date of illness onset (Fig. 2 A). IgA levels were already high at day 11 in severe cases, but still low in mild cases. The kinetics of IgA showed an increasing trend in mild cases, but declining in severe cases (Fig. 2A). The production of IgM remained at a low level in both the severe and the mild group (Fig. 2B). Surprisingly, IgG was detected in one mild case at the first week after illness onset. At the third week, 68.9% (n = 20/29) patients seroconverted in mild cases, while it was 100% (n = 7/7) in severe cases (Fig. 2C). A similar increasing trend of IgG was observed both in severe and in mild cases (Fig. 2C).
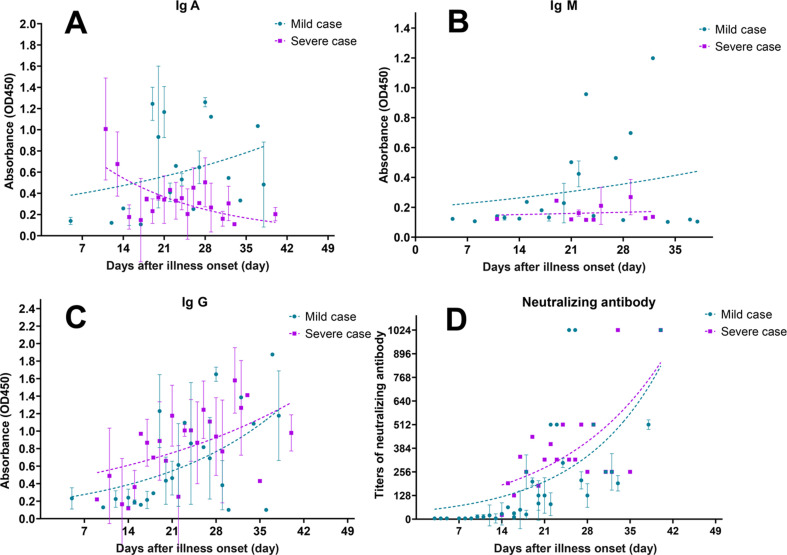
Kinetics of IgA (A), Ig M (B), Ig G (C) and neutralizing antibodies (D) to SARS-CoV-2 over time in 35 COVID-19 cases. The dashed curve draws with experiential growth model and indicates the dynamic tendency of antibodies. Each dot of ELISA presents as mean and standard deviation, and titres of neutralizing antibodies present geometric mean and standard deviation. Purple symbols denote severe cases, and cyan symbols denote mild cases.
None of the 35 cases produced neutralizing antibodies at the first week after the date of illness onset (Fig. 1E). At the second week, seven of 28 mild cases produced neutralizing antibodies with titres of 1:32 to 1:128, and the remaining cases showed no neutralizing antibodies. At the third week, all 35 cases produced neutralizing antibodies, reaching titres of 1:512 in two of 29 mild cases and one of seven severe cases. Importantly, we found no differences in the increasing trends of neutralizing antibodies between mild and severe cases. All 21 healthy close contacts tested negative for both virus RNA and neutralizing antibodies against SARS-CoV-2.
Additionally, we determined the consistency among neutralizing antibodies and IgA, IgM and IgG by calculating the Spearman correlation coefficient and kappa value between mild and severe case groups. Neutralizing antibodies correlated with IgG (r s = 0.85, p < 0.05; kappa = 0.85) and IgA (r s = 0.64, p < 0.05; kappa = 0.61) in severe but not in mild cases (IgG, r s = 0.42, kappa = 0.33; IgA, r s = 0.32, kappa = 0.22). No correlation with IgM antibodies in either severe (r s = 0.17, kappa = 0.06) or mild cases (r s = 0.27, kappa = 0.15) was seen (Table S2).
Discussion
Our data showed that the course of virus shedding and antibody development triggered by SARS-CoV-2 resembles that of SARS-CoV and MERS-CoV [4,5]. The major difference was prolonged virus shedding in upper respiratory tract samples, e.g. nasopharyngeal swab samples, which seems to mimic the manifestation of seasonal influenza [6]. Although we found higher viral load in sputum and faeces than in upper respiratory tract samples, the heterogeneity of the specimens limits its significance in explaining the differences of the ability for onward transmission by either sample type. The early production of IgG antibodies showed a similar increasing trend with neutralizing antibodies in both mild and severe cases. However, the seroconversion rate of IgM was low in the acute stage of infection in both severe and mild cases. This phenomenon was also encountered by other groups [[7], [8], [9]]. Thus, the early detection of COVID-19 through determination of IgM may not be an ideal strategy. Surprisingly, we found two cases of early secretion of IgA in serum at the first week after illness onset in the mild group, which highlights an alternative choice of target antibody for early detection of COVID-19 in clinic [10]. Moreover, we found a higher proportion of seroconversion for IgA in severe cases, which may serve as a candidate indicator of prognosis of COVID-19 in future [10]. The correlation between neutralizing antibodies and IgG suggests that IgG could be used as a marker for the production of neutralizing antibodies [11]. However, we found the titres of neutralizing antibodies in most cases remained at a low level, which may indicate an inability to clear the infection. Indeed, we found neutralizing antibodies in all severe cases. The potential mechanisms, because of either low titres or lower host immune cell-mediated response, need further investigation [12]. In conclusion, our findings may be of significance in interpreting the kinetics of viral load and antibodies to SARS-CoV-2 in both severe and mild cases, which may aid the current and future prevention and control of the global pandemic of COVID-19.
Author contributions
S.J., S.R. and K.C. designed the study. T.X., B.R. and T.J. collected the epidemiological and clinical data. L.C., Z.L., H.L., R.Y., Z.P., H.X., X.Q., P.J., C.F., K.B., S.J., L.Z. and L.J. carried out the investigations. S.J. drafted the manuscript, S.R. and K.C. revised the final manuscript.
Transparency declaration
This work was supported by grants from the Guangdong Provincial Novel Coronavirus Scientific and Technological Project (2020111107001) and Guangzhou Novel Coronavirus Scientific and Technological Project (202008040004). All authors report no conflicts of interest relevant to this article.
Footnotes
Appendix ASupplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.08.043.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figs1

References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cmi.2020.08.043
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7474805
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cmi.2020.08.043
Article citations
Precision nutrition to reset virus-induced human metabolic reprogramming and dysregulation (HMRD) in long-COVID.
NPJ Sci Food, 8(1):19, 30 Mar 2024
Cited by: 2 articles | PMID: 38555403 | PMCID: PMC10981760
Review Free full text in Europe PMC
Characterizing Infections in Two Epidemic Waves of SARS-CoV-2 Omicron Variants: A Cohort Study in Guangzhou, China.
Viruses, 16(4):649, 22 Apr 2024
Cited by: 2 articles | PMID: 38675989 | PMCID: PMC11053513
Performance Analysis of Serodiagnostic Tests to Characterize the Incline and Decline of the Individual Humoral Immune Response in COVID-19 Patients: Impact on Diagnostic Management.
Viruses, 16(1):91, 06 Jan 2024
Cited by: 2 articles | PMID: 38257792 | PMCID: PMC10820597
Nasopharyngeal Viral Load Is the Major Driver of Incident Antibody Immune Response to SARS-CoV-2 Infection.
Open Forum Infect Dis, 10(12):ofad598, 02 Dec 2023
Cited by: 2 articles | PMID: 38111750 | PMCID: PMC10727195
A unique cytotoxic CD4+ T cell-signature defines critical COVID-19.
Clin Transl Immunology, 12(8):e1463, 28 Aug 2023
Cited by: 0 articles | PMID: 37645435 | PMCID: PMC10461786
Go to all (35) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Screening for SARS-CoV-2 antibodies in convalescent plasma in Brazil: Preliminary lessons from a voluntary convalescent donor program.
Transfusion, 60(12):2938-2951, 16 Sep 2020
Cited by: 43 articles | PMID: 32935877 | PMCID: PMC7756544
Analytical and clinical validation of an ELISA for specific SARS-CoV-2 IgG, IgA, and IgM antibodies.
J Med Virol, 93(2):803-811, 27 Jul 2020
Cited by: 56 articles | PMID: 32667733 | PMCID: PMC7405491
Closing the serological gap in the diagnostic testing for COVID-19: The value of anti-SARS-CoV-2 IgA antibodies.
J Med Virol, 93(3):1436-1442, 21 Aug 2020
Cited by: 29 articles | PMID: 32790181 | PMCID: PMC7436746
[SARS-CoV-2 and Microbiological Diagnostic Dynamics in COVID-19 Pandemic].
Mikrobiyol Bul, 54(3):497-509, 01 Jul 2020
Cited by: 8 articles | PMID: 32755524
Review
Funding
Funders who supported this work.
Grant ID: 2020111107001
Grant ID: 202008040004