Abstract
Free full text

Saliva Sampling and Its Direct Lysis, an Excellent Option To Increase the Number of SARS-CoV-2 Diagnostic Tests in Settings with Supply Shortages
ABSTRACT
As part of any plan to lift or ease the confinement restrictions that are in place in many different countries, there is an urgent need to increase the capacity of laboratory testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Detection of the viral genome through reverse transcription-quantitative PCR (RT-qPCR) is the gold standard for this virus; however, the high demand of the materials and reagents needed to sample individuals, purify the viral RNA, and perform the RT-qPCR has resulted in a worldwide shortage of several of these supplies. Here, we show that directly lysed saliva samples can serve as a suitable source for viral RNA detection that is less expensive and can be as efficient as the classical protocol, which involves column purification of the viral RNA. In addition, it bypasses the need for swab sampling, decreases the risk of the health care personnel involved in the testing process, and accelerates the diagnostic procedure.
INTRODUCTION
With the worldwide COVID-19 health emergency, there is an urgent need for rapid and reliable methods of diagnosis for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The accepted gold standard for detection of this virus is the amplification of regions of the viral genome by reverse transcription-quantitative PCR (RT-qPCR) in nasopharyngeal and oropharyngeal swabs (1, 2). Unfortunately, given the enormous demand for the reagents needed to collect the biological samples and to purify the viral RNA, there have been shortages of many of the reagents needed for the diagnostic tests. Swabs, viral transport media, and kits for viral RNA extraction are among the most common consumables that have become scarce, compromising the number of tests that can be done in many parts of the world.
Recently, several reports have demonstrated the possibility of using saliva instead of oral and nasal swabs to detect the genome of SARS-COV-2 (3,–5). Saliva collection also has many collateral benefits, including self-collection, which decreases the risk to health care workers in charge of taking the swabs and does not require the use of personal protective equipment (PPE), which has also become a scarce item in this pandemic (6, 7). In addition, the methods to extract the RNA from biological samples require the use of purification kits, whose availability has also become limited due to the heavy worldwide demand.
In this study, we compared the RT-qPCR results from 253 paired samples obtained from saliva and swabs of ambulatory patients; the RNA in the swab samples was extracted using a commercial RNA purification kit, and the saliva samples were directly mixed with a lysis buffer, boiled, and used for the RT-qPCR protocol. We found a very good correlation of results between the two types of samples, and we propose that saliva sampling and direct lysis, which together simplify the sampling of patients and accelerate the preparation of RNA for RT-qPCR, represent an excellent alternative that will facilitate sampling and diagnosis of a larger number of persons at a reduced cost.
MATERIALS AND METHODS
Sample collection.
A total of 253 paired samples from oropharyngeal and nasopharyngeal swabs (OPSs and NPSs, respectively) and saliva were collected during a span of 30 days (from 2 May to 31 May) by health care workers from the epidemiology department of the health ministry of the state of Morelos, Mexico (Secretaría de Salud Morelos [SSM]). All but 3 samples were from ambulatory patients; the 3 exceptions were collected from hospitalized patients.
days (from 2 May to 31 May) by health care workers from the epidemiology department of the health ministry of the state of Morelos, Mexico (Secretaría de Salud Morelos [SSM]). All but 3 samples were from ambulatory patients; the 3 exceptions were collected from hospitalized patients.
Swab sampling.
Oropharyngeal and nasopharyngeal swabs were taken from 71 patients, while a single oropharyngeal swab was taken from each of 182 patients. After their collection, swabs were placed in 2.5 ml of viral transport medium.
ml of viral transport medium.
Saliva collection.
Saliva was self-collected by patients that were asked to spit on several occasions into sterile urine cup containers until completing roughly 2 to 3 ml of saliva. No viral transport media, or stabilizing agents, were added to the saliva samples.
ml of saliva. No viral transport media, or stabilizing agents, were added to the saliva samples.
After collection, both swab and saliva samples were stored at 4°C until transported to the Institute of Biotechnology/UNAM for their analysis, which was within 24 to 36 h after sample collection.
h after sample collection.
Nucleic acid extraction and SARS-CoV-2 detection by RT-qPCR.
Total RNA was extracted from swab samples using the QIAamp viral RNA minikit (Qiagen) following the manufacturer’s protocol, using 140 μl of viral transport medium from each swab, and the purified RNA was eluted in 60
μl of viral transport medium from each swab, and the purified RNA was eluted in 60 μl of elution buffer.
μl of elution buffer.
Saliva samples were treated with Quick Extract DNA extraction solution (QE; Lucigen) by mixing 50 μl of saliva with 50
μl of saliva with 50 μl of the QE reagent and heating for 5 min at 95°C; the mixture was then cooled on ice and kept at 4°C until use (within 1
μl of the QE reagent and heating for 5 min at 95°C; the mixture was then cooled on ice and kept at 4°C until use (within 1 h of QE treatment). For saliva samples that had high viscosity, 1 volume of sterile phosphate-buffered saline (PBS) was added and mixed by repeated pipetting, and the diluted saliva sample was extracted as mentioned above.
h of QE treatment). For saliva samples that had high viscosity, 1 volume of sterile phosphate-buffered saline (PBS) was added and mixed by repeated pipetting, and the diluted saliva sample was extracted as mentioned above.
SARS-CoV-2 detection was performed using the Berlin protocol, using the reported oligonucleotides and probes for viral gene E and for human RNase P (8). The RT-qPCRs were performed using the StarQ one-step RT-qPCR (Genes 2 Life) kit, using 5 μl of the column-extracted total RNA in 20
μl of the column-extracted total RNA in 20 μl of reaction mixture or 2.5
μl of reaction mixture or 2.5 μl of the QE-treated saliva in 22.5
μl of the QE-treated saliva in 22.5 μl of reaction mixture. Samples were analyzed in an ABI Prism 7500 sequence detector system (Applied Biosystems) with the following thermal protocol: 50°C for 15
μl of reaction mixture. Samples were analyzed in an ABI Prism 7500 sequence detector system (Applied Biosystems) with the following thermal protocol: 50°C for 15 min, 95°C for 2
min, 95°C for 2 min, and then 45 cycles of 95°C for 15 s and 60°C for 30 s. All samples with a threshold cycle (CT) value equal to or less than 38 were classified as positive.
min, and then 45 cycles of 95°C for 15 s and 60°C for 30 s. All samples with a threshold cycle (CT) value equal to or less than 38 were classified as positive.
Determination of viral copy number.
To determine the viral copy number in a sample, a standard curve was generated using a 10-fold serial dilution of an in vitro T7 RNA transcript that carries the sequence recognized by oligonucleotides and probe for gene E. Briefly, the logarithm of concentration of each dilution was plotted against the CT and the viral copy number from unknown samples was determined by extrapolating the CT value onto the corresponding standard curve.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software Inc.) as described in Results.
RESULTS
Detection of SARS-CoV-2 in paired swab and saliva samples.
To evaluate if saliva is a good source of viral RNA for the RT-qPCR, we determined the presence of the SARS-CoV-2 genome in paired saliva and swab samples from 253 ambulatory patients. All patients had two or more symptoms related to COVID-19 (8, 9); 116 (45.4%) were male and 137 (54.1%) female, with a median age of 41 (±14.4) years. Samples were taken from ambulatory patients in the respiratory triage of the Tlaltenango health center in Cuernavaca, Morelos, Mexico. The RT-qPCR Berlin protocol was used to detect SARS-CoV-2, using only the primers and probe for gene E, since previous studies have shown a weak detection of viral RNA when the RNA-dependent RNA polymerase (RdRp) gene is probed (9, 10). As an internal control of RNA content in the samples, the RNase P gene was detected. Total RNA was purified from swabs using the QIAamp viral RNA minikit; the RNA in saliva was directly obtained using the QE lysis buffer (Lucigen) and boiling for 5 min, as reported previously (11).
min, as reported previously (11).
During the course of the study, and due to the shortage of swabs, the health center shifted temporarily from collecting two swabs per person (nasopharyngeal swab [NPS] plus oropharyngeal swab [OPS]) to only one swab (OPS) per individual. For the 253 patients included in this study, two swabs were used in 71 (28%) of the cases, while a single OPS was taken from the other 182 (72%) irrespective of the number of swabs collected, saliva samples were taken from all patients.
Of the 182 patients with a single swab collected, 80 (43.9%) were positive for SARS-CoV-2 as determined by either the swab or saliva samples. Of these, 41 (51.2%) were positive as determined by both types of samples, while 28 (35%) were positive only by saliva and not by the swab sample and 11 (13.7%) were positive only by the OPS. In total, out of the 80 individuals found to be positive for the virus, 69 (86.2%) were correctly identified using saliva, while only 52 (65%) were identified with the OPS (Table 1 and Fig. 1).
TABLE 1
Summary of results obtained from parallel testing of swab and saliva samples from patients suspected of having COVID-19
| Result for saliva | No. of patients with indicated result for single swab (OPS) | Total | No. of patients with indicated result for double swabs (OPS + NPS) | Total | ||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||
| Positive | 41 | 28 | 69 | 19 | 6 | 25 |
| Negative | 11 | 102 | 113 | 9 | 37 | 46 |
    Total Total | 52 | 130 | 182 | 28 | 43 | 71 |
On the other hand, 34 (47.8%) of the 71 patients with two swabs collected were found to be positive for SARS-CoV-2 by either the swabs or the saliva samples. Of these, 19 (55.8%) were positive by both swabs and saliva, while 6 (17.6%) were positive only by saliva and 9 (26.4%) were positive only by the two swab samples. In total, in this group of patients, of the 34 individuals identified as positive for the virus, 25 (73.5%) were identified by testing saliva, while 28 (82.3%) were positive by the swabs (Table 1 and Fig. 1).
Quantitation of viral RNA.
When the numbers of viral genome copies in the single OPS and saliva samples were compared, a significant difference in the geometric mean was detected, with saliva samples having a titer 1.9 log10 higher than that observed in the swabs (P <
< 0.0024 [Fig. 2A]). This can be better appreciated when the viral copy numbers in paired swabs and saliva from the same patient are plotted and represented as connecting lines (Fig. 2B); for 31 of the paired samples, the number of viral copies was higher in saliva samples than in swabs. Human RNase P was used as an internal control of sampling quality; of interest, the comparison between the mean of CT values obtained from OPS and saliva samples showed a difference of at least 6.8 CT units between both types of samples (Fig. 2C), indicating that there is more cellular material in saliva, as reported in other studies (12). The viral genome copy numbers in the double-swab and saliva samples were not statistically different, although a larger set of data would be needed to confirm these results (Fig. 2D).
0.0024 [Fig. 2A]). This can be better appreciated when the viral copy numbers in paired swabs and saliva from the same patient are plotted and represented as connecting lines (Fig. 2B); for 31 of the paired samples, the number of viral copies was higher in saliva samples than in swabs. Human RNase P was used as an internal control of sampling quality; of interest, the comparison between the mean of CT values obtained from OPS and saliva samples showed a difference of at least 6.8 CT units between both types of samples (Fig. 2C), indicating that there is more cellular material in saliva, as reported in other studies (12). The viral genome copy numbers in the double-swab and saliva samples were not statistically different, although a larger set of data would be needed to confirm these results (Fig. 2D).
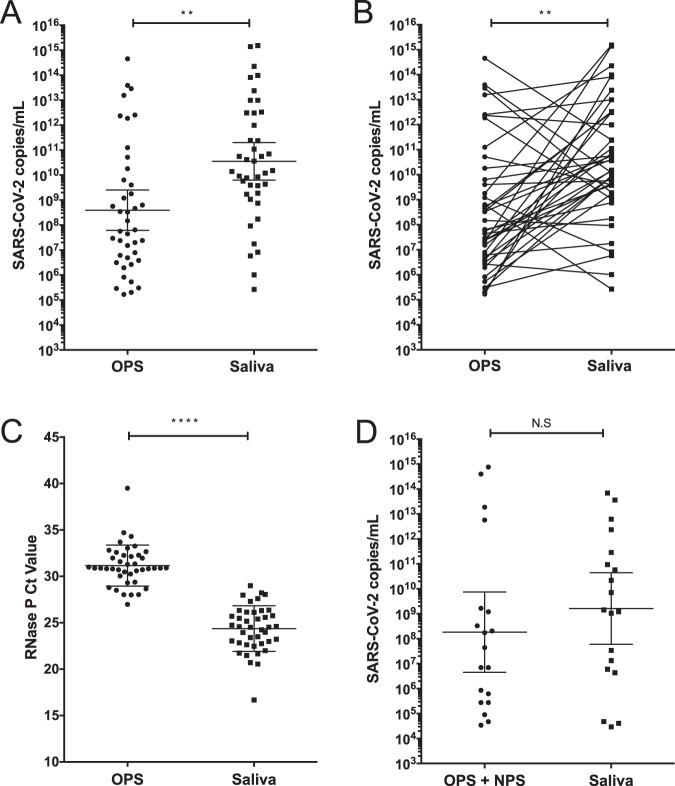
A high SARS-CoV-2 genome copy number is detected in saliva samples. (A) Viral titer detected in paired OPS and saliva samples. (B) Viral titer detected in paired OPS and saliva samples are represented by lines connecting both samples. Data were compared by a Wilcoxon test (**, P <
< 0.0024). (C) RT-PCR cycle CT values for RNase P detected in OPS and saliva samples. Data were compared by Wilcoxon test (****, P
0.0024). (C) RT-PCR cycle CT values for RNase P detected in OPS and saliva samples. Data were compared by Wilcoxon test (****, P <
< 0.0001). (D) Viral titer detected in paired double (NPS plus OPS) and saliva samples. Data were compared by Wilcoxon test; no statistical significance (N.S) was found (P
0.0001). (D) Viral titer detected in paired double (NPS plus OPS) and saliva samples. Data were compared by Wilcoxon test; no statistical significance (N.S) was found (P <
< 0.6226). Bars represent the geometric medians and 95% confidence intervals.
0.6226). Bars represent the geometric medians and 95% confidence intervals.
DISCUSSION
In this study, we analyzed 253 paired samples from either a single OPS compared to saliva or a double OPS and NPS and saliva. RNA purified from swabs using commercial column kits was compared with saliva samples directly lysed with QE buffer (surpassing the RNA extraction protocol) as a source for the RT-qPCR assay. Although the coincidence rate between the single OPS and saliva samples was relatively low (51.2%), the saliva samples were clearly more efficient in detecting the virus than single OPS samples (86.2% versus 65%). On the other hand, the efficiency of detection of the virus in saliva compared to the double OPS and NPS was slightly lower (73.5% versus 82.3%), with a coincidence rate of 55.8%.
Taken together, these results suggest that that saliva is a good source for SARS-CoV-2 detection, especially compared with a single OPS. Furthermore, it can be implemented for diagnostic tests using a simple QE buffer-based sample preparation in place of the column-based RNA purification method that is currently employed for swab analyses.
The reason for the low coincidence in the positive results obtained with swab and saliva samples is not clear. The failure of identification of SARS-CoV-2 in swabs when the saliva samples were positive for the virus could be due to bad swab sampling, what can be corroborated by the higher CT values of RNase P detected in these samples (Fig. 2C), with the consequent low viral copy number. This is a major concern, since the medical personnel in charge of taking the samples frequently do not do it correctly for the risk associated with this process. It has been reported that oropharyngeal swabs have a lower viral titer than nasopharyngeal swabs (1); thus, this could contribute to the discrepancies observed. Furthermore, it has also been previously reported that nasopharyngeal swabs have a lower viral titer than saliva samples (12), which could also contribute to explain our findings. On the other hand, the false negatives in saliva could be due either to the absence or undetectable levels of virus in the saliva samples or to unknown problems during the collection, transport, and/or storage of the sample before its arrival to the laboratory.
SARS-CoV-2 has been detected in saliva at higher titers during the first days after the onset of symptoms, with the viral titer declining over time. It is not clear how long after symptom onset the viral RNA can be detected in saliva, although some reports suggest a short period of detection (~13 days) compared with that for nasopharyngeal swabs (~19
days) compared with that for nasopharyngeal swabs (~19 days) (13). However, other reports have recently demonstrated the detection of viral RNA in saliva for longer periods (~20
days) (13). However, other reports have recently demonstrated the detection of viral RNA in saliva for longer periods (~20 days or longer) (4, 14). The patients included in this study were ambulatory and according to their clinical interviews were between 1 and 7
days or longer) (4, 14). The patients included in this study were ambulatory and according to their clinical interviews were between 1 and 7 days (median of 4
days (median of 4 days) of the onset of symptoms. We did not find a significant difference between the onset of symptoms and the cases in which positivity was determined only with saliva versus those that were detected only with swabs.
days) of the onset of symptoms. We did not find a significant difference between the onset of symptoms and the cases in which positivity was determined only with saliva versus those that were detected only with swabs.
Direct lysis of nasopharyngeal or oropharyngeal swab samples in viral transport medium using the QE buffer has been reported as a suitable method for direct RT-qPCR for SARS-CoV-2 detection, with rates similar to those of methods based on column purification (11, 15). However, we have found a great variability in the results obtained using the QE lysis protocol when applied to swab samples, most likely due to variations in the material of the swabs used and to variations in the preparation of the viral transport medium employed (data not shown). In this regard, it has recently been reported that the composition of viral transport media can affect the detection of RNA from SARS-CoV-2 and other viruses (16), and due to the scarcity of these media, several laboratories have started to prepare their own transport media, introducing an additional confusion factor. A similar situation occurs with swabs, since in view of the scarcity of suitable materials, other materials are being employed, despite the fact that some of them are known to inhibit RT-PCR (17).
The use of saliva samples offers the advantage that no additives or transport media need to be used for their preservation or analysis if stored in cold temperatures and analyzed up to 36 h after their collection. Our results indicate that a rapid processing of saliva using direct lysis with QE buffer offers an excellent alternative to the current swab analysis that uses RNA column purification, since it is a sensitive, fast, and inexpensive method that can be used for massive screening, in particular in those settings where common supplies needed for the classical methods are in shortage.
ACKNOWLEDGMENTS
We are grateful to the health care workers of Servicios Estatales de Salud de Morelos for their invaluable help in collecting the samples and to the personnel of the Laboratorio Estatal de Salud Pública del Estado de Morelos for their support in the preparation and transport of the samples. The work of P. Gaytán, E. López, and J. Yañez from the DNA sequencing and synthesis unit is also acknowledged.
Some of the reagents used in this study were provided by the Instituto Nacional de Diagnóstico y Referencia Epidemiológica, supported by INSABI. This work was supported by grant 314343 from CONACyT. J.M.-C. was the recipient of a scholarship from CONACyTs.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.01659-20
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7512180
Citations & impact
Impact metrics
Article citations
An Open One-Step RT-qPCR for SARS-CoV-2 detection.
PLoS One, 19(1):e0297081, 25 Jan 2024
Cited by: 0 articles | PMID: 38271448 | PMCID: PMC10810446
Comparing SARS-CoV-2 Viral Load in Human Saliva to Oropharyngeal Swabs, Nasopharyngeal Swabs, and Sputum: A Systematic Review and Meta-Analysis.
Can J Infect Dis Med Microbiol, 2023:5807370, 10 Aug 2023
Cited by: 1 article | PMID: 37600753 | PMCID: PMC10435302
Review Free full text in Europe PMC
Salivary SARS-CoV-2 RNA for diagnosis of COVID-19 patients: a systematic revisew and meta-analysis of diagnostic accuracy.
Jpn Dent Sci Rev, 21 Jun 2023
Cited by: 3 articles | PMID: 37360001 | PMCID: PMC10284464
Review Free full text in Europe PMC
Critical evaluation of strategies to achieve direct real-time PCR detection of swine pathogens in oral fluids.
J Vet Diagn Invest, 35(5):521-527, 20 Jun 2023
Cited by: 0 articles | PMID: 37337714 | PMCID: PMC10467463
A simple agglutination system for rapid antigen detection from large sample volumes with enhanced sensitivity.
Anal Chim Acta, 1277:341674, 29 Jul 2023
Cited by: 0 articles | PMID: 37604625 | PMCID: PMC10777812
Go to all (43) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits.
J Clin Virol, 129:104519, 23 Jun 2020
Cited by: 39 articles | PMID: 32629187 | PMCID: PMC7309780
Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19.
J Infect Chemother, 27(1):126-129, 30 Sep 2020
Cited by: 50 articles | PMID: 33060046 | PMCID: PMC7524660
Value of swab types and collection time on SARS-COV-2 detection using RT-PCR assay.
J Virol Methods, 286:113974, 16 Sep 2020
Cited by: 23 articles | PMID: 32949663 | PMCID: PMC7493793
Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: a Systematic Review and Meta-analysis.
J Clin Microbiol, 59(5):e02881-20, 20 Apr 2021
Cited by: 182 articles | PMID: 33504593 | PMCID: PMC8091856
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Consejo Nacional de Ciencia y Tecnología (1)
Grant ID: 314343

 a
a




