Abstract
Background
Tumour mutational status is an important determinant of the response of metastatic colorectal cancer to targeted treatments. However, the genotype of the tissue obtained at the time of diagnosis might not accurately represent tumour genotype after multiple lines of treatment. This retrospective exploratory analysis investigated the clinical activity of regorafenib in biomarker subgroups of the CORRECT study population defined by tumour mutational status or plasma protein levels.Methods
We used BEAMing technology to identify KRAS, PIK3CA, and BRAF mutations in DNA obtained from the plasma of 503 patients with metastatic colorectal cancer who enrolled in the CORRECT trial. We quantified total human genomic DNA isolated from plasma samples for 503 patients using a modified version of human long interspersed nuclear element-1 (LINE-1) quantitive real-time PCR. We also measured the concentration of 15 proteins of interest-angiopoietin 2, interleukin 6, interleukin 8, placental growth factor, soluble TIE-1, soluble VEGFR1, VEGF-A, VEGF-C, VEGF-D, VEGF-A isoform 121, bone morphogenetic protein 7, macrophage colony-stimulating factor, stromal cell-derived factor-1, tissue inhibitor of metalloproteinase 2, and von Willebrand factor-in plasma samples from 611 patients. We did correlative analyses of overall survival and progression-free survival in patient subgroups based on mutational status, circulating DNA concentration, and protein concentrations. The CORRECT trial was registered with ClinicalTrials.gov, number NCT01103323.Findings
Tumour-associated mutations were readily detected with BEAMing of plasma DNA, with KRAS mutations identified in 349 (69%) of 503 patients, PIK3CA mutations in 84 (17%) of 503 patients, and BRAF mutations in 17 (3%) of 502 patients. We did not do correlative analysis based on BRAF genotype because of the low mutational frequency detected for this gene. Some of the most prevalent individual hot-spot mutations we identified included: KRAS (KRAS G12D, 116 [28%] of 413 mutations; G12V, 72 [17%]; and G13D, 67 [16%]) and PIK3CA (PIK3CA E542K, 27 [30%] of 89 mutations; E545K, 37 [42%]; and H1047R, 12 [14%]). 41 (48%) of 86 patients who had received anti-EGFR therapy and whose archival tumour tissue DNA was KRAS wild-type in BEAMing analysis were identified as having KRAS mutations in BEAMing analysis of fresh plasma DNA. Correlative analyses suggest a clinical benefit favouring regorafenib across patient subgroups defined by KRAS and PIK3CA mutational status (progression-free survival with regorafenib vs placebo: hazard ratio [HR] 0·52, 95% CI 0·35-0·76 for KRAS wild-type; HR 0·51, 95% CI 0·40-0·65 for KRAS mutant [KRAS wild type vs mutant, pinteraction=0·74]; HR 0·50, 95% CI 0·40-0·63 for PIK3CA wild-type; HR 0·54, 95% CI 0·32-0·89 for PIK3CA mutant [PIK3CA wild-type vs mutant, pinteraction=0·85]) or circulating DNA concentration (progression-free survival with regorafenib vs placebo: HR 0·53, 95% CI 0·40-0·71, for low circulating DNA concentrations; HR 0·52, 95% CI 0·40-0·70, for high circulating DNA concentrations; low vs high circulating DNA, pinteraction=0·601). With the exception of von Willebrand factor, assessed with the median cutoff method, plasma protein concentrations were also not associated with regorafenib activity in terms of progression-free survival. In univariable analyses, the only plasma protein that was associated with overall survival was TIE-1, high concentrations of which were associated with longer overall survival compared with low TIE-1 concentrations. This association was not significant in multivariable analyses.Interpretation
BEAMing of circulating DNA could be a viable approach for non-invasive analysis of tumour genotype in real time and for the identification of potentially clinically relevant mutations that are not detected in archival tissue. Additionally, the results show that regorafenib seems to be consistently associated with a clinical benefit in a range of patient subgroups based on mutational status and protein biomarker concentrations.Funding
Bayer HealthCare Pharmaceuticals.Free full text

Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial
Associated Data
Summary
Background
Tumour mutational status is an important determinant of the response of metastatic colorectal cancer to targeted treatments. However, the genotype of the tissue obtained at the time of diagnosis might not accurately represent tumour genotype after multiple lines of treatment. This retrospective exploratory analysis investigated the clinical activity of regorafenib in biomarker subgroups of the CORRECT study population defined by tumour mutational status or plasma protein levels.
Methods
We used BEAMing technology to identify KRAS, PIK3CA, and BRAF mutations in DNA obtained from the plasma of 503 patients with metastatic colorectal cancer who enrolled in the CORRECT trial. We quantified total human genomic DNA isolated from plasma samples for 503 patients using a modified version of human long interspersed nuclear element-1 (LINE-1) quantitive real-time PCR. We also measured the concentration of 15 proteins of interest—angiopoietin 2, interleukin 6, interleukin 8, placental growth factor, soluble TIE-1, soluble VEGFR1, VEGF-A, VEGF-C, VEGF-D, VEGF-A isoform 121, bone morphogenetic protein 7, macrophage colony-stimulating factor, stromal cell-derived factor-1, tissue inhibitor of metalloproteinase 2, and von Willebrand factor—in plasma samples from 611 patients. We did correlative analyses of overall survival and progression-free survival in patient subgroups based on mutational status, circulating DNA concentration, and protein concentrations. The CORRECT trial was registered with ClinicalTrials.gov, number NCT01103323.
Findings
Tumour-associated mutations were readily detected with BEAMing of plasma DNA, with KRAS mutations identified in 349 (69%) of 503 patients, PIK3CA mutations in 84 (17%) of 503 patients, and BRAF mutations in 17 (3%) of 502 patients. We did not do correlative analysis based on BRAF genotype because of the low mutational frequency detected for this gene. Some of the most prevalent individual hot-spot mutations we identified included: KRAS (KRAS G12D, 116 [28%] of 413 mutations; G12V, 72 [17%]; and G13D, 67 [16%]) and PIK3CA (PIK3CA E542K, 27 [30%] of 89 mutations; E545K, 37 [42%]; and H1047R, 12 [14%]). 41 (48%) of 86 patients who had received anti-EGFR therapy and whose archival tumour tissue DNA was KRAS wild-type in BEAMing analysis were identified as having KRAS mutations in BEAMing analysis of fresh plasma DNA. Correlative analyses suggest a clinical benefit favouring regorafenib across patient subgroups defined by KRAS and PIK3CA mutational status (progression-free survival with regorafenib vs placebo: hazard ratio [HR] 0·52, 95% CI 0·35–0·76 for KRAS wild-type; HR 0·51, 95% CI 0·40–0·65 for KRAS mutant [KRAS wild type vs mutant, pinteraction=0·74]; HR 0·50, 95% CI 0·40–0·63 for PIK3CA wild-type; HR 0·54, 95% CI 0·32–0·89 for PIK3CA mutant [PIK3CA wild-type vs mutant, pinteraction=0·85]) or circulating DNA concentration (progression-free survival with regorafenib vs placebo: HR 0·53, 95% CI 0·40–0·71, for low circulating DNA concentrations; HR 0·52, 95% CI 0·40–0·70, for high circulating DNA concentrations; low vs high circulating DNA, pinteraction=0·601). With the exception of von Willebrand factor, assessed with the median cutoff method, plasma protein concentrations were also not associated with regorafenib activity in terms of progression-free survival. In univariable analyses, the only plasma protein that was associated with overall survival was TIE-1, high concentrations of which were associated with longer overall survival compared with low TIE-1 concentrations. This association was not significant in multivariable analyses.
Interpretation
BEAMing of circulating DNA could be a viable approach for non-invasive analysis of tumour genotype in real time and for the identification of potentially clinically relevant mutations that are not detected in archival tissue. Additionally, the results show that regorafenib seems to be consistently associated with a clinical benefit in a range of patient subgroups based on mutational status and protein biomarker concentrations.
Introduction
Standard treatment for metastatic colorectal cancer includes drugs that target the molecular drivers of colorectal cancer pathogenesis, such as VEGF and EGFR. These drugs have improved overall survival, progression-free survival, and response in metastatic colorectal cancer.1–9
Tumour genotype plays an important part in drug resistance in patients with metastatic colorectal cancer. For example, use of anti-EGFR-antibody treatment is restricted to patients with RAS-wild-type tumours, some of whom acquire RAS mutations during treatment as a mechanism of drug resistance.10,11 Genotyping of tumour tissue can help with the selection of patients with tumours amenable to treatment; however, the value of testing a tumour sample is limited by intertumour and intratumour heterogeneity. Moreover, archival tissue will not show genotypic changes that have occurred since the sample was obtained.
Regorafenib is a multikinase inhibitor that inhibits various protein kinases implicated in oncogenesis, angiogenesis, and the tumour microenvironment.12 Treatment with regorafenib has shown significant benefits for overall survival and progression-free survival in patients with previously treated metastatic colorectal cancer in two placebo-controlled phase 3 trials, CORRECT13 and CONCUR.14
Investigation of the effect of genotype on treatment outcomes in patients treated with regorafenib is important in view of the association between tumour genotype and response to treatment that exists with other drugs used for metastatic colorectal cancer. In this retrospective exploratory analysis, we investigated the clinical activity of regorafenib in biomarker subgroups of the study population of CORRECT, as defined by tumour mutation status, plasma DNA concentration, or plasma protein concentrations.
Methods
Study design and participants
CORRECT13 was an international, multicentre, randomised, placebo-controlled phase 3 study in patients with histologically or cytologically documented metastatic adenocarcinoma of the colon or rectum who had received all approved standard therapies available locally (including a fluoropyrimidine, oxaliplatin, irinotecan, bevacizumab, and, if appropriate, cetuximab or panitumumab), with disease progression during, or within 3 months after the last administration of the last standard therapy. 760 patients were randomly assigned 2:1 to receive oral regorafenib 160 mg or matching placebo once per day for the first 3 weeks of each 4-week cycle and were treated until disease progression, death, unacceptable toxic effects, withdrawal of patient consent, or decision by the treating physician that discontinuation would be in the patient’s best interest. All patients also received best supportive care.
The primary endpoint was overall survival, with secondary endpoints including progression-free survival, objective tumour response rate, disease control rate, and safety. Results of the efficacy and safety analyses have been reported previously.13 At study entry, all patients had previously received bevacizumab, and about 51% had received an anti-EGFR treatment.13 Biomarkers were assessed in a planned substudy, with permission from the ethics review boards at each participating study centre, and genotypic analyses were performed only for samples collected from patients who provided separate written, informed consent. Genetic biomarker analyses were done by BEAMing (beads, emulsions, amplification, and magnetics) of DNA obtained from fresh plasma collected at baseline just before initiation of treatment and archival tumour tissue. Mutation data from historical testing of archival tumour tissue was also collected. Protein biomarker analyses were done with fresh plasma collected at baseline.
Procedures
Mutational analysis was done with BEAMing,15 a technique based on emulsion PCR that allows detection of one mutant allele in 10 000 wild-type alleles.16 Results from previous studies have supported the concordance of genotyping in patient-matched plasma and tumour tissue samples by use of BEAMing.17 All BEAMing experiments were done by Inostics GmbH (Hamburg, Germany). Circulating DNA was isolated from roughly 2 mL of fresh plasma obtained from patients in the CORRECT trial, just before the initiation of treatment. Plasma samples were thawed at room temperature for 10 min and cell debris was pelleted by centrifugation before DNA purification using a QIAamp DNA purification kit (Qiagen, Venlo, Netherlands). To reduce the possibility of false-positive results, the sensitivity cutoff for the BEAMing assay of plasma DNA was set at a lower limit of 0·02% mutant alleles based on the contract research organisation’s experience, although the assay is capable of detecting 0·01% mutant alleles. For the mutational analysis of tumour tissue DNA, archival tumour tissue specimens were examined by a pathologist, and samples with fewer than 5% tumour cells present were deemed unevaluable (unless a mutation was subsequently detected). One to three tissue sections were scraped from glass slides and DNA was isolated using a QuickExtract formalin-fixed paraffin-embedded (FFPE) DNA Extraction kit (Epicentre, Madison, WI, USA). For the BEAMing assay of DNA from tumour tissue, the lower cutoff limit was set at 1·0% mutant alleles to avoid potential false-positive results caused by interpatient contamination resulting from tissue sectioning. The appendix shows the genes and mutations analysed by BEAMing (appendix p 2). All KRAS, PIK3CA, and BRAF mutations examined have been reported to be oncogenically activating.18–22 Next generation sequencing was done by Foundation Medicine (Cambridge, MA, USA) using the DNA isolated from archival FFPE tumour tissue samples on the Foundation ONE panel.
We measured the total amount of human genomic DNA isolated from plasma samples using a modified version of human long interspersed nuclear element-1 (LINE-1) quantitive real-time (qRT)-PCR.23 We designed primers to amplify a 96 base-pair region within the abundant consensus region of the human LINE-1 family. We did the qRT-PCR in the presence of SYBR Green I dye (Life Technologies, Carlsbad, CA, USA). We used a small aliquot (a few microlitres) of plasma DNA (the precise volume was not recorded) as a template for the qRT-PCR. We ran dilutions of normal human genomic DNA in parallel on each plate as reference standards for the quantification. Each sample and reference standard was run in duplicate. PCR reactions were done in a real-time cycler (Eppendorf, Hamburg, Germany) and threshold cycle number was determined according to the manufacturer’s instructions with background fluorescence subtracted. We chose a human LINE-1 sequence-based assay, rather than an individual gene-based assay, because this assay needed a much smaller amount of starting material. We report DNA amount in genome equivalents, with one genome equivalent being one haploid human genome weighing 3·3 pg.
We determined the concentration of 15 proteins of interest in plasma samples using multiplex immuno-assays or ELISAs. We did all assays in duplicate, and the mean coefficient of variation calculated for all proteins was less than the pre-set mean coefficient of variation of 15%. Assays for protein concentration were done by AssayGate (Ijamsville, MD, USA). We selected the proteins of interest on the basis of their roles in angiogenesis and colorectal cancer pathogenesis.24–34 Biomarkers associated with angiogenesis were angiopoietin 2, interleukin 6, interleukin 8, placental growth factor (PlGF), soluble TIE-1, soluble VEGFR1, VEGF-A, VEGF-C, VEGF-D, and VEGF-A isoform 121. Biomarkers associated with colorectal cancer pathogenesis were bone morphogenetic protein 7, macrophage colony-stimulating factor, stromal cell-derived factor-1, tissue inhibitor of metalloproteinase 2, and von Willebrand factor. We defined patient subgroups on the basis of high or low concentrations of each protein using three methods to define the cutoff: median (high values defined as greater than or equal to the median); best-fit (high values defined as greater than or equal to the optimum cutoff point based on a Cox model); and receiver operating characteristic (ROC) curve (high values defined as greater than or equal to the optimum cutoff based on ROC analysis).
Statistical analysis
The biomarker analyses reported here are retrospective and exploratory, and all findings are considered hypothesis-generating rather than confirmatory, and should be interpreted with caution. CORRECT was not specifically powered to assess the clinical activity of regorafenib in any biomarker-defined subgroups, and we did no sample size calculations for our analyses. We did no adjustments for multiple testing. We did all statistical analyses with SAS version 9.1 and R version 3.02.
The primary objective of the statistical analysis of DNA was to establish whether the treatment effects of regorafenib on overall survival and progression-free survival were affected by baseline tumour mutation status (mutant or wild-type). We tested these differences directly using a Cox regression model with treatment effect, mutation status, and an interaction between the two, with the interaction as the main test of effect. Additionally, we investigated the potential prognostic value of KRAS and PIK3CA mutation status, circulating DNA concentrations, KRAS mutant allele frequency in circulating DNA, and KRAS mutant molecule concentration in circulating DNA by constructing Cox regression models and Kaplan–Meier survival estimates for the placebo group, with overall survival and progression-free survival as the dependent variables, and biomarker concentrations as the independent variables. To avoid the potential confounding effects of regorafenib treatment, we only investigated the prognostic effects of DNA biomarkers in the placebo group.
The primary objective of the statistical analysis of plasma proteins was to establish whether the treatment effects of regorafenib on overall survival and progression-free survival were affected by the concentrations of any one of the 15 proteins that we measured in baseline plasma samples. We identified optimum cutoff points for the best-fit method by starting at the 11th smallest observation and assessing every cutoff point up to the 10th largest observation; we then generated a Cox model to predict each outcome. The model with the best fit was the one with largest log-likelihood. For the ROC curve analysis, the optimal cutoff was the point on the ROC curve with the largest average sensitivity and specificity. We calculated the potential predictive value of each protein biomarker using a Cox regression model of treatment effect, protein concentration, and an interaction between the two, with the interaction as the primary test of effect. We also assessed the potential prognostic value of each protein biomarker by constructing Cox regression models and Kaplan-Meier survival estimates for the placebo group, with overall survival and progression-free survival as the dependent variables and protein concentrations as the independent variables. Models for the protein biomarker subgroups were also assessed formally with log-rank tests. As with the DNA biomarker analyses, we only investigated the prognostic effects of protein biomarkers in the placebo group to avoid the potential confounding effects of regorafenib treatment. The CORRECT trial is registered with ClinicalTrials.gov, number NCT01103323.
Role of the funding source
The study sponsor provided the study drug and collaborated with the investigators on protocol design, data collection, analysis, and interpretation, and preparation of this report. An independent data monitoring committee, composed of three oncologists and a statistician, supervised the overall integrity of the trial and safety of participants. JT and H-JL had full access to the data and had final responsibility for the content of this report and the decision to submit for publication. JT had the final responsibility to submit the manuscript. All authors had access to the study data and reviewed this report. The sponsor paid for writing assistance.
Results
In CORRECT, 760 patients were randomly assigned (2:1) to receive treatment with regorafenib (505 patients) or placebo (255 patients) between April 30, 2010, and March 22, 2011. The trial was stopped by the data monitoring committee at the second planned interim analysis on Oct 22, 2011, after 432 deaths, because the hazard ratio (HR) for overall survival for regorafenib versus placebo crossed the prespecified overall survival efficacy boundary (HR 0∙77, 95% CI 0∙64–0∙94, one-sided p=0∙0052). The database cutoff date used for the present analysis was July 21, 2011.13 The median duration of follow-up was 4·9 months (IQR 3·2–7·0).
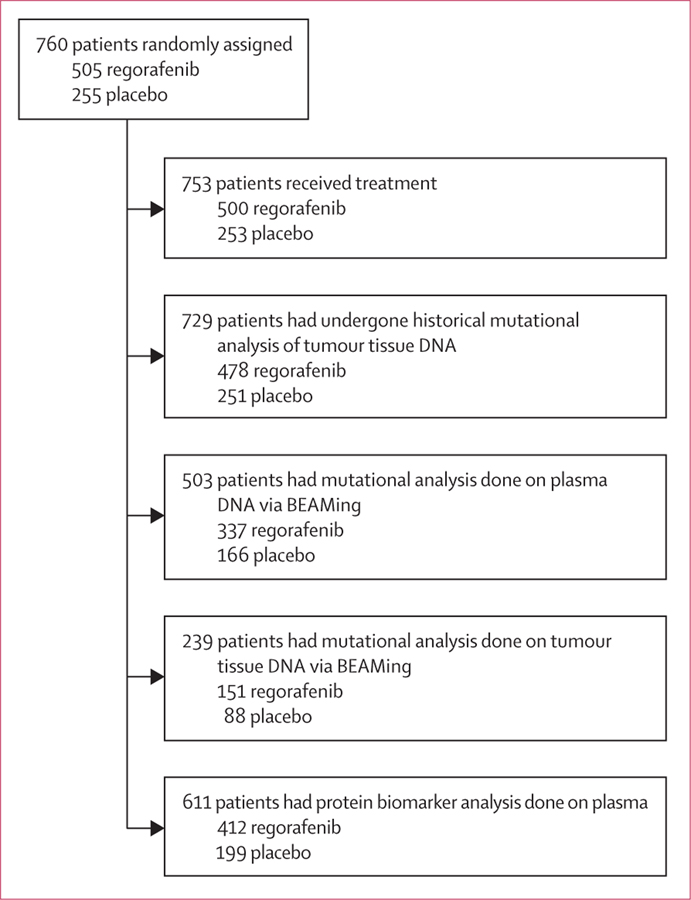
Subsets of enrolled patients were available for biomarker analysis (figure 1). Patient demographics and disease characteristics in the patient subgroups defined by genetic and protein biomarkers seemed to be consistent with those of the overall trial population (table 1).
Table 1:
Baseline patient demographics and disease characteristics
| Overall population of CORRECT | Population subgroup with genetic biomarker data | Population subgroup with protein biomarker data | ||||
|---|---|---|---|---|---|---|
| Regorafenib (n=505) | Placebo (n=255) | Regorafenib (n=337) | Placebo (n=166) | Regorafenib (n=412) | Placebo (n=199) | |
| Age (years) | 61 (54–67) | 61 (54–68) | 61 (54–69) | 61 (55–67) | 61 (54–68) | 61 (55–67) |
| Sex | ||||||
 Men Men | 311 (62%) | 153 (60%) | 214 (64%) | 102 (61%) | 261 (63%) | 124 (62%) |
 Women Women | 194 (38%) | 102 (40%) | 123 (36%) | 64 (39%) | 151 (37%) | 75 (38%) |
| Ethnic origin | ||||||
 White White | 392 (78%) | 201 (79%) | 254 (75%) | 130 (78%) | 321 (78%) | 160 (80%) |
 Black Black | 6 (1%) | 8 (3%) | 5 (1%) | 4 (2%) | 5 (1%) | 6 (3%) |
 Asian Asian | 76 (15%) | 35 (14%) | 51 (15%) | 23 (14%) | 60 (15%) | 25 (13%) |
 Other or not specified Other or not specified | 31 (6%) | 11 (4%) | 27 (8%) | 9 (5%) | 26 (6%) | 8 (4%) |
| Region | ||||||
 North America, western Europe, Israel, and Australia North America, western Europe, Israel, and Australia | 420 (83%) | 212 (83%) | 281 (83%) | 135 (81%) | 346 (84%) | 166 (83%) |
 Asia Asia | 69 (14%) | 35 (14%) | 45 (13%) | 23 (14%) | 54 (13%) | 25 (13%) |
 Eastern Europe Eastern Europe | 16 (3%) | 8 (3%) | 11 (3%) | 8 (5%) | 12 (3%) | 8 (4%) |
| ECOG performance status | ||||||
 0 0 | 265 (52%) | 146 (57%) | 179 (53%) | 94 (57%) | 229 (56%) | 114 (57%) |
 1 1 | 240 (48%) | 109 (43%) | 158 (47%) | 72 (43%) | 183 (44%) | 85 (43%) |
| Primary site of disease* | ||||||
 Colon Colon | 323 (64%) | 172 (67%) | 214 (64%) | 117 (70%) | 264 (64%) | 138 (69%) |
 Rectum Rectum | 151 (30%) | 69 (27%) | 104 (31%) | 44 (27%) | 123 (30%) | 52 (26%) |
 Colon and rectum Colon and rectum | 30 (6%) | 14 (5%) | 18 (5%) | 5 (3%) | 24 (6%) | 9 (5%) |
| KRAS mutation† | ||||||
 No No | 205 (41%) | 94 (37%) | 142 (42%) | 61 (37%) | 169 (41%) | 75 (38%) |
 Yes Yes | 273 (54%) | 157 (62%) | 179 (53%) | 104 (63%) | 221 (54%) | 120 (60%) |
 Unknown Unknown | 27 (5%) | 4 (2%) | 16 (5%) | 1 (1%) | 22 (5%) | 4 (2%) |
| Number of previous systemic anticancer treatments (on or after diagnosis of metastatic disease) | ||||||
 1–2 1–2 | 135 (27%) | 63 (25%) | 93 (28%) | 42 (25%) | 114 (28%) | 55 (28%) |
 3 3 | 125 (25%) | 72 (28%) | 83 (25%) | 45 (27%) | 105 (25%) | 53 (27%) |
 ≥4 ≥4 | 245 (49%) | 120 (47%) | 161 (48%) | 79 (48%) | 193 (47%) | 91 (46%) |
| Previous panitumumab, cetuximab, or both | ||||||
 No No | 241 (48%) | 134 (53%) | 159 (47%) | 90 (54%) | 197 (48%) | 101 (51%) |
 Yes Yes | 264 (52%) | 121 (47%) | 178 (53%) | 76 (46%) | 215 (52%) | 98 (49%) |
| Time from diagnosis of metastases to randomisation (months) | 31·0 (20·6–43·3) | 29·9 (20·2–46·4) | 30·6 (20·1–42·4) | 28·7 (20·1–42·8) | 29·9 (20·1–42·7) | 28·3 (19·9–43·1) |
 <18 months <18 months | 91 (18%) | 49 (19%) | 61 (18%) | 32 (19%) | 77 (19%) | 38 (19%) |
 ≥18 months ≥18 months | 414 (82%) | 206 (81%) | 276 (82%) | 134 (81%) | 335 (81%) | 161 (81%) |
Data are n (%) or median (IQR). ECOG=Eastern Cooperative Oncology Group.
We analysed cancer-associated hot-spot mutations in KRAS, PIK3CA, and BRAF (appendix p 2) by BEAMing, using DNA isolated from fresh plasma and archival tumour tissue. Table 2 shows the availability of plasma and archival tissue specimens for BEAMing analysis of KRAS, PIK3CA, and BRAF mutational status, and the frequencies of KRAS, PIK3CA, and BRAF mutations. The frequency of individual KRAS and PIK3CA mutations in plasma DNA was generally consistent with data reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (table 3). BRAF mutations were only identified in 14 patients in the regorafenib group and three patients in the placebo group; no further analyses in patient subgroups based on BRAF status were done because of the small number of mutations detected.
Table 2:
Summary of genotyping analyses done in the CORRECT trial
| DNA source for genotyping | Genotyping technology used | Biomarker sampling frequency (n=760) | Mutation frequency | |
|---|---|---|---|---|
| KRAS | Plasma* | BEAMing | 503 (66%) | 349/503 (69%) |
| Archival tumour tissue | BEAMing | 239 (31%) | 140/239 (59%) | |
| Archival tumour tissue | Historical† | 729 (96%) | 430/729 (59%) | |
| PIK3CA | Plasma* | BEAMing | 503 (66%) |  84/503 (17%) 84/503 (17%) |
| Archival tumour tissue | BEAMing | 236 (31%) |  29/236 (12%) 29/236 (12%) | |
| BRAF | Plasma* | BEAMing | 502 (66%) |  17/502 (3%) 17/502 (3%) |
| Archival tumour tissue | BEAMing | 269 (35%) |  4/269 (1%) 4/269 (1%) |
Data are n (%) or n/N (%). Biomarker sampling frequency is the proportion of patients in CORRECT for whom mutation data were generated; for some patients, DNA samples were of insufficient quality or quantity to examine all three genes. Mutation frequency is the proportion of patients in CORRECT with mutation data who were identified as having a mutation in the specified gene.
Table 3:
Frequency of individual hot-spot mutations detected by BEAMing of plasma DNA in the CORRECT trial
| Frequency in COSMIC*35 | Plasma DNA BEAMing in CORRECT | |
|---|---|---|
| KRAS | ||
| G12D | 5783/16 453 (35%) | 116/413 (28%) |
| G12V | 3677/16 453 (22%) | 72/413 (17%) |
| G12C | 1334/16 453 (8%) | 35/413 (8%) |
| G12A | 1039/16 453 (6%) | 36/413 (9%) |
| G12S | 963/16 453 (6%) | 16/413 (4%) |
| G12R | 203/16 453 (1%) | 9/413 (2%) |
| G13D | 3235/16 453 (20%) | 67/413 (16%) |
| Q61H | 126/16 453 (1%) | 50/413 (12%) |
| A146T | 93/16 453 (1%) | 12/413 (3%) |
| PIK3CA | ||
| E542K | 273/1098 (25%) | 27/89 (30%) |
| E545K | 412/1098 (38%) | 37/89 (42%) |
| E545G | 36/1098 (3%) | 9/89 (10%) |
| H1047R | 306/1098 (28%) | 12/89 (13%) |
| H1047L | 47/1098 (4%) | 3/89 (3%) |
| H1047Y | 24/1098 (2%) | 1/89 (1%) |
In total, we identified 413 KRAS mutations and 89 PIK3CA mutations. BRAF is not shown because only a single mutation (V600E) was assessed for this gene. COSMIC=Catalogue of Somatic Mutations in Cancer.
Comparison of patient-matched fresh plasma and archival tumour samples showed concordant mutation status in 161 (76%) of 211 patients for KRAS, 183 (88%) of 207 patients for PIK3CA, and 230 (97%) of 236 patients for BRAF (denominators are the number of patients with matched samples tested for each mutation in archival tissue and by BEAMing of plasma DNA). Because rates of discordance for KRAS were unexpectedly high compared with PIK3CA and BRAF, we decided to investigate the subgroup of patients with discordant KRAS status further. Most of the discordance in KRAS status was because of the detection of a mutation in plasma but not in the patient-matched archival tumour sample (41 [82%] of 50 patients with discordant results). For 30 (73%) of the 41 patients with discordant KRAS status caused by identification of KRAS mutations in fresh plasma, the KRAS wild-type status of the archival tissue was confirmed by BEAMing analysis (appendix p 3). Sufficient archival tissue was available for 19 (63%) of these 30 patients to additionally confirm the KRAS wild-type status of the tumour tissue by next-generation sequencing. In our study, the 41 patients with KRAS mutations detected in plasma, but not in archival tissue, made up 48% of the 86 patients who had received anti-EGFR therapy and whose archival tumour tissue DNA was KRAS wild-type on BEAMing analysis. KRAS mutations detected in the plasma DNA of the subgroup with discordant plasma and tissue results had some notable features compared with the mutational profile of patients with KRAS mutations who had concordant plasma and tissue results (appendix pp 3–7): first, a high representation of the KRAS Q61H mutation; second, multiple KRAS mutations in single plasma DNA samples; and third, a low frequency of mutant alleles detected.
Additionally, mutations were detected by BEAMing analysis from both fresh plasma and archival tumour tissue in 108 patient-matched samples for KRAS and 16 patient-matched samples for PIK3CA (appendix pp 4–7). Comparison of the mutations identified between patient-matched plasma and tumour tissue shows that the hot-spot mutation identified was identical in 105 (97%) of 108 cases for KRAS and in 15 (94%) of 16 cases for PIK3CA. In these samples, the mean mutant allele frequency detected in plasma DNA was 11·05% (range 0·03–57·93) for KRAS and 8·23% (0·13–28·99) for PIK3CA.
Eight patients identified as having KRAS mutations by BEAMing analysis of plasma and tumour tissue had previously been identified as having wild-type KRAS by historical testing (appendix p 8). Tumour tissue BEAMing results were confirmed in five patients who had sufficient archival tumour tissue available for next-generation sequencing. Thus, these eight patients seem to have been incorrectly classified as having KRAS-wild-type tumours on historical analysis because of issues with assay sensitivity or testing for a limited repertoire of KRAS mutations. Because these eight patients represent only a portion of patients in CORRECT for whom BEAMing results could be confirmed with DNA from matched plasma and tumour tissue, additional patients might have had their tumours incorrectly identified as KRAS wild-type based on historical test results.
The clinical benefit of regorafenib in terms of overall survival and progression-free survival in the KRAS-mutant and KRAS-wild-type subgroups defined by historical testing in the CORRECT trial13 was supported by the plasma DNA genotyping results (figures 2 and and3).3). Furthermore, the clinical benefit of regorafenib was shown in PIK3CA-mutant and PIK3CA-wild-type subgroups, as defined by BEAMing analysis of plasma (figure 3). Interaction testing showed no significant differences between wild-type or mutant subgroups for either KRAS or PIK3CA (figure 3), suggesting a clinical benefit of regorafenib in all subgroups tested. Evaluation of overall survival and progression-free survival in the patient subgroup in which BEAMing of plasma DNA was done shows that this subgroup is representative of the overall CORRECT population (figure 3).
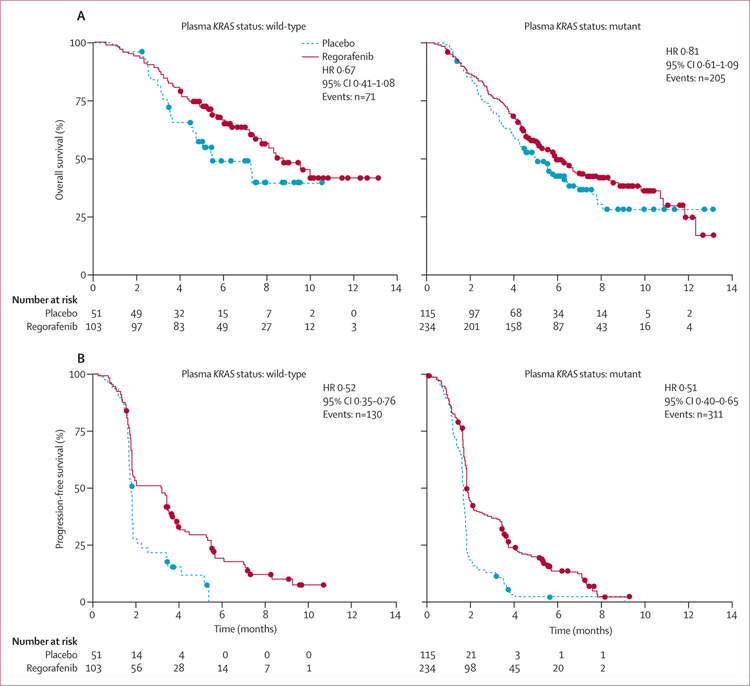
(A) Overall survival. (B) Progression-free survival. HR=hazard ratio.
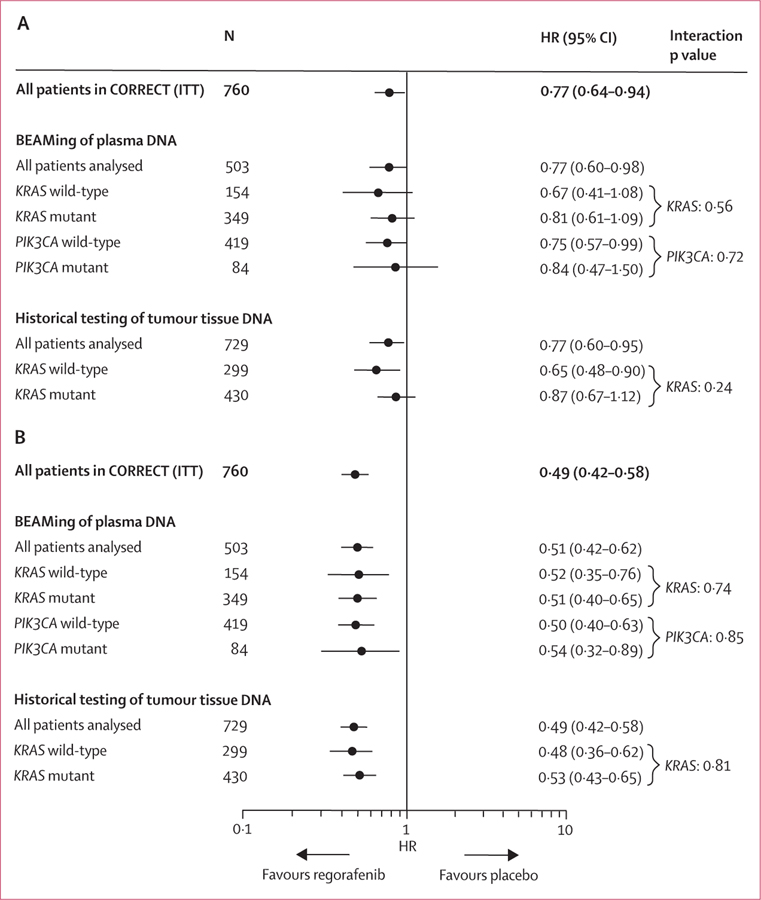
HR=hazard ratio. ITT=intention to treat.
We did not do correlative analyses based on BRAF mutational status identified with BEAMing of plasma DNA because of the small number of patients identified as having BRAF mutations. Correlative analyses of mutational status identified from genotyping of archival tumour tissue with BEAMing were not informative, because tumour mutational data were available from only 239 (31%) of 760 patients (figure 1), and this biomarker subgroup was not representative of the overall trial population in terms of clinical outcomes (data not shown).
We identified no prognostic effect of KRAS mutation status in the 251 patients who received placebo and for whom historical mutation data were available (mutant vs wild-type: HR for overall survival 0·92, 95% CI 0·67–1·27; HR for progression-free survival 1·14, 95% CI 0·87–1·48). The prognostic effects of KRAS mutation status based on BEAMing of archival tissue DNA are not reported because of the small number of patients in this subgroup (n=88), compared with 251 patients in the historical data subgroup.
We measured total circulating DNA in fresh plasma samples obtained at the time of enrolment into the trial to assess potential associations between circulating DNA concentration and clinical outcome. Circulating DNA concentrations from 503 patients showed high interpatient variability (mean 33 447 genome equivalents of DNA per mL, SD 78 859, median 10 653 genome equivalents of DNA per mL, range 1114–804 450). Analysis of overall survival and progression-free survival in patients with baseline plasma DNA concentrations more than (high) or less than (low) the median suggested that the clinical benefit of regorafenib compared with placebo was not significantly affected by baseline plasma DNA concentrations (overall survival with regorafenib vs placebo: HR 0·97, 95% CI 0·63–1·52 for low circulating DNA concentrations; HR 0·70, 95% CI 0·52–0·95 for high circulating DNA concentrations; pinteraction=0·199; progression-free survival with regorafenib vs placebo: HR 0·53, 95% CI 0·40–0·71 for low circulating DNA concentrations; HR 0·52, 95% CI 0·40–0·70 for high circulating DNA concentrations; pinteraction=0·601). However, our analysis of the potential prognostic effect of plasma DNA concentrations in the placebo group showed that a high baseline circulating DNA concentration was associated with shorter median overall and progression-free survival compared with patients with a low baseline circulating DNA concentration (figure 4, appendix p 9). We also identified an association between high baseline circulating DNA concentration and reduced median overall survival compared with low baseline circulating DNA concentrations when we analysed patients in the regorafenib treatment group (HR 0·34, 95% CI 0·25–0·47). High baseline KRAS mutant allele frequency in circulating DNA (appendix p 10) and a high plasma concentration of KRAS mutant molecules (appendix p 11) were also associated with a shorter median overall survival compared with that seen for low baseline KRAS mutant allele frequency and plasma concentration in placebo-treated patients.
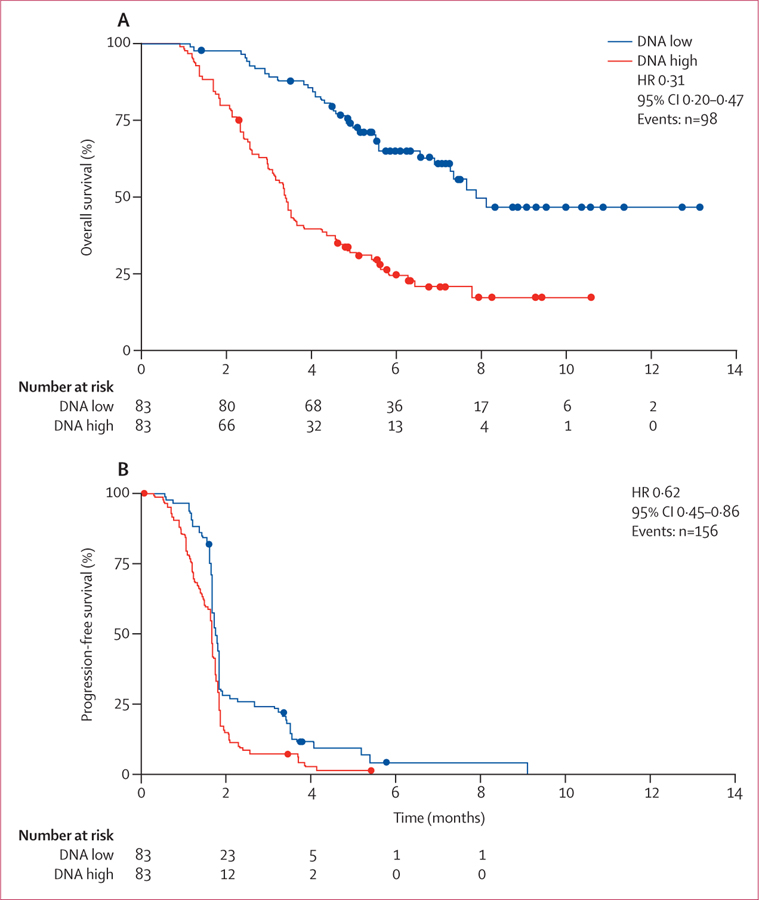
HR=hazard ratio.
We measured the concentrations of 15 proteins associated with angiogenesis or colorectal cancer pathogenesis in baseline plasma samples from 611 patients in CORRECT. Univariable analysis identified soluble TIE-1 as the only protein for which the HR for regorafenib versus placebo differed between subgroups (appendix pp 12–15); a high concentration of circulating TIE-1 was associated with improved overall survival, but not progression-free survival, compared with a low concentration. This association was not significant in multivariable analyses (data not shown). We identified a significant association between TIE-1 concentration and the overall survival benefit of regorafenib in our best-fit and ROC curve analyses (both pinteraction=0.035), but not when we used the median cutoff method (pinteraction=0.248; appendix pp 12–15).
Our assessment of the potential prognostic effect of the plasma concentration of these proteins in placebo patients identified an association between protein concentration and clinical outcome for interleukin 8 and PlGF, both of which retained significance in multivariable analysis for overall survival after controlling for various clinical factors (appendix pp 16–17). For both of these proteins, higher plasma concentrations were associated with poorer clinical outcomes compared with those seen in patients with lower concentrations (appendix pp 16–17). The effect of plasma concentration on progression-free survival was significant for interleukin 8, but not for PlGF (appendix pp 16–17). We also identified a prognostic effect for overall survival in a univariable analysis, but not in a multivariable analysis, for angiopoietin 2, soluble TIE-1, interleukin 6, and macrophage colony-stimulating factor (appendix pp 16–17).
Discussion
Our analysis shows that regorafenib provides a consistent overall and progression-free survival benefit in a range of subgroups of patients with metastatic colorectal cancer based on tumour mutation status and plasma protein biomarker concentrations. Furthermore, our results support the use of circulating tumour DNA to establish tumour genotype at the time of treatment.
To our knowledge, this is the largest study published so far to address the genetic evolution of metastatic colorectal cancer by integrating genomic data from tumour specimens obtained at diagnosis and plasma specimens collected after patients had received all available cytotoxic and targeted agents for the treatment of metastatic colorectal cancer. The samples we used for our analyses were obtained during a large, international, randomised, placebo-controlled phase 3 trial of a cancer type for which the importance of tumour genotyping is well established in clinical decision making. Another strength of the study is the fact that all analyses were done with the same technology (BEAMing), thus providing an opportunity to study clonal variation of oncogenes in response to treatment. Tumour genotyping using circulating plasma-derived DNA offers potential advantages over archival tumour DNA testing, including convenience, non-invasive sample collection, and more accurate representation of a tumour’s current mutational status. Such a method of monitoring tumour genotype could be useful to establish a patient’s suitability for a particular treatment. For example, the emergence of RAS mutations during treatment could provide the rationale for a switch to an alternative drug that has been shown to be active against colorectal cancer tumours with such gene alterations.
The robustness of our genotyping results is supported by the 100% concordance between the KRAS mutation status detected by BEAMing and the results from next-generation sequencing of tumour tissue from 24 patients (appendix p 3 and p 8), and the consistency of individual KRAS and PIK3CA mutation frequencies we obtained with those reported in COSMIC. Additionally, KRAS or PIK3CA mutations identified in patient-matched plasma and tumour tissue DNA were nearly always identical.
The genes assessed in our study are the most frequently mutated proto-oncogenes in colorectal cancer.20–22,36–38 We did not assess NRAS mutations because of the low frequency of this mutation in colorectal cancer (4% in COSMIC),35 which was confirmed by next-generation sequencing of a subset of archival tumour tissue specimens from patients in CORRECT (2% NRAS mutational frequency).13 The frequency of KRAS mutant tumours in CORRECT (59% in archival tumour tissue and 69% in plasma DNA) was higher than has previously been reported in colorectal cancer (36% in COSMIC).35 A possible explanation for this high mutation frequency is that patients with KRAS-wild-type tumours are eligible to receive additional lines of treatment, whereas patients with KRAS mutant tumours have fewer therapeutic options. By contrast, patients with BRAF mutations might have been under-represented because of their poor prognoses39 (3·4% in CORRECT vs 10·7% in COSMIC).35 Furthermore, 220 (29%) of 760 patients in CORRECT had rectal tumours,13 which have a low frequency of BRAF mutations.40 The frequency of PIK3CA mutations in CORRECT (17% via plasma BEAMing) was consistent with that usually reported in colorectal cancer (16% in COSMIC).35
BEAMing analysis of plasma and tumour DNA from patients in the CORRECT trial revealed consistent trends toward a clinical benefit with regorafenib defined by KRAS and PIK3CA mutational status. These findings support and extend previous findings,13,14 with results from both trials showing evidence of a trend for regorafenib benefit irrespective of historical KRAS mutational status; this is consistent with the antiangiogenic mechanism of action of regorafenib.12,41,42
In our study, the concordance of KRAS and PIK3CA mutations detected by BEAMing analysis in patient-matched plasma and tumour tissue samples was not 100%. This finding is not unexpected in view of the fact that the plasma samples were obtained at trial entry, whereas tumour tissue specimens were archival and often several years old. The notion of tumour evolution is consistent with the identification of a subgroup of patients for whom archival tumour tissue was KRAS wild-type, but plasma DNA was KRAS mutant. Acquisition of KRAS mutations as a mechanism of resistance to anti-EGFR-antibody therapy has been previously reported in patients with metastatic colorectal cancer;10,11 this hypothesis is further supported by the fact that all patients with apparent acquired KRAS mutations had received anti-EGFR-antibody treatment before enrolment in CORRECT.
We identified a use for circulating DNA beyond tumour genotyping: baseline circulating DNA concentrations were prognostic for clinical outcome, with shorter median survival observed in patients with a higher plasma DNA concentration than in patients with a lower plasma DNA concentration. We detected this association in both the placebo and regorafenib treatment groups. Circulating DNA concentrations have been reported to be higher in patients with colorectal cancer than in healthy people,43,44 which supports a working hypothesis whereby the amount of tumour-derived DNA in the circulation represents the level of metastatic burden and associated effect on clinical outcome. However, the process whereby metastatic lesions liberate DNA into the circulation is poorly understood, and the effect of factors such as tumour location is unknown.
Measurement of plasma protein concentrations showed that regorafenib had a greater effect on overall survival in patients who had a high concentration of TIE-1 than in patients with a low concentration. This finding is not unexpected because TIE-1 can interact with the TIE-2 angiopoietin receptor, and deletion of TIE-1 in mice decreases tumour-associated endothelial-cell survival and inhibits tumour growth.24 Moreover, TIE-1 is expressed by some tumours, including colorectal cancer,25 and findings from a previous trial suggested that a high concentration of TIE-1 was associated with superior regorafenib response.45 Regorafenib has some inhibitory activity on both TIE-146 and TIE-212 kinases, and we suggest that tumours dependent on these receptors will be especially responsive to regorafenib. However, this hypothesis remains to be investigated, and whether a high concentration of circulating TIE-1 can identify such tumours is unknown. Notably, a benefit was not seen for progression-free survival, and the effect on overall survival was not significant in multivariable analysis. Furthermore, although regorafenib had particularly robust clinical activity in patients with high circulating TIE-1 concentrations, a beneficial trend was also evident in patients with low circulating TIE-1 concentrations (appendix p 12).
High concentrations of interleukin 8 and PlGF were prognostic for poor clinical outcome in patients who received placebo. Interleukin 8 is a chemokine that might function in the growth and progression of colorectal cancer,27 and PlGF is a member of the VEGF family of ligands.28,47 Consistent with the results of our study, high concentrations of these proteins have previously been reported to be associated with poor outcome in patients with colorectal cancer.27,48
In summary, regorafenib seems to be active in various subgroups of patients with metastatic colorectal cancer defined by genotypic and protein biomarkers, thus supporting and extending previous findings from the CORRECT trial.13 However, since the CORRECT trial was not specifically powered to assess the clinical activity of regorafenib in biomarker-defined subgroups, and because we did this analysis retrospectively, the results from these analyses should be considered exploratory and hypothesis generating. The findings from this biomarker study also support BEAMing analysis of circulating DNA as a viable approach to obtain real-time tumour-associated genotypic information in a non-invasive way.
Acknowledgments
The CORRECT trial was sponsored by Bayer HealthCare AG. We thank the patients, their families, and the study investigators. We also thank Lei Xu and Frank Cihon of Bayer HealthCare for data analysis support; Robert Gerwien for data analysis support; Scott Wilhelm of Bayer HealthCare and Chetan Lathia for input on the biomarker study design; Bernard Kerr of Succinct Medical Communications for editorial assistance in the preparation of this manuscript, with financial support from Bayer HealthCare AG. The authors retained editorial control over the content.
Funding Bayer HealthCare Pharmaceuticals.
Declaration of interests
AA has received payment for consultancy work and received payment for lectures from Bayer, and his institution has received grants from Bayer. AF has received payment for consultancy work for Bayer, Amgen, Roche, Merck Serono, Eli-Lilly and Sanofi. AG has had grants, consulting fees and support for travel to meetings paid to his institution by Bayer, has had money paid to his institution for consultancy work for Amgen, Genentech, BMS, and Eli-Lilly, and his institution has received grants from Genentech, Eli-Lilly, Pfizer, Boston Biomedicals, Eisai, and Sanofi. AS has received payment for consultancy work for Bayer, Amgen, Celgene, Merck, Roche, and Sanofi, and his institution has received payment for lectures from Amgen, Bayer, Celgene, Merck, Roche, and Sanofi. CB has received payment for consultancy work for Bayer, Merck, and Roche, has received grants from Novartis and Pfizer, and has received payment for lectures from Bayer, Merck, Roche, and Celgene. DJS has received payment for consultancy work and received support for travel from Bayer. EVC has received payment for consultancy work for Bayer and has received grants from Bayer. H-JL has received payment for consultancy work, received grants, and received support for travel to meetings from Bayer, Roche, and Merck Serono. JT has received fees from Bayer for participation in review activities, and has received payment for consultancy work for Amgen, Imclone, Eli-Lilly, Merck Serono, Millennium, Novartis, Roche, Sanofi, Celgene, Chugai, Taiho, Bayer, and Merck. LM has received payment for consultancy work and received payment for development of educational presentations from Bayer. MY has received payment for consultancy work for Bayer, and his institution has received grants from Bayer. OB has received payment for consultancy work for Roche and Merck Serono, and has received payment for educational presentations from Bayer. RMG has received payment from Bayer relating to other clinical studies of regorafenib, has received payment for consultancy work for Biothera, Kanghong Pharma, Eli-Lilly, Taiho, Baxter, and Novo Nordisk, has received payment for lectures from Sanofi, and his institution has received money from Bayer, Sanofi, Medimmune, and Merck. SS has received payment for consultancy work for Bayer, Amgen, Roche, Ignyta, and Sanofi, and holds stocks in Ignyta. TY and his institution have received grants from Bayer, Daiichi-Sankyo, Taiho, Eli Lilly, Pfizer, Yakult, Honsha, Chugai, and Dainippon Sumitomo, and has received payment for lectures from Merck Serono, Chugai, and Takeda. YH has had money paid to his institution by Bayer for data collection in the CORRECT study and by Merck Serono for consultancy work. AW, DL, MJ, and MT are employees of Bayer. DL holds stock in Bayer.
Footnotes
See Online for appendix
For the Catalogue of Somatic Mutations in Cancer database see http://www.cancer.sanger.ac.uk/cosmic
Contributor Information
Josep Tabernero, Vall d’Hebron University Hospital and Institute of Oncology, Universitat Autònoma de Barcelona, Barcelona, Spain.
Heinz-Josef Lenz, University of Southern California/Norris Comprehensive Cancer Center, Keck School of Medicine, Los Angeles, CA, USA.
Salvatore Siena, Niguarda Cancer Center, Ospedale Niguarda Ca’ Granda, Milan, Italy. Università di Milano, Milan, Italy.
Alberto Sobrero, IRCCS San Martino IST, Genoa, Italy.
Alfredo Falcone, University Hospital, Pisa, Italy.
Marc Ychou, ICM–Val d’Aurelle, Montpellier, France.
Yves Humblet, St-Luc University Hospital, Université Catholique de Louvain, Brussels, Belgium.
Olivier Bouché, Centre Hospitalier Universitaire Reims, Robert Debré Hospital, Reims, France.
Laurent Mineur, Gastrointestinal and Liver Oncology Unit, Institut Sainte Catherine, Avignon, France.
Carlo Barone, Catholic University of Sacred Heart, Rome, Italy.
Antoine Adenis, Centre Oscar Lambret, Lille, France.
Takayuki Yoshino, National Cancer Center Hospital East, Kashiwa, Japan.
Richard M Goldberg, Ohio State University School of Medicine, James Cancer Hospital and Solove Research Institute, Columbus, OH, USA.
Daniel J Sargent, Mayo Clinic, Rochester, MN, USA.
Andrea Wagner, Bayer Pharma AG, Berlin, Germany.
Dirk Laurent, Bayer Pharma AG, Berlin, Germany.
Axel Grothey, Mayo Clinic, Rochester, MN, USA.
Eric Van Cutsem, University Hospital Gasthuisberg/Leuven and KU Leuven, Leuven, Belgium.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/s1470-2045(15)00138-2
Read article for free, from open access legal sources, via Unpaywall:
https://air.unimi.it/bitstream/2434/338910/2/2015%20Tabernero%20J%20et%20al%2c%20Lancet%20Oncol%20.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
TRIM21 induces selective autophagic degradation of c-Myc and sensitizes regorafenib therapy in colorectal cancer.
Proc Natl Acad Sci U S A, 121(42):e2406936121, 10 Oct 2024
Cited by: 0 articles | PMID: 39388269
SHR-8068 combined with adebrelimab and bevacizumab in the treatment of refractory advanced colorectal cancer: study protocol for a single-arm, phase Ib/II study.
Front Immunol, 15:1450533, 09 Oct 2024
Cited by: 0 articles | PMID: 39445023 | PMCID: PMC11496094
Exploratory Study Identifies Matrix Metalloproteinase-14 and -9 as Potential Biomarkers of Regorafenib Efficacy in Metastatic Colorectal Cancer.
Cancers (Basel), 16(16):2855, 15 Aug 2024
Cited by: 0 articles | PMID: 39199626 | PMCID: PMC11352555
Stage-Specific Plasma Metabolomic Profiles in Colorectal Cancer.
J Clin Med, 13(17):5202, 02 Sep 2024
Cited by: 0 articles | PMID: 39274416 | PMCID: PMC11396754
Regorafenib plus FOLFIRINOX as first-line treatment for patients with RAS-mutant metastatic colorectal cancer (FOLFIRINOX-R trial): a dose-escalation study.
Cancer Chemother Pharmacol, 94(3):443-452, 10 Jul 2024
Cited by: 0 articles | PMID: 38987363 | PMCID: PMC11420384
Go to all (184) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Clinical Trials
- (2 citations) ClinicalTrials.gov - NCT01103323
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis.
Lancet Oncol, 11(8):753-762, 08 Jul 2010
Cited by: 1281 articles | PMID: 20619739
The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis.
Acta Oncol, 53(7):852-864, 25 Mar 2014
Cited by: 220 articles | PMID: 24666267
Review
Biomarkers of benefit from cetuximab-based therapy in metastatic colorectal cancer: interaction of EGFR ligand expression with RAS/RAF, PIK3CA genotypes.
BMC Cancer, 13:49, 02 Feb 2013
Cited by: 70 articles | PMID: 23374602 | PMCID: PMC3599697
Promising biomarkers for predicting the outcomes of patients with KRAS wild-type metastatic colorectal cancer treated with anti-epidermal growth factor receptor monoclonal antibodies: a systematic review with meta-analysis.
Int J Cancer, 133(8):1914-1925, 13 Jul 2013
Cited by: 49 articles | PMID: 23494461
Review
Funding
Funders who supported this work.
Bayer HealthCare
NCI NIH HHS (1)
Grant ID: P30 CA014089