Abstract
Free full text

Predictors of COVID-19 severity: a systematic review and meta-analysis
Associated Data
Version Changes
Revised. Amendments from Version 1
In the revised version of our current article, we provided the revision of method, the limitation, and the clinical implication of our study.
Peer Review Summary
| Review date | Reviewer name(s) | Version reviewed | Review status |
|---|---|---|---|
| 2021 Jan 19 | Morteza Arab-Zozani | Version 2 | Approved |
| 2020 Nov 2 | Annelies Wilder-Smith | Version 1Version 1 | Approved |
| 2020 Sep 21 | Morteza Arab-Zozani | Version 1Version 1 | Approved |
Abstract
Background: The unpredictability of the progression of coronavirus disease 2019 (COVID-19) may be attributed to the low precision of the tools used to predict the prognosis of this disease.
Objective: To identify the predictors associated with poor clinical outcomes in patients with COVID-19.
Methods: Relevant articles from PubMed, Embase, Cochrane, and Web of Science were searched as of April 5, 2020. The quality of the included papers was appraised using the Newcastle-Ottawa scale (NOS). Data of interest were collected and evaluated for their compatibility for the meta-analysis. Cumulative calculations to determine the correlation and effect estimates were performed using the Z test.
Results: In total, 19 papers recording 1,934 mild and 1,644 severe cases of COVID-19 were included. Based on the initial evaluation, 62 potential risk factors were identified for the meta-analysis. Several comorbidities, including chronic respiratory disease, cardiovascular disease, diabetes mellitus, and hypertension were observed more frequent among patients with severe COVID-19 than with the mild ones. Compared to the mild form, severe COVID-19 was associated with symptoms such as dyspnea, anorexia, fatigue, increased respiratory rate, and high systolic blood pressure. Lower levels of lymphocytes and hemoglobin; elevated levels of leukocytes, aspartate aminotransferase, alanine aminotransferase, blood creatinine, blood urea nitrogen, high-sensitivity troponin, creatine kinase, high-sensitivity C-reactive protein, interleukin 6, D-dimer, ferritin, lactate dehydrogenase, and procalcitonin; and a high erythrocyte sedimentation rate were also associated with severe COVID-19.
Conclusion: More than 30 risk factors are associated with a higher risk of severe COVID-19. These may serve as useful baseline parameters in the development of prediction tools for COVID-19 prognosis.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a global crisis across health, economic, and educational dimensions 1, 2. The disease has spread rapidly, can cause severe illness, and is characterized by a high mortality rate in certain groups. Mortality is particularly high in the absence of proven effective standard management measures 3. One of the problems with the management of this disease is the absence of standardized methods for diagnosis and the inability to estimate prognosis based on clinical features. Certain reports have shown that poor prognostic prediction has correlated with high mortality among patients with COVID-19 4, 5. Among patients with similar clinical characteristics and with similar treatment regiments, there may be a diversity in clinical outcomes 6. Therefore, the development and use of an accurate predictor for COVID-19 prognosis will be beneficial for the clinical management of patients with COVID-19, and will help reduce the mortality rate. Successful implementation of such a prediction mechanism could have a large public health impact. Better understanding of clinical progression could also improve public health messaging, particularly as many individuals may consider COVID-19 to not be severe.
Prognostic tools for the prediction of COVID-19 severity in patients have been in development since January 2020. At least nine studies proposed the use of prognostic tools for the prediction of COVID-19 severity 7– 15. However, a recent systematic review and critical appraisal study evaluated the accuracy of these tools using prediction model risk of bias assessment tool (PROBAST) and reported a high risk of bias 16. The establishment of a prediction model for the estimation of disease prognosis may help health workers segregate patients according to prediction status. However, the high risk of bias in these prediction tools might lead to inaccurate prediction of COVID-19 severity. A comprehensive study of the identification of risk factors that might play a significant role in determining the severity of patients with COVID-19 is necessary. We performed a systematic review and meta-analysis to assess the risk factors associated with poor clinical outcomes among patients with COVID-19. To the best of our knowledge, this is the first meta-analysis to assess the comprehensive risk factors that might affect the severity of COVID-19 in patients. The results of our study might serve as preliminary data for the compilation or improvement of the scoring system in the prediction of COVID-19 severity.
Methods
Study design
We performed a systematic review and meta-analysis to evaluate potential risk factors that might influence the severity of COVID-19. These risk factors include comorbidities, clinical manifestations, and laboratory findings. Accordingly, we searched the relevant studies from major scientific websites and databases to collect the data of interest, and determined the association and effect estimates by calculating the combined odds ratio (OR) and 95% confidence intervals (95% CI). The protocols for the systematic review and meta-analysis were similar to those used in previous studies 17– 23, as well as to those recommended by Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) 24.
Eligibility criteria
Studies were included in this review if they met the following inclusion criteria: (1) assessed the clinical manifestations and laboratory findings of patients with mild to severe COVID-19; (2) provided adequate data for the calculation of OR and 95% CI (event per sample size or mean ± SD in both case and control groups), (3) written inEnglish. Review articles, articles with non-standard data presentation, and duplicate publications were excluded.
Search strategy and data extraction
Major scientific databases (PubMed, Embase, Cochrane, and Web of Science) were searched for articles as of April 5, 2020. Moreover, we also searched in google scholar for the additional database. A comprehensive initial search was performed to identify the potential predictors, and a final search was performed to identify the relevant papers that could be included in the meta-analysis. We used the keywords adapted from medical subject headings: ["COVID-19" or "Coronavirus disease-19" or "SARS-CoV-2"] and ["mild" or "severe" or "prognosis" or "clinical outcome"] and ["clinical manifestation" or "morbidity" or "laboratory findings"]. Only studies written in English were included. If a duplicate publication was found, the article with the larger sample size was included. We also searched for relevant studies from the reference lists in the articles. During data extraction, the following information of interest was extracted: (1) first author name; (2) publication year; (3) country of origin, (4) sample size of mild and severe cases, (5) clinical manifestations, (6) morbidities, and (7) laboratory findings. Data extraction was performed by two independent investigators (JKF and MI) using a pilot form. If the disagreement was found, we performed a discussion to resolve the disagreement.
Assessment of the methodological quality
Before inclusion in the meta-analysis, the methodological quality of the articles was assessed using the New Castle-Ottawa scale (NOS). NOS scores range from 0 to 9 and consider three items: selection of patients (4 points), comparability of the groups (2 points), and ascertainment of exposure (3 points). Each study was interpreted to be of low quality (for scores ≤ 4), moderate quality (for scores between 5–6), or high quality (for scores ≥ 7) 25. Articles with moderate to high quality were included in the analysis. The study assessment was conducted by two independent investigators (MI and YP) using a pilot form. The discrepancies between the findings of the two investigators were solved by consulting with another investigator (JKF).
Study measures
The outcome measure of the study was the severity of COVID-19 (mild vs. severe). The risk factors or predictors included three major groups: comorbidities, clinical manifestations, and laboratory parameters. Comorbid factors such as chronic kidney disease, chronic liver disease, chronic respiratory disease, cerebrovascular accident, cardiovascular disease, diabetes mellitus, hypertension, and malignancy were compatible with the analysis. For clinical manifestations, fever, cough, dry cough, expectoration, sore throat, dyspnea, diarrhea, myalgia, nasal congestion, anorexia, abdominal pain, fatigue, dizziness, headache, fever, heart rate, respiratory rate, systolic blood pressure, and diastolic blood pressure were included in this study. Among laboratory characteristics, the presence of leukocytosis, leukocytopenia, anemia, lymphocytopenia; the levels or the counts of white blood cell (WBC), hemoglobin, neutrophil, lymphocyte, monocyte, platelet, activated partial thromboplastin time (aPTT), partial thromboplastin time (PTT), aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, albumin, serum creatinine, blood urea nitrogen (BUN), high-sensitivity (Hs)-troponin I, creatine kinase, high-sensitivity C-reactive protein (Hs-CRP), C-reactive protein (CRP) >8 mg/L, interleukin 6 (IL-6), glucose, D-dimer, serum ferritin, sodium, potassium, lactate dehydrogenase, and procalcitonin, CD4 and CD8; erythrocyte sedimentation rate (ESR); elevated IL-16; and elevated ESR were all included.
Statistical analysis
The significant risk factors that might govern the severity of COVID-19 were determined by the calculation of a pooled OR and 95% CI. The significance of the pooled ORs was determined using the Z test (p<0.05 was considered statistically significant). Prior to identification of the significant risk factors, data were evaluated for heterogeneity and potential publication bias. The heterogeneity among included studies was evaluated using the Q test. If heterogeneity existed (p<0.10), a random effect model was adopted; if not, a fixed effect model was adopted. Egger’s test and a funnel plot were used to assess the reporting or publication bias (p<0.05 was considered statistically significant). Furthermore, we performed a moderator analysis to identify the independent predictors of poor clinical outcomes among patients with COVID-19. The data were analyzed using Review Manager version 5.3 (Revman Cochrane, London, UK). To prevent analytical errors, statistical analysis was performed by two authors (JKF and MI). The cumulative calculation was presented in a forest plot.
Results
Eligible studies
Our searches yielded 6,209 potentially relevant studies, of which 6,170 studies were excluded after assessment of the titles and abstracts. Subsequently, further review of the complete texts was performed for 39 potential studies. In the full text review, we excluded 20 studies because they were reviews articles (n = 9), inadequacy of data for the calculation of OR and 95% CI (n = 7), and poor quality (n = 4). Eventually, 19 papers were included in our meta-analysis 26– 42 The paper selection process adopted in our study is summarized in Figure 1, and the characteristics of studies included in our analysis are outlined in Table 1.
Figure 1.
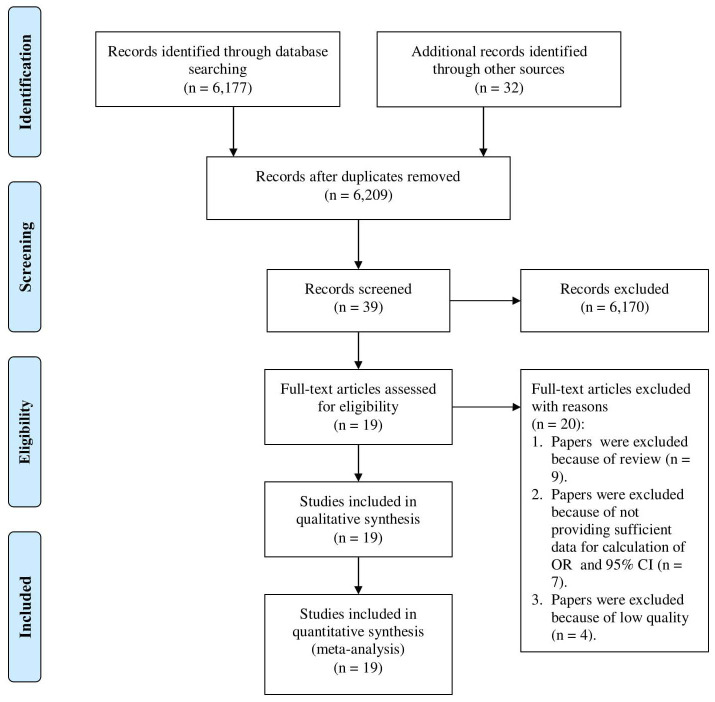
Table 1.
| Author & year | Country | City | Hospital | Sample size | Outcome measure | NOS | |
|---|---|---|---|---|---|---|---|
| Severe | Mild | ||||||
| Bai et al. 2020 26 | China | Wuhan | Jinyintan Hospital | 91 | 36 | Died vs. cured | 7 |
| Cai et al. 2020 27 | China | Shenzen | Third people's Hospital | 58 | 240 | Severe vs. non severe | 9 |
| Chen et al. 2020 28 | China | Wuhan | Tongji hospital | 11 | 10 | Severe vs. moderate | 9 |
| Chen et al. 2020 29 | China | Mixed | Multicenter | 50 | 241 | Severe vs. mild-moderate | 9 |
| Chen et al. 2020 30 | China | Wuhan | Zhongnan Hospital | 14 | 11 | Viral clearance vs. without
viral clearance | 9 |
| Duan et al. 2020 31 | China | Wuhan | Wuhan Pulmonary Hospital | 44 | 72 | Uncured vs. cured | 9 |
| Gao et al. 2020 32 | China | Fuyang | Second People's Hospital | 15 | 28 | Severe vs. mild | 7 |
| Guan et al. 2020 33 | China | Guangdong | National Health Commision
of China | 926 | 173 | Severe vs. non-severe | 7 |
| Huang et al. 2020 34 | China | Wuhan | Jinyintan hospital | 13 | 28 | ICU vs. non-ICU | 9 |
| Jian-Ya et al. 2020 35 | China | Chongqing | Three Gorges Hospital | 7 | 44 | Severe vs. non severe | 9 |
| Liu et al. 2020 36 | China | Wuhan | Union Hospital | 69 | 69 | Severe vs. non severe | 7 |
| Shi et al. 2020 37 | China | Wuhan | Renmin Hospital | 48 | 53 | Died <3 d vs. >3 d | 9 |
| Wang et al. 2020 38 | China | Mixed | Multicenter | 50 | 115 | CT imaging score >11 vs. <11 | 8 |
| Wang et al. 2020 39 | China | Wuhan | Wuhan First People's
Hospital | 22 | 283 | Survivor vs. non-survivor | 8 |
| Wang et al. 2020 43 | China | Wuhan | Zhongnan Hospital | 36 | 102 | ICU vs. non-ICU | 9 |
| Xu et al. 2020 40 | China | Mixed | Multicenter | 25 | 44 | Severe vs. mild | 8 |
| Zhang et al. 2020 41 | China | Wuhan | Zhongnan Hospital | 55 | 166 | Severe vs. non-severe | 9 |
| Zhang et al. 2020 44 | China | Wuhan | Wuhan Seventh Hospital | 56 | 82 | Severe vs. non-severe | 7 |
| Zhou et al. 2020 42 | China | Wuhan | Wuhan Pulmonary Hospital | 54 | 137 | Survivor vs. non-survivor | 8 |
Note: ICU, intensive care unit; CT, computed tomography; NOS, Newcastle Ottawa Scale.
Risk factors of severe COVID-19
We found that eight comorbidities, 19 clinical manifestations, and 35 laboratory parameters were available for the meta-analysis ( Table 2 and Table 3). Among the comorbid factors, chronic respiratory disease (OR: 2.48; 95% CI: 1.44, 4.27), cardiovascular disease (OR: 1.70; 95% CI: 1.05, 2.78), diabetes mellitus (OR: 2.10; 95% CI: 1.33, 3.34), and hypertension (OR: 2.33; 95% CI: 1.42, 3.81) were associated with a greater risk of severe COVID-19 ( Figure 2A–D).
Table 2.
| Clinical characteristics | NS | Model | Value | pE | pHet | p | OR | 95%CI | |
|---|---|---|---|---|---|---|---|---|---|
| Severe | Mild | ||||||||
| Comorbids | |||||||||
| Chronic kidney disease | 6 | Random | 14 [3.94] | 15 [1.68] | 1.3430 | 0.0280 | 0.1910 | 2.56 | 0.63-10.45 |
| Chronic liver disease | 6 | Fixed | 16 [4.82] | 26 [4.04] | <0.0001 | 0.3220 | 0.3220 | 1.45 | 0.70-3.01 |
| Chronic respiratory disease | 10 | Fixed | 31 [5.47] | 31 [1.66] | 0.7060 | 0.1020 | 0.0010 | 2.48 | 1.44-4.27 |
| Cerebrovascular accident | 5 | Random | 20 [5.54] | 30 [2.09] | 0.9110 | 0.0380 | 0.1850 | 2.02 | 0.71-5.70 |
| Cardiovascular disease | 13 | Random | 76 [10.45] | 94 [4.95] | 0.5400 | 0.0580 | 0.0310 | 1.70 | 1.05-2.78 |
| Diabetes mellitus | 17 | Random | 156 [19.24] | 194 [8.40] | 0.7040 | <0.0001 | 0.0020 | 2.10 | 1.33-3.34 |
| Hypertension | 15 | Random | 269 [35.54] | 369 [16.79] | 0.7680 | <0.0001 | 0.0010 | 2.33 | 1.42-3.81 |
| Malignancy | 11 | Fixed | 29 [4.43] | 40 [2.23] | 0.6150 | 0.1430 | 0.5330 | 1.18 | 0.70-1.99 |
| Symptoms | |||||||||
| Fever | 16 | Random | 599 [79.34] | 1932 [80.84] | 0.9220 | <0.0001 | 0.1730 | 1.51 | 0.83-2.74 |
| Cough | 12 | Random | 377 [64.33] | 1120 [ 54.05] | 0.9560 | <0.0001 | 0.1890 | 1.53 | 0.81-2.90 |
| Dry cough | 4 | Fixed | 75 [44.38] | 178 [55.97] | 0.3130 | 0.1880 | 0.0360 | 0.66 | 0.44-0.97 |
| Expectoration | 10 | Fixed | 136 [26.67] | 438 [29.05] | <0.0001 | 0.8370 | 0.4970 | 1.09 | 0.85-1.39 |
| Sore throat | 10 | Random | 59 [10.57] | 196 [10.96] | 0.7860 | 0.0040 | 0.6350 | 1.18 | 0.59-2.37 |
| Dyspnea | 13 | Random | 286 [42.56] | 318 [16.51] | 0.6340 | <0.0001 | <0.0001 | 3.28 | 2.09-5.15 |
| Diarrhea | 13 | Random | 65 [9.62] | 134 [6.68] | 0.5180 | 0.0690 | 0.8030 | 1.07 | 0.67-1.69 |
| Myalgia | 11 | Fixed | 105 [17.89] | 283 [15.70] | <0.0001 | 0.7330 | 0.5160 | 1.10 | 0.831-1.44 |
| Nasal congestion | 4 | Fixed | 15 [5.02] | 53 [4.34] | 0.9350 | 0.1000 | 0.7590 | 1.12 | 0.55-2.29 |
| Anorexia | 9 | Random | 103 [25.37] | 143 [15.10] | 0.6960 | 0.0040 | 0.0490 | 1.83 | 1.00-3.34 |
| Abdominal pain | 5 | Fixed | 15 [6.07] | 6 [0.95] | <0.0001 | 0.5650 | 0.0040 | 3.91 | 1.53-10.02 |
| Fatigue | 13 | Random | 310 [46.48] | 694 [34.49] | 0.6790 | <0.0001 | 0.0040 | 2.00 | 1.25-3.20 |
| Dizziness | 4 | Fixed | 13 [10.08] | 24 [5.02] | 0.6510 | 0.1950 | 0.0180 | 2.67 | 1.18-6.01 |
| Headache | 11 | Fixed | 56 [10.45] | 197 [11.58] | 0.5070 | 0.1110 | 0.9950 | 1.00 | 0.71-1.41 |
| Signs | |||||||||
| Temperature >38°C | 5 | Random | 200 [57.97] | 738 [50.14] | 0.6090 | 0.0020 | 0.2660 | 1.44 | 0.76-2.73 |
| Heart rate (x/min) | 4 | Fixed | 269 ± 35.54 | 87.88 ± 13.30 | <0.0001 | 0.4070 | 0.0010 | 1.79 | 1.25-2.56 |
| Respiratory rate (x/min) | 5 | Random | 22.6 ± 4.80 | 20.36 ± 2.00 | 0.8080 | <0.0001 | 0.0100 | 2.85 | 1.28-6.33 |
| SBP (mmHg) | 5 | Fixed | 132.57 ± 23.16 | 123.88 ± 14.37 | 0.3340 | 0.1560 | <0.0001 | 1.84 | 1.31-2.60 |
| DBP (mmHg) | 3 | Random | 76.50 ± 10.61 | 75.59 ± 9.89 | 0.5350 | 0.0260 | 0.7190 | 1.14 | 0.56-2.32 |
Note, Value, data were presented in number [%] or mean ± SD; NS, number of studies; pE, p Egger; pHet, p heterogeneity; OR, odd ratio; CI, confidence interval; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Table 3.
| Clinical
characteristics | NS | Model | Value | pE | pHet | p | OR | 95%CI | |
|---|---|---|---|---|---|---|---|---|---|
| Severe | Mild | ||||||||
| Complete Blood Count | |||||||||
| WBC (10^9/L) | 14 | Random | 7.32 ± 3.84 | 5.17 ± 2.04 | 1.4980 | <0.0001 | <0.0001 | 4.92 | 2.12-11.31 |
| Leukocytosis | 6 | Fixed | 62 [26.00] | 40[6.03] | 0.0000 | 0.5940 | <0.0001 | 5.38 | 3.36-8.62 |
| Leukopenia | 6 | Fixed | 44 [18.00] | 206 [31.07] | 0.2890 | 0.2480 | 0.0160 | 0.59 | 0.41-0.87 |
| Neutrophil count
(10^9/L) | 12 | Random | 5.96 ± 3.62 | 3.84 ± 2.12 | 1.6380 | <0.0001 | 0.0010 | 5.45 | 2.04-14.54 |
| Lymphocyte count
(10^9/L) | 15 | Random | 0.74 ± 0.36 | 1.03 ± 0.44 | 0.6440 | <0.0001 | <0.0001 | 0.34 | 0.23-0.50 |
| Lymphocytopenia | 6 | Random | 158 [59.00] | 40 [6.03] | 0.8270 | <0.0001 | <0.0001 | 3.19 | 1.14-7.07 |
| Monocyte count
(10^9/L) | 6 | Random | 0.38 ± 0.17 | 0.36 ± 0.15 | 0.5860 | 0.0100 | 0.5100 | 1.22 | 0.68-2.20 |
| Hemoglobin (g/L) | 9 | Fixed | 129.11 ± 16.98 | 132.02 ± 17.50 | 0.0900 | 0.4000 | 0.0460 | 0.76 | 0.58-1.00 |
| Anaemia | 2 | Random | 18 [17.00] | 39 [10.32] | 0.7640 | 0.0660 | 0.4730 | 1.58 | 0.45-5.56 |
| Platelet count (10^9/L) | 12 | Random | 172.58 ± 69.19 | 183.21 ± 62.50 | 0.5550 | 0.0010 | 0.8200 | 0.82 | 0.55-1.23 |
| Physiological function | |||||||||
| AST (U/L) | 11 | Random | 56.20 ± 35.83 | 28.67 ± 11.18 | 0.6930 | <0.0001 | <0.0001 | 4.91 | 2.96-8.12 |
| ALT (U/L) | 12 | Random | 38.65 ± 22.90 | 25.60 ± 14.71 | 0.8060 | <0.0001 | <0.0001 | 3.23 | 1.90-5.52 |
| Total bilirubin (μmol/L) | 7 | Random | 15.80 ± 9.50 | 13.46 ± 4.62 | 1.6600 | <0.0001 | 0.5800 | 1.46 | 0.41-5.21 |
| Albumin (g/L) | 6 | Random | 32.39 ± 3.64 | 35.53 ± 3.71 | 2.3900 | <0.0001 | 0.0950 | 0.19 | 0.03-1.34 |
| aPTT (s) | 7 | Random | 31.23 ± 5.02 | 33.13 ± 3.66 | 1.1900 | <0.0001 | 0.3420 | 0.58 | 0.19-1.79 |
| PTT (s) | 11 | Random | 13.45 ± 1.86 | 12.53 ± 1.31 | 0.7700 | <0.0001 | 0.2430 | 0.56 | 0.21-1.48 |
| Serum creatinine
(μmol/L) | 13 | Random | 82.04 ± 31.69 | 70.25 ± 20.87 | 0.6670 | <0.0001 | 0.0010 | 2.14 | 1.37-3.33 |
| BUN (mmol/L) | 10 | Random | 6.71 ± 2.70 | 4.74 ± 1.38 | 1.0220 | <0.0001 | <0.0001 | 6.15 | 3.05-12.37 |
| Hs-Troponin I (pg/ml) | 6 | Random | 31.9 ± 61.55 | 3.55 ± 3.71 | 1.1290 | <0.0001 | <0.0001 | 9.25 | 3.51-24.37 |
| Creatine kinase (U/L) | 10 | Random | 121.13 ± 115.63 | 77.47 ± 56.26 | 0.4860 | 0.0030 | <0.0001 | 2.44 | 1.65-3.62 |
| Inflammation markers | |||||||||
| Hs-CRP (mg/L) | 10 | Random | 73.25 ± 49.97 | 29.96 ± 24.40 | 1.5600 | <0.0001 | <0.0001 | 14.27 | 5.13-39.71 |
| CRP >8 mg/L | 3 | Random | 147 [83.10] | 254 [52] | 1.1590 | 0.0050 | 0.0060 | 8.34 | 1.85-37.62 |
| ESR (mm/h) | 4 | Random | 50.60 ± 27.25 | 29.19 ± 26.52 | 0.4200 | 0.0710 | <0.0001 | 4.45 | 2.56-7.76 |
| Elevated ESR | 2 | Fixed | 73 [68.00] | 214 [44.49] | <0.0001 | 0.8060 | <0.0001 | 2.80 | 1.78-4.39 |
| IL-6 (pg/ml) | 8 | Random | 30.45 ± 31.29 | 11.06 ± 10.89 | 0.9120 | <0.0001 | <0.0001 | 6.68 | 3.20-13.94 |
| Elevated IL-6 | 2 | Fixed | 44 [66] | 115 [46.56] | <0.0001 | 0.7160 | 0.0200 | 1.98 | 1.12-3.52 |
| CD4 count(10^9/L) | 3 | Random | 217.19 ± 118.56 | 337.87 ± 149.93 | 1.5920 | 0.0010 | 0.2760 | 0.34 | 0.05-2.39 |
| CD8 count (10^9/L) | 3 | Random | 178.80 ± 95.77 | 224.17 ± 76.36 | 1.4260 | 0.0030 | 0.1420 | 0.26 | 0.04-1.57 |
| Others | |||||||||
| Glucose (mmol/L) | 3 | Random | 7.04 ± 1.83 | 6.45 ± 1.33 | 0.9480 | 0.0030 | 0.3340 | 1.80 | 0.55-5.90 |
| D-dimer (pg/mL) | 15 | Random | 111.34 ± 145.12 | 38.88 ± 28.93 | 0.6070 | <0.0001 | <0.0001 | 6.19 | 4.22 - 9.08 |
| Serum Ferritin (μg/L) | 4 | Fixed | 1062.90 ± 868.19 | 600.67 ± 758.61 | 0.4310 | 0.1070 | 0.0310 | 1.96 | 1.06-3.62 |
| Sodium (mmol/L) | 3 | Random | 137.40 ± 3.13 | 92.39 ± 1.77 | 3.2770 | <0.0001 | 0.2840 | 11.93 | 0.13-1109.37 |
| Potassium (mmol/L) | 3 | Random | 4.12 ± 0.61 | 4.00 ± 0.54 | 0.9630 | 0.0010 | 0.7470 | 1.21 | 0.32-0.75 |
| Lactate dehydrogenase
(U/L) | 9 | Random | 381.85 ± 159.44 | 283. 03 ± 89.40 | 0.6840 | <0.0001 | <0.0001 | 8.28 | 4.75-14.46 |
| Procalcitonin (ng/mL) | 10 | Random | 0.40 ± 0.29 | 0.12 ± 0.07 | 0.9880 | <0.0001 | <0.0001 | 6.62 | 3.32-13.21 |
Note: Value, data were presented in number [%] or mean ± SD; NS, number of studies; pE, p Egger; pHet, p heterogeneity; OR, odd ratio; CI, confidence interval; CBC, complete blood count; WBC, white blood cells; AST, aspartate transaminase; ALT, alanine transaminase; aPTT, activated partial thromboplastin time; PTT, partial thromboplastin time; BUN, blood urea nitrogen; Hs-CRP, high sensitivity C reactive protein; ESR, erythrocyte sedimentation rate; IL, interleukin.
Figure 2.
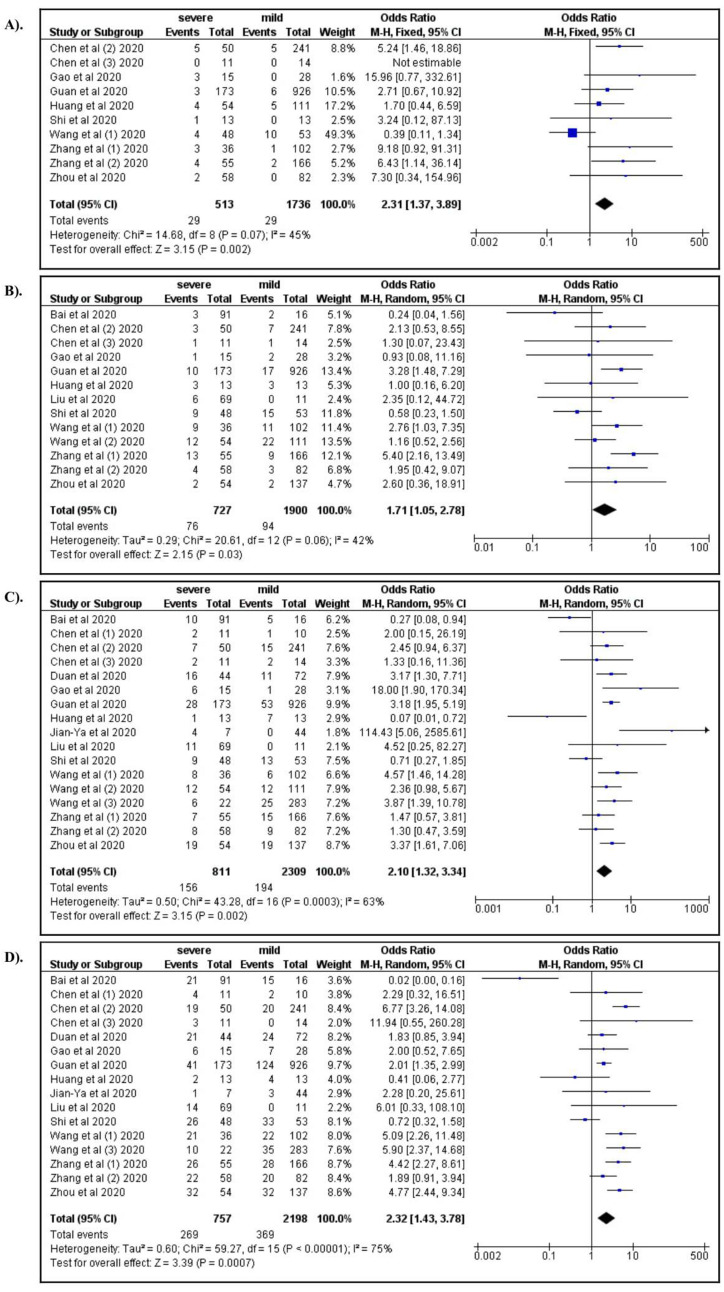
A) Chronic respiratory disease; B) Cardiovascular diease; C) Diabetes mellitus; D) Hypertension.
Among the clinical manifestations, dyspnea (OR: 3.28; 95% CI: 2.09, 5.15), anorexia (OR: 1.83; 95% CI: 1.00, 3.34), fatigue (OR: 2.00; 95% CI: 1.25, 3.20), and dizziness (OR: 2.67; 95% CI: 1.18, 6.01) were associated with severe COVID-19 ( Figure 3A–D). In addition, increased respiratory rate (OR: 2.85; 95% CI: 1.28, 6.33) and increased systolic blood pressure (OR: 1.84; 95% CI: 1.31, 2.60) were also associated with severe COVID-19 ( Figure 4A and B). Compared to productive cough, dry cough was associated with a lower risk of severe COVID-19 (OR: 0.66; 95% CI: 0.44, 0.97).
Figure 3.
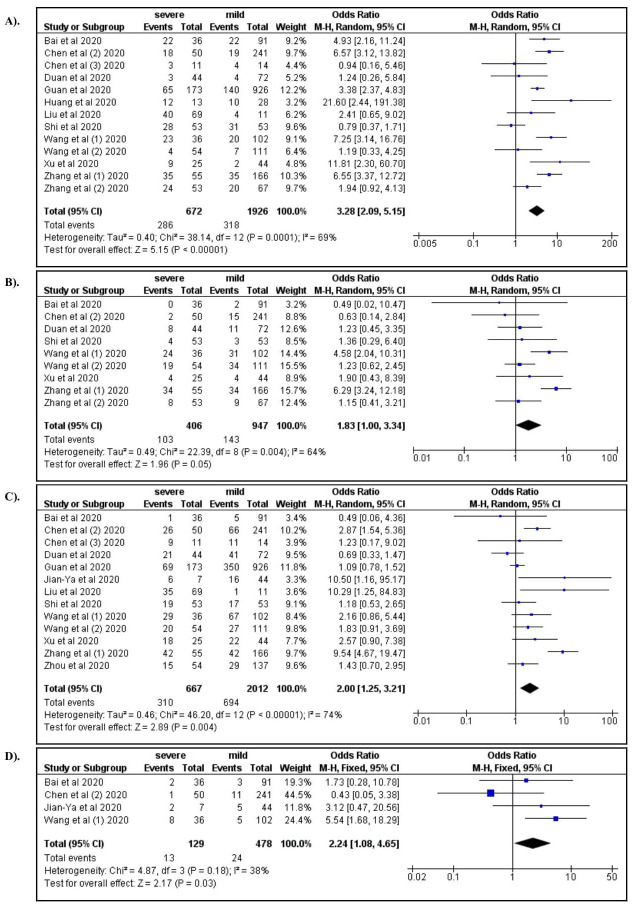
A) Dyspnea; B) Anorexia; C) Fatique; D) Dizziness.
Figure 4.
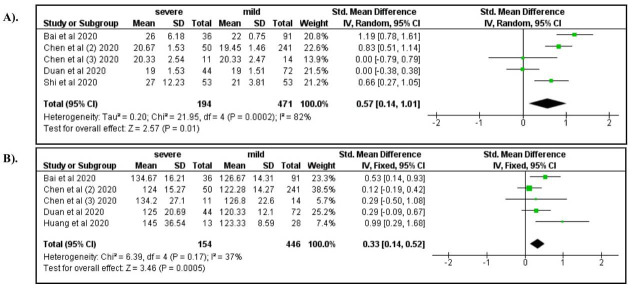
A) Respiratory rate; B) Systolic blood pressure.
Among laboratory characteristics, severe COVID-19 was associated with elevated WBC count (OR: 4.92; 95% CI: 2.12, 11.31), increased neutrophil count (OR: 5.45; 95% CI: 2.04, 14.54), lymphocytopenia (OR: 3.19; 95% CI: 1.14, 7.07), and decreased hemoglobin levels (OR: 0.76; 95%CI: 0.58, 1.00) ( Figure 5A–D). Elevated levels of AST, ALT, and serum creatinine increased the risk for severe manifestations of COVID-19 (ORs 4.91, 3.23, and 2.14, respectively; Figure 6A–C). Elevated levels of BUN (OR: 6.15; 95% CI: 3.05, 12.37), Hs-troponin I (OR: 9.25; 95% CI: 3.51, 24.37), creatine kinase (OR: 2.44; 95% CI: 1.65, 3.62), Hs-CRP (OR: 14.27; 95% CI: 5.13, 39.71), IL-6 (OR: 6.68; 95% CI: 3.20, 13.94), D-dimer (OR: 6.19; 95% CI: 4.22, 9.08), ferritin (OR: 1.96; 95% CI: 1.06, 3.62), lactate dehydrogenase (OR: 8.28; 95% CI: 4.75, 14.46), procalcitonin (OR: 6.62; 95% CI: 3.32, 13.21), ESR (OR: 4.45; 95% CI: 2.56, 7.76), and CRP >8 (OR: 8.34; 95% CI: 1.85, 37.62) were also associated with severe COVID-19 ( Figure 7– Figure 9). A low risk of severe COVID-19 was associated with low leukocyte levels (OR: 0.59; 95% CI: 0.41, 0.87) and elevated lymphocyte levels (OR: 0.34; 95% CI: 0.23, 0.50).
Figure 5.
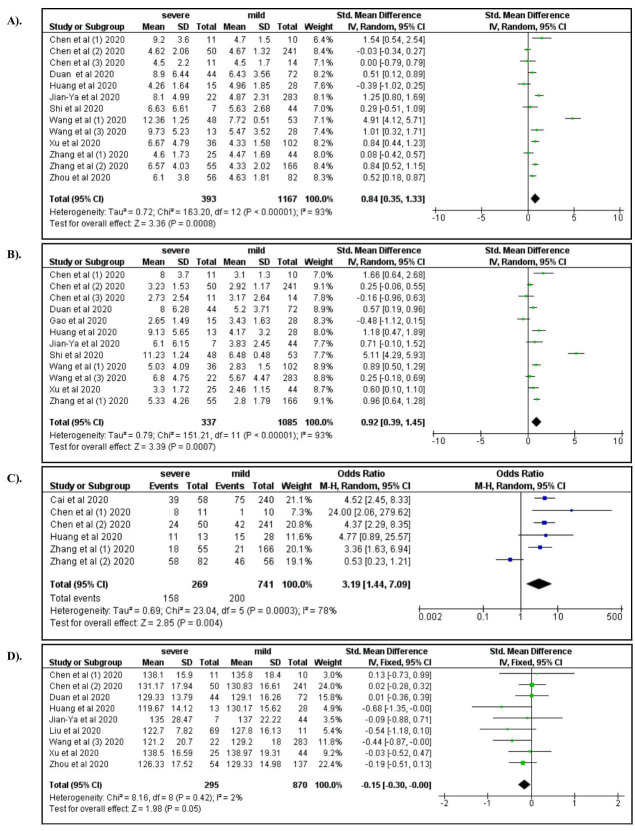
A) White blood cells; B) Neutrophil count; C) Lymphocytopenia; D) Hemoglobin.
Figure 6.
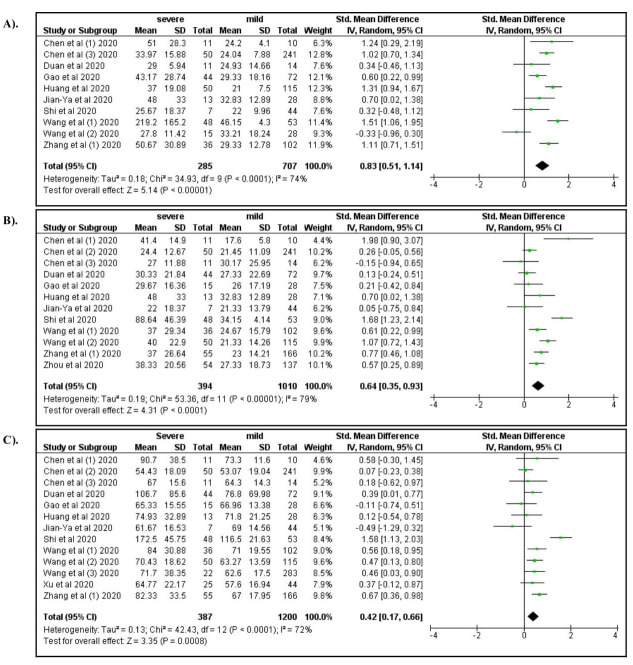
A forest plot of the association between the risk of severe COVID-19 and the levels of AST ( A), ALT ( B), and serum creatinine ( C).
Figure 7.
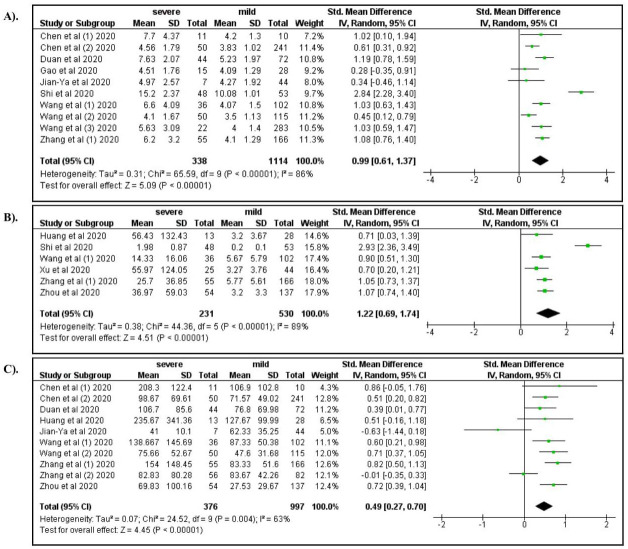
A forest plot of the association between the risk of severe COVID-19 and the levels of BUN ( A), Hs-troponin ( B), and creatine kinase ( C).
Figure 8.
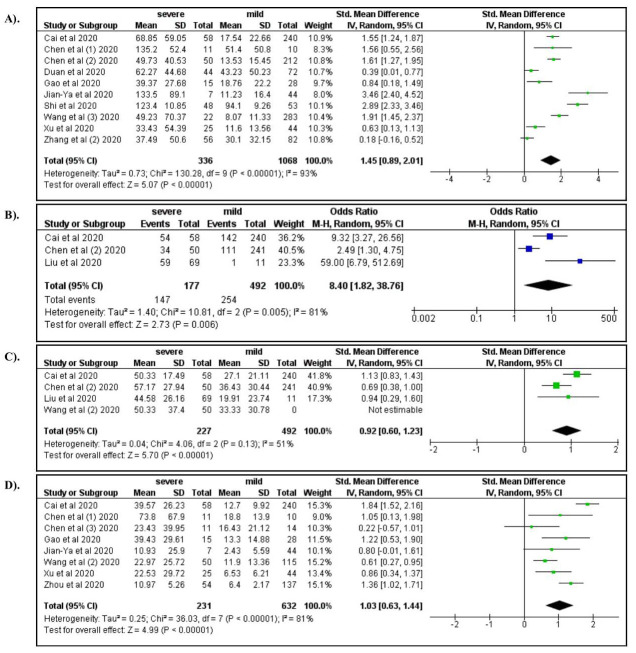
A forest plot of the association between the risk of severe COVID-19 and the levels of CRP ( A), Hs-CRP ( B), ESR ( C), and IL-6 ( D).
Figure 9.
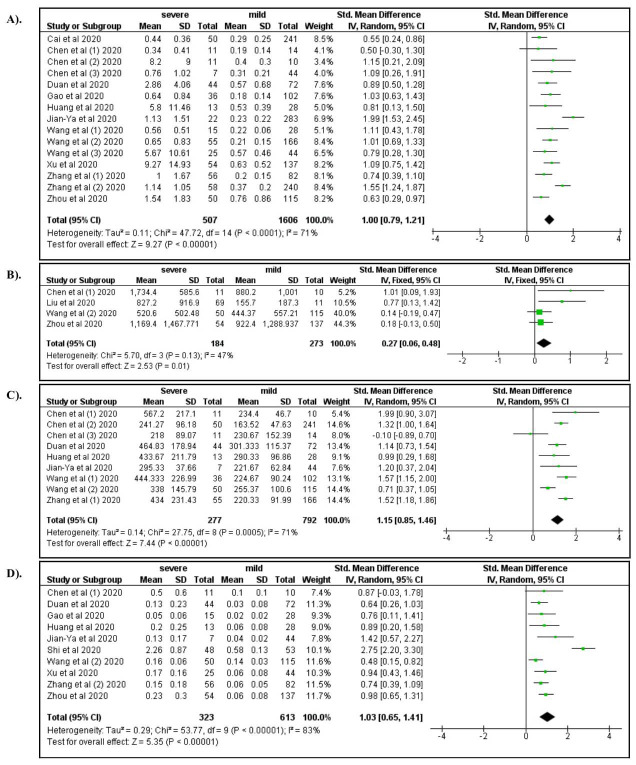
A forest plot of the association between the risk of severe COVID-19 and the levels of D-dimer ( A), serum ferritin ( B), lactate dehydrogenase ( C), and procalcitonin ( D).
Source of heterogeneity
Heterogeneity was detected in the data of chronic kidney disease, cerebrovascular disease, cardiovascular disease, diabetes mellitus, hypertension, and malignancy among the comorbid factors analyzed. Therefore, we used the random effect model to analyze the data. The fixed effect model was used to analyze the data on chronic liver disease and chronic respiratory disease, as there was no evidence of heterogeneity. For clinical manifestations, the data on fever, cough, sore throat, dyspnea, diarrhea, anorexia, fatigue, temperature >38°C, respiratory rate, and diastolic blood pressure were analyzed using the random effect model while the rest of clinical manifestation data were analyzed using the fixed effect model.
Among laboratory parameters, evidence of heterogeneity was found in count of WBC, neutrophil, monocyte, lymphocyte, platelet, CD4, and CD8; the presence of lymphocytopenia and anemia; the levels of AST, ALT, total bilirubin, albumin, aPTT, PTT, serum creatinine, BUN, Hs-Troponin I, creatine kinase, IL-6, Hs-CRP, glucose, D-dimer, sodium, potassium, lactate dehydrogenase, and procalcitonin; elevated CRP; and ESR. Accordingly, the data were analyzed using the random effect model. The data for the remaining parameters were analyzed using the fixed effect model.
Potential publication bias
We used Egger's test to assess the potential publication bias. Our cumulative calculation revealed that reporting or publication bias (p<0.05) existed with respect to chronic liver disease, expectoration, myalgia, abdominal pain, heart rate, leukocytosis, elevated ESR, and elevated IL-6 levels.
Discussion
Our data suggest that comorbidities, such as chronic respiratory disease, cardiovascular disease, diabetes, and hypertension, were associated with a higher risk of severe COVID-19, among which, hypertension was the strongest risk factor. These results are consistent with those of previous meta-analyses 43, 44 that indicated that chronic respiratory disease, cardiovascular disease, diabetes, and hypertension are significantly associated with higher COVID-19 mortality. Hypertension and diabetes are also associated with higher mortality among patients with dengue fever, West Nile virus infection, Zika virus infection, and yellow fever 45. To date, no study has reported details of the primary mechanism underlying the association between severe COVID-19 and comorbid factors. However, immune responses might be the most crucial factor underlying this association. Patients with comorbidities such as cardiovascular disease, chronic respiratory disease, hypertension, and diabetes were observed to have a lower immunity status than healthy individuals 46– 48. Since COVID-19 primarily affects the respiratory tract 49, patients with chronic respiratory diseases might be at a higher risk of contracting severe COVID-19. In addition, endothelial dysfunction might also play a pivotal role 50.
COVID-19 is a novel disease, and the immune response of this disease is not completely understood. Our data suggest that elevated leukocyte and neutrophil levels and reduced lymphocyte levels are associated with severe COVID-19. In other viral infections, such as influenza, elevated leukocyte and neutrophil levels serve as important predictors of disease severity 51. The role of leukocytes in the pathogenesis of COVID-19 is conflicting. In most cases, viral infections have been observed to cause leukopenia 52. Furthermore, a study also reported that leukopenia was observed at a significantly higher frequency among COVID-19 patients than among non-COVID-19 patients 53. However, in our present study, we did not compare COVID-19 and non-COVID-19 patients. The major factor that seemed to affect our findings was the occurrence of cytokine storm in patients. In COVID-19, there is an immune system overreaction, which results in a cytokine storm. In this condition, leukocytes might be over-activated, which might lead to the release of high levels of cytokines 54. Consistent with our data, a study has confirmed that cytokine storm is significantly associated with severe COVID-19 55. The theory underlying the role of neutrophils in COVID-19, as reported in our study, remains unclear. The speculations might be attributed to the involvement of neutrophil extracellular traps (NETs). While no study has assessed the precise role of NETs in COVID-19 pathogenesis, certain researchers speculate that SARS-CoV-2 might stimulate neutrophils to produce NETs, similar to several other viral pathogens 56. Furthermore, this might lead to neutrophil infiltration in pulmonary capillaries, organ damage, and the development of acute respiratory distress syndrome 57.
Low lymphocyte levels were observed in patients with severe COVID-19 compared with those with mild COVID-19. In the context of the immunological mechanism, our results might be contradictory. Lymphocyte subsets are known to play an important role in the action against bacterial, viral, fungal, and parasitic infections 58; therefore, the levels of circulating lymphocytes should increase. The immunological response in COVID-19 is unique and remains unclear. However, certain propositions might help describe our findings. First, coronaviruses infect human cells through ACE2 receptors 59. Since ACE2 receptors are also expressed by lymphocytes 60, the coronaviruses may enter lymphocytes and induce apoptosis. Second, the feedback mechanism between pro-inflammatory cytokines (such as IL-6) and lymphocytes might also explain our results. A study revealed that elevation in the levels of pro-inflammatory cytokines correlated with reduction in the levels of lymphocytes 61. Moreover, our findings also confirmed the significant elevation in the levels of IL-6. Third, ACE2 receptors are expressed by cells from various organs, including the thymus and spleen 62. As coronaviruses infect human cells through the ACE2 receptors, the spleen and thymus might also be damaged in patients with COVID-19, which would lead to lower levels of lymphocyte production. Fourth, lymphocyte proliferation requires a balanced metabolism, and metabolic disorders such as hyperlactic acidemia have been reported to disturb lymphocyte proliferation 63. Hyperlactic acidemia has been observed in patients with severe COVID-19 64.
The studies included in this systematic review also suggest that the levels of D-dimer were significantly higher in patients with severe COVID-19. Coagulation in patients with COVID-19 has been a major concern, and the lack of reliable data and meta-analyses prevents a holistic comparison. Certain infectious diseases that cause abnormal coagulation have been associated with poor clinical outcomes 65. The theory behind this mechanism is not understood clearly. It is widely known that ACE2 receptors are important for the infection of host cells by SARS-CoV-2, and ACE2 receptors are expressed in various cells in the human body, including endothelial cells 66. Consequently, a massive inflammatory reaction may occur in endothelial cells owing to SARS-CoV-2 infection 67, which may lead to increased coagulation, disseminated intravascular coagulation 68, and increased fibrin degradation 69. High fibrin degradation leads to elevated levels of fibrinogen and D-dimer 70, which might also explain the occurrence of venous thromboembolism in critical patients of COVID-19 71. In addition, a study with a short follow-up period also reported the existence of a dynamic correlation between the D-dimer levels and the severity of COVID-19 72. Furthermore, pulmonary embolism and deep vein thrombosis were also observed in patients with severe COVID-19 73, 74, which suggests that D-dimer might play a prominent role in governing the severity of COVID-19 patients.
We also observed that inflammatory markers, including elevated levels of CRP, ESR, and IL-6, were found both in patients with severe and mild COVID-19, with a significant increase detected in patients with severe COVID-19. Other variables associated with adverse outcomes, such as ferritin, lactate dehydrogenase, and procalcitonin levels, were found to be elevated predominantly in patients with severe COVID-19. Our findings were consistent with those of a previous meta-analysis 75, and indicated that high levels of CRP, lactate dehydrogenase, and ESR were associated with adverse outcomes in COVID-19. Another meta-analysis had also confirmed that elevated levels of IL-6 were observed in patients with COVID-19 who exhibited poor clinical outcomes 76. Therefore, the levels of CRP, ESR, IL-6, ferritin, procalcitonin, and lactate dehydrogenase can serve as potential markers for the evaluation of COVID-19 prognosis.
The high mortality rate and treatment failure in patients with COVID-19 can be attributed to the fact that COVID-19 affects multiple organs, including the lung, heart, kidney, and liver 77. Our data suggest that elevated levels of urea and creatinine, and not chronic kidney disease, were associated with severe COVID-19, which indicates that acute inflammation might be caused by SARS-CoV-2 infection. Previous meta-analyses have also reported findings consistent with our results 78, 79. Moreover, anatomical studies have reported significant renal inflammation in patients with severe COVID-19 75, 80, 81. There might be two mechanisms by which SARS-CoV-2 induces renal inflammation. First, SARS-CoV-2 might directly infect renal tubular epithelial cells and podocytes through ACE2 receptors, which facilitates the targeted infection of certain cells by the virus. Consequently, acute tubular necrosis, podocytopathy, microangiopathy, and collapsing glomerulopathy might occur owing to the massive inflammation in renal tubular epithelial cells and podocytes 82, 83. Second, the binding between SARS-CoV-2 and ACE2 receptors might activate angiotensin II and induce cytokine production, which may lead to hypercoagulopathy and microangiopathy, and eventually cause renal hypoxia 84, 85.
Conversely, with respect to liver function, we observed that the levels of liver enzymes were higher in patients with severe COVID-19. Previous studies in this context have elucidated that ACE2 receptors are highly expressed in bile duct cells; therefore, infection of these cells by coronaviruses might lead to abnormalities in the levels of liver enzymes 86. However, a recent anatomical study on liver biopsy specimens from patients with severe COVID-19 revealed that moderate microvascular steatosis and mild lobular and portal activities were observed 87. These data suggest that it cannot be determined clearly whether the elevated levels of liver enzymes in patients with severe COVID-19 are caused by direct infection or by drug-induced liver injury. Therefore, further studies are required to elucidate the precise mechanism underlying the elevation of liver enzymes levels in patients with severe COVID-19.
Meta-analyses on this topic have been performed previously 43, 44, 75, 76, 88– 91. However, compared to previous studies, our study has the following strengths. The previous studies only reported limited factors, such as clinical manifestations 43, 88, 90, 91, laboratory findings 76, 89, or a combination of only clinical manifestations and laboratory findings 75. In our study, we included all comorbidities, clinical manifestations, and laboratory characteristics. Additionally, compared to previous studies, this study has a larger sample size; the data on 1,934 patients with mild and 1,644 patients with severe COVID-19 treated across 19 hospitals were retrieved. However, this study also has certain limitations. Certain crucial factors that might play an important role in the pathogenesis of COVID-19, including secondary infection, treatment, and immunological status were not controlled for. Our current findings should be interpreted with caution because the majority of studies included were cross-sectional, and the samples corresponding to the data analyzed originated only in China. Moreover, in our study, we did not perform the sub-group analysis according to the transmission area. As already reported, the transmission of COVID-19 in China was also affected by the transmission area 92. Therefore, this limitation might also affect the final findings of our study. Longitudinal studies may reveal more long-term impacts of SARS-CoV-2 infection 93.
Conclusion
COVID-19 is an emergent infectious disease, and the major problem associated with it is the unknown pattern of disease development. We identified 34 factors that are associated with severe COVID-19. This might improve our understanding of COVID-19 progression and provide baseline data to compile or improve the prediction models for the estimation of COVID-19 prognosis. Moreover, our current findings may also contribute to guide the prioritization of high-risk target populations for vaccination.
Data availability
Underlying data
All data underlying the results are available as part of the article and no additional source data are required.
Reporting guidelines
Figshare: PRISMA checklist for ‘Predictors of COVID-19 severity: a systematic review and meta-analysis’, https://doi.org/10.6084/m9.figshare.12813683.v1 94
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Notes
[version 2; peer review: 2 approved]
Funding Statement
We thank to Lembaga Pengelola Dana Pendidikan (LPDP) Republik Indonesia for supporting this project.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
The authors responded to my comments clearly and the manuscript is accepted for publication in your journal.
Are the rationale for, and objectives of, the Systematic Review clearly stated?
Yes
Is the statistical analysis and its interpretation appropriate?
Yes
Are sufficient details of the methods and analysis provided to allow replication by others?
Partly
Are the conclusions drawn adequately supported by the results presented in the review?
Yes
Reviewer Expertise:
NA
I confirm that I have read this submission and believe that I have an appropriate level of expertise to confirm that it is of an acceptable scientific standard.
The strength of this paper is the meta-analysis in terms of effect estimates. The weakness is the focus of data from China, while we should learn more from global data including the comparison between HIC and LMIC.
In China, severity was also found to correlate with the force of infection, eg those in high transmission areas had more severe disease outcomes than those from lower transmission areas in China, see: Exposure to SARS-CoV-2 in a hightransmission setting increases the risk of severe COVID-19 compared with exposure to a low transmission setting?
Chen D, Hu C, Su F, Song Q, Wang Z. J Travel Med. 2020 Aug 20;27(5):taaa094. 10.1093/jtm/taaa094. 1
The authors highlight the need for a scoring system for the prediction of severity. There is another reason why it is important to identify risk factors for severe disease: to guide prioritization of high risk target populations for vaccination
Are the rationale for, and objectives of, the Systematic Review clearly stated?
Yes
Is the statistical analysis and its interpretation appropriate?
Yes
Are sufficient details of the methods and analysis provided to allow replication by others?
Yes
Are the conclusions drawn adequately supported by the results presented in the review?
Yes
Reviewer Expertise:
COVID-19, Zika and dengue
I confirm that I have read this submission and believe that I have an appropriate level of expertise to confirm that it is of an acceptable scientific standard.
References
In this meta-analysis, you investigated the predictors of COVID-19 severity through the literature. You considered a topic of interest and provided a well-written manuscript. However, there are some things that will improve your reporting.
Abstract, method section, please insert detail about critical/quality appraisal of the included studies.
Abstract, method section, line 1, please remove " and extracted" from the text. It maybe causes a misunderstanding between this step and the data extraction step.
Method section, please remove line five. "the protocols for the ...". Mentioning the PRISMA is enough.
Method section, eligibility criteria, (2) please mention the type of data for adequate data. what is adequate data?
Method section, search strategy, why is Scopus not searched? You may have missed some articles that are only indexed in Scopus.
Method section, search strategy, this sentence not related to this section. If you limit the search to EN publication then you need to change the verb. If not this sentence related to inclusion criteria.
Method section, search strategy, based on PRISMA, add at least one search strategy for one database as a supplement.
Method section, data extraction, please added the country of origin for each study. The predictors may be different from one setting to another setting.
Method section, data extraction, please add details about how resolved disagreement between reviewers.
Method section, how did you handle the publication bias?
Result section, there is some problem in figure 1. Please fill it considering other related studies. The number for "record screened" is incorrect.
Result section, table 1, all studies are from China. If all studies are from China it is better to change the title. these are a predictor of severity in China. In my opinion, this is a limitation of your study.
Cheers
Are the rationale for, and objectives of, the Systematic Review clearly stated?
Yes
Is the statistical analysis and its interpretation appropriate?
Yes
Are sufficient details of the methods and analysis provided to allow replication by others?
Partly
Are the conclusions drawn adequately supported by the results presented in the review?
Yes
Reviewer Expertise:
Systematic review and meta-analysis in health and medical intervention
I confirm that I have read this submission and believe that I have an appropriate level of expertise to confirm that it is of an acceptable scientific standard.
Articles from F1000Research are provided here courtesy of F1000 Research Ltd
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.12688/f1000research.26186.2
Article citations
The severity assessment and nucleic acid turning-negative-time prediction in COVID-19 patients with COPD using a fused deep learning model.
BMC Pulm Med, 24(1):515, 14 Oct 2024
Cited by: 0 articles | PMID: 39402509 | PMCID: PMC11476205
Prospective Variation of Cytokine Trends during COVID-19: A Progressive Approach from Disease Onset until Outcome.
Int J Mol Sci, 25(19):10578, 01 Oct 2024
Cited by: 0 articles | PMID: 39408907 | PMCID: PMC11477561
Obesity Parameters as Predictor of Poor Outcomes in Hospitalized Patients with Confirmed Mild-to-Moderate COVID-19.
Infect Dis Rep, 16(5):894-905, 12 Sep 2024
Cited by: 0 articles | PMID: 39311212 | PMCID: PMC11417886
Shock index and shock index, pediatric age-adjusted as predictors of mortality in pediatric patients with trauma: A systematic review and meta-analysis.
PLoS One, 19(7):e0307367, 18 Jul 2024
Cited by: 0 articles | PMID: 39024206 | PMCID: PMC11257222
Review Free full text in Europe PMC
More severe comorbidities, advanced age, and incomplete vaccination increase the risk of COVID-19 mortality.
Narra J, 4(2):e949, 14 Aug 2024
Cited by: 0 articles | PMID: 39280314 | PMCID: PMC11391969
Go to all (122) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Meta-analysis investigating the relationship between clinical features, outcomes, and severity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pneumonia.
Am J Infect Control, 49(1):82-89, 12 Jun 2020
Cited by: 84 articles | PMID: 32540370 | PMCID: PMC7292004
Review Free full text in Europe PMC
Laboratory features of severe vs. non-severe COVID-19 patients in Asian populations: a systematic review and meta-analysis.
Eur J Med Res, 25(1):30, 03 Aug 2020
Cited by: 161 articles | PMID: 32746929 | PMCID: PMC7396942
Review Free full text in Europe PMC
Biomarkers of cytokine storm as red flags for severe and fatal COVID-19 cases: A living systematic review and meta-analysis.
PLoS One, 16(6):e0253894, 29 Jun 2021
Cited by: 84 articles | PMID: 34185801 | PMCID: PMC8241122
Review Free full text in Europe PMC
The Clinical Characteristics and Risk Factors of Severe COVID-19.
Gerontology, 67(3):255-266, 06 Jan 2021
Cited by: 58 articles | PMID: 33406518 | PMCID: PMC7900480
Funding
Funders who supported this work.




