Abstract
Free full text

Genomic copy‐number alterations of MYC and FHIT genes are associated with survival in esophageal squamous‐cell carcinoma
Abstract
Esophageal squamous‐cell carcinoma (ESCC) is one of the most common cancers and is associated with a poor prognosis. Studies are warranted on the clinical relevance of its genomic copy‐number alterations (CNA) as prognosticators for ESCC. In the present study, we first screened recurrent CNA by array‐based comparative genomic hybridization using an in‐house focused bacterial artificial chromosome‐based array for 108 loci in 45 ESCC specimens. We detected 14 regions showing recurrent (>20%) CNA (4 losses and 10 gains) by array‐based comparative genomic hybridization in the first cohort. Among them, loss of 3p14.2 and gain of 8q24.21 for the FHIT and MYC genes, respectively, and the accumulation of those two CNA (higher FM‐CNA scores) were significantly associated with a worse overall survival (OS) in the first cohort (P = 0.0273, P = 0.0356 and P = 0.0089, respectively). In the independent validation cohort of 92 resected ESCC cases, loss of FHIT, gain of MYC and higher FM‐CNA scores determined by a quantitative genomic PCR‐based copy‐number analysis were associated with a worse OS (P = 0.0011, P = 0.0104 and P = 0.0008, respectively) and disease‐free survival (P = 0.0038, P = 0.0132 and P = 0.0021, respectively). In addition, the Cox model showed the presence of either CNA to be an independent prognosticator for OS and disease‐free survival in the validation cohort (P = 0.0120 and P = 0.0255, respectively). These results suggest that CNA of MYC and FHIT are poor prognostic markers, and risk stratification based on the copy‐number status of those genes is useful to select the optimal treatment strategy in resected ESCC patients. (Cancer Sci, 10.1111/j.1349‐7006.2012.02329.x, 2012)
Esophageal squamous‐cell carcinoma (ESCC) is one of the most common forms of cancer and is associated with a poor prognosis.1 Despite recent advances in diagnostic techniques and combined treatment modalities, patients with ESCC still have extremely poor survival rates, even after extended surgery. Currently, pathologic stage (pStage) is the most reliable prognostic factor for ESCC and a few molecules have been applied in a clinical setting as therapeutic and/or diagnostic biomarkers. Therefore, the significance of detecting a novel biomarker using reasonable molecules should be emphasized.
Chromosomal aberration is one of several mechanisms that can lead to gene dysregulation and has long been known to play a critical role in the pathogenesis of cancers.2 The characterization of those alterations linked to ESCC might provide information relevant to a refined prognosis. Conventional and array‐based comparative genomic hybridization (CGH and aCGH, respectively) analyses have demonstrated genetic complexity in ESCC and identified some recurrent copy‐number alterations (CNA) associated with clinical parameters, particularly with survival: gain of 1p36.32, 3q11.2, 3q22.3, 5p, 5p15, 7q, 11q13.2, 12p and 19p13.3 and loss of 3p, 4p, 7q34, 9p, 9q34.3, 10q11.21, 11q and 18q21.1‐q23 have been associated with a poor prognosis in univariate and/or multivariate analyses,3, 4, 5, 6, 7 but genetic alterations and biological characteristics have so far had a limited impact on clinical prognostication and treatment. More accurate prognostic genetic markers are needed to distinguish high‐risk patients from low‐risk patients, so that optimal treatment can be used.
In the present study, we screened CNA associated with the survival of ESCC patients by aCGH using an in‐house focused bacterial artificial chromosome (BAC)‐based array, and identified one amplified and one deleted BAC probe containing the fragile histidine triad (FHIT) and MYC genes, respectively, as candidate prognosticators. Further analysis using quantitative real‐time genomic PCR (gPCR) for those genes in a larger independent group of ESCC patients validated our findings.
Materials and Methods
Patients and tissues
A total of 137 patients with ESCC undergoing tumor resection from 2002 to 2008 at the Tokyo Medical and Dental University Hospital (TMDU, Tokyo, Japan) were included in this study. Relevant clinical and survival data were available for all 137 patients. Written consent was obtained from patients after approval was granted by the local ethics committee. Disease stage (pStage) was defined in accordance with the TNM classification8 and the Japanese Classification of Esophageal Cancer.9 The median follow‐up period for the surviving patients was 31.3 (ranging from 3.0 to 111.9) months.
For the aCGH analysis, primary tumor samples and corresponding non‐cancerous esophageal mucosa of 45 of 137 cases resected from April 2005 to January 2008 (Table S1) were obtained during surgery and frozen immediately in liquid nitrogen and stored at −80°C until required. To compare results of the quantitative real‐time gPCR analysis with those of the aCGH analysis, formalin‐fixed paraffin‐embedded (FFPE) primary tumor samples and corresponding non‐cancerous esophageal mucosa were also obtained from 34 of 45 cases.
For the validation analysis using real‐time gPCR, FFPE primary tumor samples were obtained from 92 of 137 patients (Table S2), who underwent curative esophagectomy without chemotherapy or radiotherapy before surgery from January 2002 to June 2006 at TMDU and were not included in the aCGH analysis.
Genomic DNA (gDNA) was extracted from frozen and FFPE tissues using a Gentra Puregene Tissue Kit (Qiagen, Valencia, CA, USA) and a QIAamp DNA FFPE Tissue Kit (Qiagen), respectively.
Array‐based comparative genomic hybridization analysis
We newly designed and constructed an in‐house focused BAC‐based array named “MCG Cancer Array‐Mini” containing 108 BAC harboring known cancer‐related genes, such as oncogenes and tumor suppressor genes, to detect cancer‐specific copy number aberrations of those genes (Table S3). Hybridization was carried out using 0.375 μg of tumor and non‐tumorous control DNA, and hybridized arrays were analyzed as described previously, except with a global normalization using the ratio from all spots.10, 11
Quantitative real‐time genomic PCR analysis
The copy‐numbers of the FHIT and MYC genes were measured using a TaqMan Copy Number Assay Hs02758348_cn and Hs03467977_cn (Applied Biosystems, Foster City, CA, USA), respectively, which contain a FAM dye‐based assay for each target locus and a VIC dye‐based assay for the RPPH1 gene on chromosome 14 as a reference locus. PCR was carried out in duplicate using 20 ng of gDNA as a template and the ABI PRISM 7500 sequence detection System (Applied Biosystems), according to the manufacturer's instructions.
Statistical analysis
The clinicopathological variables pertaining to the corresponding patients were analyzed for statistical significance using the χ2 or Fisher's exact test. For the analysis of death and/or the probability of relapse after operation, Kaplan–Meier survival curves were constructed for groups based on univariate predictors and differences between the groups were tested using the log rank test. Univariate and multivariate survival analyses were performed using the likelihood ratio test of the stratified Cox proportional‐hazards model. For multiple group comparisons, an anova was used, followed by Scheffé's post‐hoc test. Differences were assessed using a two‐sided test, and considered significant at the P < 0.05 level.
Results
Copy‐number alterations detected with array‐based comparative genomic hybridization using the focused in‐house bacterial artificial chromosome array in esophageal squamous‐cell carcinoma
We first determined CNA in a genome‐wide manner with the MCG Cancer Array‐Mini containing 108 BAC harboring cancer‐related genes in 45 cases of ESCC for which frozen tumor tissue with corresponding non‐cancerous esophageal mucosa as a control were available (Fig. 1A). Chromosomal imbalances with one or more loci were detected in 41 (91.1%) of 45 ESCC tumors (mean, 10.5; range, 0–30), and a total of 344 gains and 124 losses were detected. CNA were detected in 93 of 108 BAC (86.1%) in 41 tumors, the most frequent (>20%) being losses of 4 regions and gains of 10 regions (Fig. 1B, Table 1).
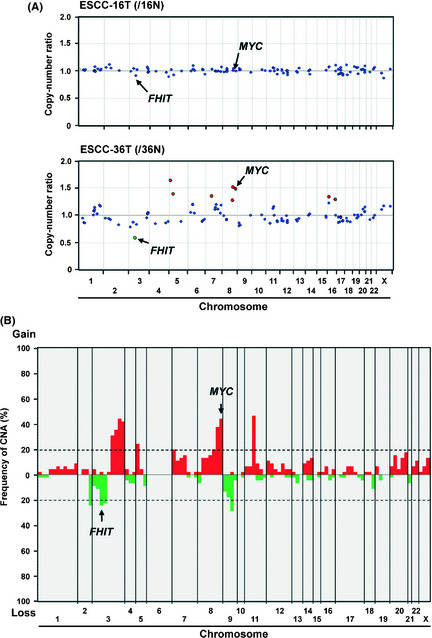
(A) Representative results of genome‐wide copy number profile detected by array‐based comparative genomic hybridization (aCGH) using the in‐house focused bacterial artificial chromosome (BAC) array (MCG Cancer Array‐Mini) for a case without (esophageal squamous‐cell carcinoma [ESCC]‐16T, upper) and with (ESCC‐36T, lower) remarkable copy‐number alterations (CNA) relative to corresponding non‐tumorous esophageal mucosa. Red and green spots represent BAC probes that showed gain and loss, respectively. (B) Genome‐wide frequency plot of DNA copy number gains (red) and losses (green) determined by aCGH using MCG Cancer Array‐Mini for all 45 ESCC tumors.
Table 1
Regions showing frequent copy‐number alterations (CNA) in array‐based comparative genomic hybridization analysis in 45 primary cases of esophageal squamous‐cell carcinoma (ESCC)
| Locus | BAC clone | Target gene | CNA | Frequency | Percent | P‐value* |
|---|---|---|---|---|---|---|
| 2q22.1 | RP11‐45N24 | LRP1B | Loss | 11/45 | 24.4 | 0.8575 |
| 3p14.1 | RP11‐215K24 | MITF | Loss | 10/45 | 22.2 | 0.2013 |
| 3p14.2 | RP11‐152A9 | FHIT | Loss | 11/45 | 26.7 | 0.0005 |
| 9p21.3 | RP11‐145E5 | CDKN2A | Loss | 13/45 | 31.1 | 0.0594 |
| 3q26.32 | RP11‐613F6 | PIK3CA | Gain | 14/45 | 31.1 | 0.0162 |
| 3q26.32 | RP11‐97J8 | GNB4 | Gain | 16/45 | 35.6 | 0.1198 |
| 3q26.33 | RP11‐43F17 | SOX2 | Gain | 20/45 | 44.4 | 0.1816 |
| 3q26.33 | RP11‐110I20 | ATP11B | Gain | 19/45 | 42.2 | 0.1764 |
| 5p15.33 | RP11‐117B23 | TERT | Gain | 11/45 | 24.4 | 0.8161 |
| 7p11.2 | RP11‐339F13 | EGFR | Gain | 9/45 | 20.0 | 0.8997 |
| 8q22.1 | RP11‐662P7 | LAPTEM4B | Gain | 9/45 | 20.0 | 0.0838 |
| 8q22.3 | RP11‐343F19 | YWHAZ | Gain | 17/45 | 37.8 | 0.0261 |
| 8q24.21 | RP11‐440N18 | MYC | Gain | 20/45 | 44.4 | 0.0050 |
| 11q13.2 | RP11‐300I6 | CCND1 | Gain | 21/45 | 46.7 | 0.1377 |
*P‐values (two‐sided) were calculated with the log‐rank test for comparisons of overall survival (OS) distributions between subgroups with and without each CNA in 45 primary cases of ESCC. BAC, bacterial artificial chromosome.
Association of chromosomal instability with overall survival in patients with esophageal squamous‐cell carcinoma
Because chromosomal imbalance is one of the hallmarks of genomic instability, a key event in the development of malignant tumors,12, 13 in neoplastic cells, we examined the association of chromosomal instability (CI) with several clinicopathologic parameters, including survival, in 45 ESCC patients. To evaluate CI, we divided the 45 ESCC patients into three groups (CI status) according to the total number of CNA: low CI (≤2 probes recognized CNA, n = 7); middle CI (3–18 probes recognized CNA, n = 29); and high CI groups (≥19 probes recognized CNA, n = 9; Fig. S1A). CI status in ESCC was not associated with age, gender, histopathological grading, pathological TNM (pTNM) categories or pStage grouping (Table S1). However, CI status seemed to be correlated with overall survival (OS, Fig. S1B), suggesting CI in tumor cells to contribute to the progression of ESCC, although the number of cases was too small to evaluate the statistical significance for predictors using the multivariate analysis.
Association of loss of FHIT and gain of MYC determined by array‐based comparative genomic hybridization with overall survival in 45 patients with esophageal squamous‐cell carcinoma
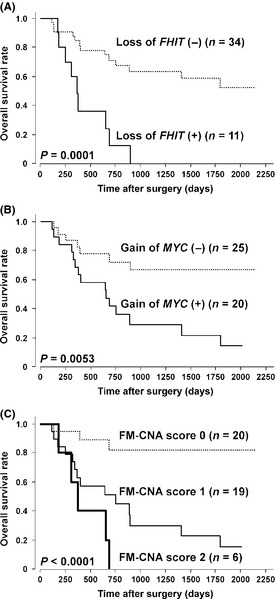
To explore regions/genes that are likely to be useful as prognosticators for ESCC, we examined the association of 14 frequently altered probes (>20%) with the OS rate in 45 ESCC patients (Table 1). Among CNA detected by 14 probes, loss of RP11‐152A9 (3p14.2, chr3:60,184,073–60,363,515 in UCSC Genome Browser on Human February 2009 [GRCh37/hg19] assembly; http://genome.ucsc.edu/cgi-bin/hgGateway) containing part of the FHIT gene (chr3:59735036–61237133) and gain of RP11‐440N18 (8q24.21, chr8:128,596,756–128,777,986) containing the whole MYC gene (chr8:128748315–128753680) showed remarkably low P‐values, suggesting these CNA to be potential prognosticators for ESCC patients (Table 1; Fig. 2A,B). Because loss of 3p14.2 (FHIT) and gain of 8q24.21 (MYC) loci were observed in a partially overlapping pattern in 45 cases of ESCC, we divided all cases into three groups based on the status of those two CNA (FM‐CNA score): FM‐CNA score of 0 (no CNA in either probe, n = 20), 1 (CNA in one of the probes, n = 19) and 2 (CNA in both probes, n = 6). FM‐CNA scores were not associated with clinicopathologic parameters such as age, gender, histopathological grading, pTNM categories and pStage grouping (Table S4), but significantly associated with OS (P = 0.0002, log rank test) in 45 cases of ESCC (Fig. 2C). Notably, CI status correlated with the loss of 3p14.2 (FHIT, P = 0.0273), gain of 8q24.21 (MYC, P = 0.0356) and FM‐CNA score (P = 0.0089), and the number of CNA detected by 108 probes positively correlated with FM‐CNA scores (Table S5). These results suggest that loss of 3p14.2 (FHIT) and/or gain of 8q24.21 (MYC) at least partly represent the genome‐wide CI status in ESCC tumors, and they retain the potential to be useful prognosticators in patients with ESCC.
Concordance between array‐based comparative genomic hybridization‐based and quantitative genomic PCR‐based copy‐number analyses of FHIT and MYC loci
To confirm the findings described above, we have to determine the CNA of two loci, 3p14.2 (FHIT) and 8q24.21 (MYC), and compare clinicopathological characteristics including prognosis. Because only FFPE samples were available for the validation analysis using a larger set of ESCC cases and quantitative gPCR is suitable to determine the copy‐number of a few targeted regions using fragmented gDNA from FFPE samples, we first tested the concordance of results obtained by different methods: aCGH using frozen tissues and quantitative gPCR14 using FFPE tissues from the same sample set.
Formalin‐fixed paraffin‐embedded samples with enough material to extract gDNA were available from 34 of the 45 ESCC tumors, including 9 cases with loss of 3p14.2 (FHIT) and 18 cases with gain of 8q24.21 (MYC) detected by aCGH. Among them, 7 samples of corresponding non‐cancerous esophageal mucosa were available in 20 cases. To normalize the genomic copy‐number in each sample, we used the RPPH1 gene on chromosome 14 as an internal control (reference) locus.
The copy‐number of the FHIT gene determined by quantitative gPCR in the 9 ESCC cases with loss of FHIT in aCGH analysis (mean ± SD = 0.40 ± 0.15) was significantly lower than that in the 20 samples of normal esophageal mucosa (mean ± SD = 0.82 ± 0.23) and 25 ESCC cases without loss of FHIT in aCGH analysis (mean ± SD = 0.69 ± 0.11, Fig. 3A). The copy‐number of the MYC gene determined by quantitative gPCR in the 18 ESCC cases with gain of MYC in aCGH analysis (mean ± SD = 3.72 ± 1.87) was significantly higher than that in the 20 samples of normal esophageal mucosa (mean ± SD = 1.89 ± 0.20) and 16 ESCC cases without gain of MYC in aCGH analysis (mean ± SD = 2.19 ± 0.57, Fig. 3A). Based on the results obtained by aCGH, we set cut‐off values for the relative copy‐number ratio of quantitative gPCR to define loss of FHIT and gain of MYC as 0.519 and 2.34, respectively, because these values showed the highest sensitivity and specificity on the receiver operating characteristic curve yielding area under the curve values of 0.929 and 0.858 for loss of FHIT and gain of MYC: the sensitivity and the specificity of quantitative gPCR were 88.9% and 92.0% for loss of FHIT and 83.3% and 81.3% for gain of MYC, respectively (Fig. 3B,C). Taken together, we conclude these cut‐off values to differentiate ESCC cases with loss of FHIT and gain of MYC effectively from those without the CNA by quantitative gPCR analysis. Loss of FHIT and gain of MYC was also confirmed by FISH using FFPE samples with BAC on the MCG Cancer Array‐Mini and corresponding control probes for both loci (data not shown).
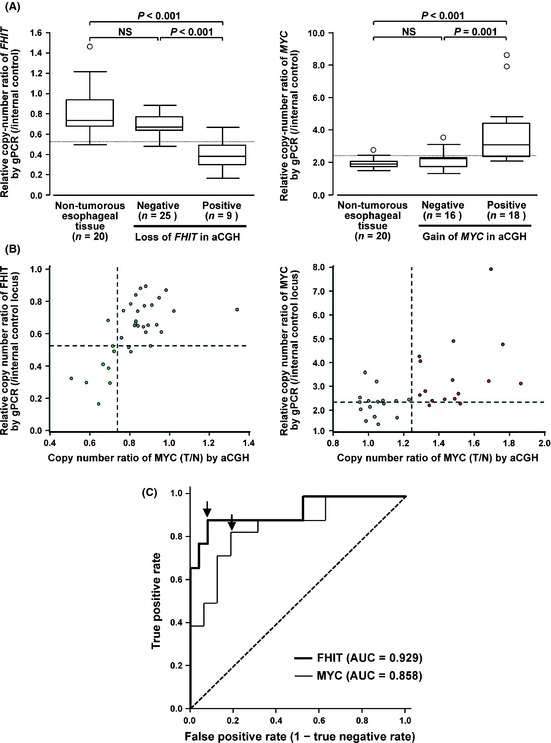
(A) Boxplot of relative copy‐numbers of the FHIT (left) and MYC (right) genes determined by quantitative genomic PCR (gPCR) in formalin‐fixed paraffin‐embedded (FFPE) tumor samples of 34 esophageal squamous‐cell carcinoma (ESCC) cases with or without loss of FHIT (left) and gain of MYC (right), respectively, and FFPE non‐tumorous tissues of 20 of those 34 cases. Dotted lines indicate cut‐off values for relative copy‐number ratio to define loss of FHIT and gain of MYC by quantitative gPCR described in C. (B) Scatter diagram for relative copy‐numbers of FHIT (left) and MYC (right) determined by array‐based comparative genomic hybridization (aCGH) (x‐axis) using frozen tumorus tissues and paired non‐tumorous tissues of ESCC and quantitative gPCR (y‐axis) using corresponding FFPE tumorous tissues. Green and red spots represent cases with loss and gain for FHIT and MYC, respectively, by aCGH in 34 cases. Dotted lines indicate cut‐off values for relative copy‐number ratio to define loss of FHIT (left) and gain of MYC (right) by aCGH (0.75 and 1.25, respectively) or quantitative gPCR described in C. (C) Receiver operating characteristic curves with respect to the sensitivity and specificity of quantitative gPCR to determine the status of loss of FHIT and gain of MYC compared with aCGH. Area under the curve (AUC) values of 0.929 and 0.858 for loss of FHIT and gain of MYC. Sensitivity and specificity of cut‐off values for relative copy‐number ratio to define loss of FHIT and gain of MYC by quantitative gPCR (0.519 and 2.34, respectively) were 88.9% and 92.0% for loss of FHIT and 83.3% and 81.3% for gain of MYC, respectively (arrows). Dotted line, random guess.
Validation of significance of loss of FHIT and gain of MYC determined by quantitative genomic PCR as prognosticators for 92 patients with esophageal squamous‐cell carcinoma
The potential significance of loss of FHIT and gain of MYC as prognosticators in ESCC determined by aCGH analysis was validated in a larger cohort of ESCC cases (92 cases) analyzed by quantitative gPCR. None of these overlapped with the 45 cases of ESCC analyzed by aCGH. Among the 92 cases, loss of FHIT and gain of MYC were detected in 51 and 30 cases, respectively. Gain of MYC was significantly correlated with deeper tumors (pT categories, P = 0.023) and lymphatic metastasis (pN categories, P = 0.021), whereas loss of FHIT was significantly correlated with younger age (P = 0.018), deeper tumors (pT categories, P = 0.007), distant metastasis (pM categories, P = 0.002) and advanced tumors (pStage, P = 0.001, Table S2). All 92 cases were classified into three categories (FM‐CNA score) based on the presence of loss of FHIT and/or gain of MYC alterations: 0 (no CNA in either gene, n = 31), 1 (CNA in one of the genes, n = 41) and 2 (CNA in both genes, n = 20). FM‐CNA scores significantly correlated with pT (P = 0.006), pN (P = 0.028), pM (P = 0.042) and pStage categories (P = 0.006, Table S2).
Kaplan–Meier survival estimates (Fig. 4) showed that loss of FHIT, gain of MYC and FM‐CNA scores were significantly associated with a worse OS and disease‐free survival (DFS) in all 92 cases (P = 0.0011 and P = 0.0038, P = 0.0104 and P = 0.0132, and P = 0.0008 and P = 0.0021, respectively [log rank test]). In the Cox proportional hazards regression model (Table 2), univariate analyses demonstrated that the pT and pN categories in the pTNM classification, pStage grouping, and copy‐number status of the FHIT and MYC genes (loss of FHIT, gain of MYC and FM‐CNA score) were significantly associated with both OS and DFS. Multivariate analyses by a Cox regression procedure using pStage and FM‐CNA scores determined by quantitative gPCR revealed that either loss of FHIT or gain of MYC (FM‐CNA score 1 and 2) and advanced tumor stage (pStages III or IV) according to the pStage grouping based on the TNM classification were independent predictive factors for a worse OS and DFS (OS, P = 0.0120 and 0.0010, respectively; DFS, P = 0.0255 and 0.0002, respectively), although neither CNA was an independent prognosticator for OS and DFS (Table 2). The presence of either loss of FHIT or gain of MYC (FM‐CNA score 1 and 2) predicted the 5‐year probability of death and recurrence with a sensitivity of 87.5% and 79.6%, a specificity of 50% and 52.6%, and an accuracy of 68.8% and 68.5%, respectively.
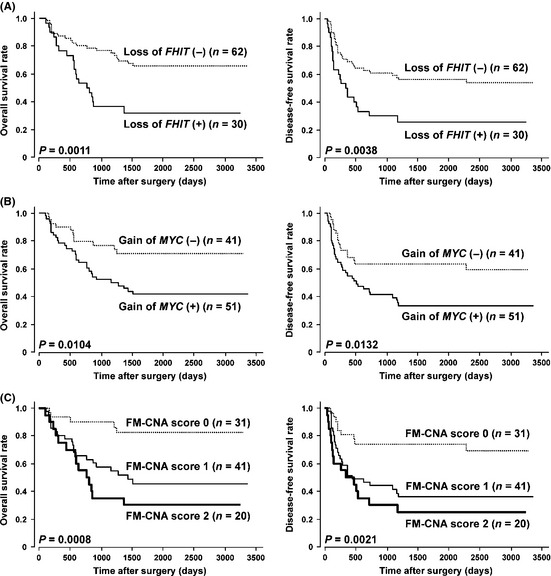
Kaplan–Meier curves for overall (left) and disease‐free (right) survival rates of 92 patients with esophageal squamous‐cell carcinoma (ESCC) according to loss of FHIT (A), gain of MYC (B) and FM‐copy‐number alterations (CNA) score (C) determined by quantitative genomic PCR using formalin‐fixed paraffin‐embedded samples.
Table 2
Cox proportional hazard regression analysis for overall and disease‐free survival (OS and DFS) in 92 patients with esophageal squamous‐cell carcinoma
| Factor | OS | DFS | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate† | Univariate | Multivariatea | |||||
| Hazard ratio (95% confidence interval) | P‐value‡ | Model 1 (P‐value)‡ | Model 2 (P‐value)‡ | Hazard ratio (95% confidence interval) | P‐value‡ | Model 1 (P‐value)‡ | Model 2 (P‐value)‡ | |
| Age (years) | ||||||||
| ≥65 versus <65 | 1.395 (0.750–2.597) | 0.2932 | X§ | X§ | 1.092 (0.627–1.905) | 0.7554 | CC | X§ |
| Histopathological grading | ||||||||
| Poor versus well–moderate | 1.372 (0.697–2.703) | 0.3595 | X§ | X§ | 1.416 (0.773–2.597) | 0.2598 | X§ | X§ |
| TNM classification | ||||||||
| pT categories | ||||||||
| pT2–4 versus pT1 | 4.902 (1.916–12.658) | 0.0009* | X§ | X§ | 5.319 (2.262–12.500) | 0.0001* | X§ | X§ |
| pN categories | ||||||||
| pN1 versus pN0 | 7.407 (2.625–20.833) | 0.0002* | X§ | X§ | 3.536 (2.778–15.385) | <0.0001* | X§ | X§ |
| pM categories | ||||||||
| pM1 versus pM0 | 1.319 (0.659–2.646) | 0.4339 | X§ | X§ | 1.520 (0.934–2.469) | 0.2722* | X§ | Xc |
| pStage | ||||||||
| III + IV versus I + II | 5.495 (2.294–13.158) | 0.0001* | 0.0012* | 0.0010* | 5.102 (2.387–10.870) | <0.0001* | 0.0002* | 0.0002* |
| Gain of MYC¶ | ||||||||
| Positive versus negative | 2.410 (1.203–4.831) | 0.0131* | 0.1394 | X§ | 2.088 (1.151–3.788) | 0.0154* | 0.2276 | X§ |
| Loss of FHIT¶ | ||||||||
| Positive versus negative | 2.740 (1.462–5.128) | 0.0016* | 0.0581 | X§ | 2.247 (1.280–3.937) | 0.0048* | 0.0968 | X§ |
| Either gain of MYC or loss of FHIT¶ | ||||||||
| Positive (FM‐CNA score 1 or 2) versus negative (FM‐CNA score 0) | 4.566 (1.779–11.765) | 0.0015* | X§ | 0.0120* | 3.185 (1.543–6.579) | 0.0017* | X§ | 0.0255* |
*Statistically significant values. †Forward‐ and backward‐stepwise analyses were used for multivariate analysis. ‡P‐values are from two‐sided tests and were statistically significant at <0.05. §Not included in multivariate analysis. ¶Copy‐number status was determined by quantitative Gpcr, as in the Materials and Methods. CAN, copy‐number alterations; FHIT, fragile histidine triad.
Discussion
Although surgical techniques and perioperative management have progressed in ESCC, the prognosis for patients with this disease remains poor. This is partly because early lymphogenous and hematogenous micrometastases may occur even in apparently localized cases.15 Combined‐modality approaches, such as primary radical surgery with adjuvant chemotherapy or chemoradiotherapy, primary definitive chemoradiotherapy, or preoperative chemoradiotherapy followed by surgery, are known to be effective for facilitating complete tumor resection and requisite for long‐term survival.16 Indices to enhance the precision of prognostication in patients who undergo resection for ESCC are critical to allow the identification of those who are most likely to benefit from this procedure and, thereby, facilitate the patient selection process and tailoring of combined‐modality approaches to optimize outcomes. A major finding of our study is that loss of FHIT and/or gain of MYC were related to an adverse prognosis in resected ESCC using a test set as well as an independent validation cohort. Because the copy‐number status of FHIT and/or MYC seems to be an independent prognosticator in univariate and multivariate analyses and a relatively accurate predictor for the 5‐year probability of death and recurrence, risk stratification based on this CNA pattern is warranted to select the optimal treatment strategy in resected ESCC patients.
In the present study, we used an in‐house focused BAC array, the MCG Cancer Array‐Mini, to detect CNA of primary ESCC tumor samples by aCGH. The MCG Cancer Array‐Mini was constructed to identify CNA in human cancers using 108 probes (Table S3) containing cancer‐related genes, which are expected to show CNA frequently in human cancers, according to information obtained from the Cancer Gene Census in the Cancer Genome Project (http://www.sanger.ac.uk/genetics/CGP/Census/),17 reports by several groups18, 19 and our own studies (e.g. on LRP1B, SMYD and PAK4).10, 11, 20, 21 As expected, this focused array efficiently detected a total of 344 gains and 124 losses in most of the ESCC tumors (41 of 45 cases). Among those alterations, recurrently detected regions with CNA in ESCC (Table 1) were part of or different from recently reported CNA regions frequently detected by aCGH in ESCC,6, 7 possibly because: (i) those reports used an oligo‐array6 or BAC‐array7 covering the entire genome, suggesting that we missed altered regions outside our probes on MCG Cancer Array‐Mini; and (ii) those reports used commercially available pooled DNA from normal human men as a control for aCGH analyses, suggesting that copy‐number variations frequently observed in recruited populations were detected as false‐positive somatic CNA in tumors. Because (i) our focused BAC array contains regions harboring genes, whose CNA have been reported and which play important roles in carcinogenesis in various human cancers and (ii) we detected true somatic CNA by aCGH using paired tumor and non‐tumorous tissues in each case, regions with CNA recurrently observed in our aCGH analysis in this study seem to be landmarks for genes biologically and clinicopathologically important to the pathogenesis of ESCC. In addition, a real‐time gPCR for the limited number of loci/genes is suitable for clinical application using small clinical samples. Based on our results, therefore, we focused each region/gene instead of whole CAN score (CI status) determined by our focused BAC array as a candidate for prognosticator in ESCC.
FHIT is a tumor suppressor gene that spans the FRA3B common fragile site of 3p14.2, a site that frequently harbors chromosomal aberrations in many tumors, including esophageal cancer. Loss of heterozygosity (LOH) in the FHIT region and reduced or lost FHIT protein expression were observed frequently in ESCC.22 FHIT was frequently methylated in early‐stage as well as advanced‐stage tumors of ESCC and is now considered a major cause of FHIT expressional loss.23, 24 Guo et al.25 report that the p16 and FHIT methylation might be one of the earliest events and an important mechanism for gene silencing in the carcinogenesis of ESCC. Notably, Kuroki et al.23 report that FHIT hypermethylation significantly correlates with LOH at FHIT in ESCC, suggesting FHIT gene alterations, such as methylation and deletion, to cooperatively play a role in the carcinogenesis of ESCC via a two‐hit mechanism for gene inactivation,26 although it remains unknown which alterations occur first and contribute more to the altered expression.27 Although it is still controversial whether loss of FHIT gene expression or methylation status of FHIT is associated with the prognosis for ESCC, our results indicate that the copy‐number status of the FHIT gene might serve as a reasonable and technically stable prognosticator of the progression of ESCC.
The MYC (c‐myc) proto‐oncogene encodes a transcription factor involved in the regulation of normal cellular proliferation, differentiation and apoptosis, and its amplification/overexpression have been shown in various human cancers.28, 29 In the present study, a frequent copy‐number increase in the MYC gene was evident in ESCC, although its clinical relevance as a prognostic marker remains unknown due to very few published reports. Bitzer et al.30 report that MYC amplification has no influence on the overall survival of ESCC patients treated either by surgery alone or by multimodal therapy. Because no association between copy‐number status and expression level of MYC is reported in ESCC,31 it is possible that MYC is not activated through its amplification, and does not exert oncogenic effects. Therefore, as shown in the present study, copy‐number status of MYC might be a good marker for the status of copy‐number instability, which contributes to the activation and inactivation of functional target oncogenes and tumor suppressor genes, respectively, in ESCC cells, and, thereby, is a reasonable indicator of the accumulation of various activated and inactivated genes involved in esophageal carcinogenesis.
In conclusion, the copy‐number status of FHIT and/or MYC is associated with the prognosis of ESCC patient cohorts, and its usefulness as an independent prognosticator for the probability of death and recurrence is confirmed, especially in resected ESCC patient cohorts. Recent trials indicate a survival benefit for preoperative chemoradiotherapy compared to surgery alone in locally advanced esophageal cancer. Therefore, this CNA pattern might be useful for risk stratification to select treatments in resected early stage ESCC patients, although no significant difference of OS and DFS depending on the FM‐CNA status was observed in N0 and stage I ESCC patients in the present study (Fig. S2). To determine the usefulness of this CNA pattern for risk stratification to select treatments in resected ESCC patients, further study using this CNA pattern in resected early stage ESCC patients, who do not need preoperative therapy, as well as in resected locally advanced ESCC patients, who are treated with preoperative therapy or surgery alone, will be needed.
Supporting information
Fig. S1. Pattern of copy‐number alterations (CNA) in 45 cases with esophageal squamous‐cell carcinoma (ESCC) analyzed by array‐based comparative genomic hybridization (aCGH) and Kaplan–Meier curves for overall survival rates of patients with primary ESCC according to chromosomal instability (CI) status.
Fig. S2. Kaplan–Meier curves for overall and disease‐free survival rates of patients with N0 and Stage I esophageal squamous‐cell carcinoma (ESCC) according to FM‐copy‐number alterations (CNA) score.
Table S1. Correlation between clinicopathological characteristics and chromosomal instability status determined by array‐based comparative genomic hybridization (aCGH) in 45 primary cases of esophageal squamous‐cell carcinoma (ESCC).
Table S2. Correlation between clinicopathological characteristics and copy‐number status of MYC and/or FHIT determined by quantitative gPCR in 92 primary cases of esophageal squamous‐cell carcinoma (ESCC).
Table S3. Probes for in‐house bacterial artificial chromosome (BAC) array “MCG Cancer Array‐Mini”.
Table S4. Correlation between clinicopathological characteristics and FM‐copy‐number alterations (CNA) score determined by array‐based comparative genomic hybridization (aCGH) in 45 primary cases of esophageal squamous‐cell carcinoma (ESCC).
Table S5. Correlation between CI status and loss of 3p14.2 (FHIT), gain of 8q24.21 (MYC), or FM‐copy‐number alterations (CNA) score determined by array‐based comparative genomic hybridization (aCGH) in 45 primary cases of esophageal squamous‐cell carcinoma (ESCC).
Acknowledgments
This study was supported in part by: Grants‐in‐aid for Scientific Research (JI and II), Scientific Research on Priority Areas and Innovative Areas (JI), and challenging Exploratory Research (II), and a Global Center of Excellence Program for International Research Center for Molecular Science in Tooth and Bone Diseases from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (JI); a Health and Labour Sciences Research Grant for the Ministry of Health, Labour and Welfare, Japan; and a grant from the New Energy and Industrial Technology Development Organization (JI). We thank Dr Shigeo Haruki for the collection of the specimens and useful information, and Ayako Takahashi and Rumi Mori for technical assistance.
References
Articles from Cancer Science are provided here courtesy of Wiley
Full text links
Read article at publisher's site: https://doi.org/10.1111/j.1349-7006.2012.02329.x
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/j.1349-7006.2012.02329.x
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1111/j.1349-7006.2012.02329.x
Article citations
Advances in prognostic biomarkers for esophageal cancer.
Expert Rev Mol Diagn, 19(2):109-119, 30 Dec 2018
Cited by: 8 articles | PMID: 30582379
Review
Overexpression of Myosin Phosphatase Target Subunit 1 (MYPT1) Inhibits Tumor Progression and Metastasis of Gastric Cancer.
Med Sci Monit, 24:2508-2517, 24 Apr 2018
Cited by: 5 articles | PMID: 29687789 | PMCID: PMC5937360
Genome-wide screening for genomic aberrations in Kazakh patients with esophageal squamous cell cancer by comparative genomic hybridization.
Int J Clin Exp Pathol, 11(1):427-437, 01 Jan 2018
Cited by: 1 article | PMID: 31938128 | PMCID: PMC6957937
Identification of abnormal nuclear and mitochondrial genes in esophageal cancer cells.
Mol Med Rep, 17(2):2817-2826, 08 Dec 2017
Cited by: 2 articles | PMID: 29257265 | PMCID: PMC5783496
The ubiquitous 'cancer mutational signature' 5 occurs specifically in cancers with deleted FHIT alleles.
Oncotarget, 8(60):102199-102211, 06 Nov 2017
Cited by: 12 articles | PMID: 29254236 | PMCID: PMC5731946
Go to all (17) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Copy number loss of FBXW7 is related to gene expression and poor prognosis in esophageal squamous cell carcinoma.
Int J Oncol, 41(1):253-259, 19 Apr 2012
Cited by: 37 articles | PMID: 22576686
Aberrant methylation of Fragile Histidine Triad gene is associated with poor prognosis in early stage esophageal squamous cell carcinoma.
Eur J Cancer, 42(7):972-980, 27 Mar 2006
Cited by: 41 articles | PMID: 16564166
[Allelic loss and down-regulation of FHIT gene expression in esophageal squamous cell carcinoma].
Ai Zheng, 23(9):992-998, 01 Sep 2004
Cited by: 3 articles | PMID: 15363189
Reduced acetylated histone H4 is associated with promoter methylation of the fragile histidine triad gene in resected esophageal squamous cell carcinoma.
Ann Thorac Surg, 82(2):396-401; discussion 401, 01 Aug 2006
Cited by: 10 articles | PMID: 16863736
 1
,
5
,
6
and
1
,
5
,
6
and