Abstract
Free full text

Glucose deprivation–induced aberrant FUT1-mediated fucosylation drives cancer stemness in hepatocellular carcinoma
Abstract
Rapidly growing tumors often experience hypoxia and nutrient (e.g., glucose) deficiency because of poor vascularization. Tumor cells respond to the cytotoxic effects of such stresses by inducing molecular adaptations that promote clonal selection of a more malignant tumor-initiating cell phenotype, especially in the innermost tumor regions. Here, we report a regulatory mechanism involving fucosylation by which glucose restriction promotes cancer stemness to drive drug resistance and tumor recurrence. Using hepatocellular carcinoma (HCC) as a model, we showed that restricted glucose availability enhanced the PERK/eIF2α/ATF4 signaling axis to drive fucosyltransferase 1 (FUT1) transcription via direct binding of ATF4 to the FUT1 promoter. FUT1 overexpression is a poor prognostic indicator for HCC. FUT1 inhibition could mitigate tumor initiation, self-renewal, and drug resistance. Mechanistically, we demonstrated that CD147, ICAM-1, EGFR, and EPHA2 are glycoprotein targets of FUT1, in which such fucosylation would consequently converge on deregulated AKT/mTOR/4EBP1 signaling to drive cancer stemness. Treatment with an α-(1,2)-fucosylation inhibitor sensitized HCC tumors to sorafenib, a first-line molecularly targeted drug used for advanced HCC patients, and reduced the tumor-initiating subset. FUT1 overexpression and/or CD147, ICAM-1, EGFR, and EPHA2 fucosylation may be good prognostic markers and therapeutic targets for cancer patients.
Introduction
Aberrant glycosylation is a hallmark of cancer and is not only a consequence but also a driver of malignant phenotypes, directly impacting key processes supporting tumor progression and metastasis, including cell adhesion, motility, invasion, and immune evasion (1–3). Glycosylation is controlled by the actions of glycosyltransferases and glycosidases on glycoproteins and/or lipids through the addition of oligosaccharides. Among approximately 10 kinds of oligosaccharide modifications, fucosylation is one of the most important types in cancer. Fucosylation, which consists of the transfer of a fucose residue to oligosaccharides and proteins, is regulated by many types of molecules, including fucosyltransferases (FUTs). Aberrant expression of FUTs and changes in fucosylation of glycoproteins are associated with various important cancer processes. Glycomics, the systematic study of glycans and glycan-binding proteins, is an emerging field in the postgenomic and postproteomic era and an area that remains largely understudied in the context of cancer biology.
Rapidly growing tumors often experience hypoxia and nutrient (e.g., glucose) deficiency because of the poor vascular supply. Tumor cells respond to the cytotoxic effects of such metabolic stresses by inducing molecular adaptations that promote survival rather than a proliferative response, resulting in clonal selection of a more malignant phenotype, especially in the innermost tumor regions (4, 5). The core of the bulk tumor is believed to be poorly vascularized, with glucose levels decreasing from the tumor periphery to the interior. Early studies in rats with tumor xenografts showed very low glucose content in tumor interstitial fluid (0.12 ± 0.10 g/L) compared with subcutaneous interstitial fluid (1.20 ± 0.20 g/L) and aortic serum (1.72 ± 0.06 g/L) (6). Hepatocellular carcinoma (HCC), a primary liver cancer, is characterized by rapid tumor growth resulting in large tumor masses. However, advanced infiltrative HCCs seldom show hypervascularity, suggesting that this type of cancer cell can generate signals that enable it to survive in nutrient-deprived conditions. Common palliative HCC therapies, including hepatic artery ligation (HAL) and transcatheter arterial (chemo) embolization (TAE/TACE), which are initially intended to restrict HCC growth through blood (nutrient) supply obstruction, often result in therapeutic resistance and tumor recurrence. Indeed, accumulating evidence has demonstrated the critical role of the tumor microenvironment in maintaining the immature phenotype of tumor-initiating cells (TICs) to promote cancer (7). In HCC and other solid tumors, liver TIC subsets and TIC functional readouts, including sphere formation and tumor-initiating potential, are all enriched in microenvironmental conditions associated with poor vascular supply (8, 9). Understanding the mechanisms that drive cancer stemness, which we now know represents the root of therapy failure and tumor recurrence, is fundamental to the design of improved therapeutic strategies.
In this study, we found that low glucose availability, commonly observed in the microenvironment of large growing tumors such as HCC, could enrich for a TIC phenotype and enhance PERK-mediated ATF4 expression to drive fucosyltransferase 1 (FUT1) expression, which work hand-in-hand to promote tumor initiation and drug resistance. Glucose-restriction stress activates the PERK-mediated unfolded protein response (UPR) to enhance the expression of ATF4, which then binds to the FUT1 promoter to drive its transcriptional activity. Blocking FUT1 in cancer cells strongly restrained HCC tumor initiation and self-renewal in vivo. More excitingly, inhibition of α-(1,2) fucosylation by a specific inhibitor of fucoglycoprotein synthesis, 2-deoxy-D-galactose (2DGal), significantly improved the efficiency and efficacy of the molecularly targeted drug sorafenib ex vivo and in vivo. Finally, integrated fucosylated-peptide screening and protein profiling revealed fucosylation of membrane-bound proteins, including intercellular adhesion molecule 1 (ICAM-1), epidermal growth factor receptor (EGFR), CD147, and ephrin type A receptor 2 (EPHA2), by FUT1 to drive cancer stemness via altered AKT/mTOR/4EBP1 signaling. Our study has 3 important findings. First, we disclose a mechanism by which glucose restriction–mediated FUT1 overexpression in HCC enhances cancer stemness and drug resistance. Second, we highlight the important role of posttranslational modification by FUT1-mediated fucosylation in promoting HCC. Third, we showed as a proof of concept a therapeutic approach that shows synergism with molecularly targeted therapy in HCC, the second deadliest cancer that to date has no promising curative treatments.
Results
Glucose restriction promotes a liver TIC phenotype.
To determine whether glucose restriction influences the TIC phenotype, we exposed HCC cells to media containing a high (4.5 g/L) or restricted/low (0.45 g/L) concentration of glucose. The clinical equivalent of the glucose concentration in medium is 450 mg/dL, which is above the physiological range (70–144 mg/dL). Early studies in rats with tumor xenografts detected very low glucose levels in tumor interstitial fluid (0.12 ± 0.10 g/L) compared with subcutaneous interstitial fluid (1.20 ± 0.20 g/L) and aortic serum (1.72 ± 0.06 g/L) (6). Together, these results indicate that the “restricted” glucose concentration used for the present study is representative of physiological conditions, whereas high-glucose cell culture conditions correspond to severe hyperglycemia. As TICs are best defined functionally, we first examined the effects of low glucose using an in vitro limiting-dilution assay. Huh7 and CLC13 HCC cells cultured under low glucose have an increased capacity to form tumor spheres, a phenotype associated with self-renewal and poorer clinical outcomes in HCC. The frequency of TICs capable of forming spheres increased 4-fold after culture under glucose restriction (Figure 1A). As the ability to propagate tumors in vivo remains the gold standard for TIC function, we next determined whether culture in low glucose influenced tumorigenic potential using a limiting-dilution and serial transplantation subcutaneous xenograft model. Low-glucose culture significantly increased in vivo tumor growth relative to high-glucose culture, as demonstrated by increased tumor incidence, expedited tumor latency, and enhanced frequency of TICs (Figure 1B and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI143377DS1). Using flow cytometry, we also observed a significant increase in the percentage of cells with putative liver TIC immunophenotypes, including the cell surface marker CD133 and aldehyde dehydrogenase (ALDH) activity, after exposure to low glucose (Supplemental Figure 1). Immunohistochemistry of harvested in vivo tumors formed with HCC cells cultured in high or low glucose concentrations, shown in Figure 1B, similarly showed enhanced CD133 and ALDH1L1 expression in HCC tumors formed with HCC cells cultured in low glucose (Supplemental Figure 2). Collectively, these data demonstrated that glucose deprivation promotes a liver TIC phenotype.

Huh7 and CLC13 HCC cells were cultured in high (4.5 g/L) or restricted/low (0.45 g/L) glucose. (A) In vitro limiting-dilution assays showed that the frequency of tumor-initiating cells increased 4-fold after culturing in low glucose (pairwise tests for differences in stem cell frequencies). (B) In vivo limiting-dilution assays found that the cells cultured in low glucose displayed an enhanced tumor incidence, expedited tumor latency, and a higher frequency of tumor-initiating cells (primary implantation, n = 15 per group; secondary implantation, n = 5 per group). (C) Strategy for mRNA profiling to identify altered transcriptomes of HCC cells grown in high- or low-glucose conditions. (D) Gene set enrichment analysis (GSEA) of differentially expressed genes identified by RNA-seq found that the PERK-mediated unfolded protein response was highly enriched in HCC cells cultured under low-glucose conditions. (E) Western blot analysis also found that GRP78, p-PERK, p-eIF2α, ATF4, and FUT1 levels were enhanced in glucose-restricted conditions and that the addition of 1 μM PERK inhibitor (PERKi) for 48 hours reversed the expression of p-PERK, p-eIF2α, ATF4, and FUT1. HG, high glucose; LG, low glucose. (F) In vitro limiting-dilution assays showed that the frequency of tumor-initiating cells increased after culture in low glucose and decreased when HCC cells cultured in low glucose were treated with PERKi (1 μM) (pairwise tests for differences in stem cell frequencies). The data shown in A, E, and F are representative of 3 independent experiments. CSC, cancer stem cell; FDR, false discovery rate. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To determine the biological mechanism through which glucose deprivation enriches liver TICs, we performed RNA sequencing (RNA-seq) to compare the mRNA profiles of HCC cells cultured under high- or low-glucose conditions (Figure 1C). We subjected differentially expressed genes, defined by a fold-change cutoff of greater than 5, to DAVID Gene Ontology (GO) analysis and gene set enrichment analysis (GSEA), where the PERK-mediated UPR was found to be significantly enriched when cells were cultured under low glucose (Figure 1D and Supplemental Figure 3). Consistently, the levels of critical molecular players in the PERK-mediated UPR signaling cascade, including GRP78, p-PERK, p-eIF2α, and ATF4, were significantly enhanced under glucose restriction (Figure 1E), as demonstrated by Western blot analysis. The addition of the PERK inhibitor (PERKi) GSK2656157 to cells cultured under glucose restriction reversed this change in p-PERK, p-eIF2α, and ATF4 expression, confirming that PERK, eIF2α, and ATF4 are important effectors of PERK-mediated UPR signaling. The addition of the PERKi to HCC cells cultured under low glucose also attenuated the ability of cells to initiate tumors and self-renew, as demonstrated by in vitro limiting-dilution spheroid formation assays (Figure 1F). These data demonstrated that in response to glucose restriction, HCC tumors activate the cellular stress response PERK-mediated UPR signaling pathway to enhance ATF4.
The PERK-mediated UPR activates FUT1 promoter activity via ATF4 induction under glucose restriction.
Further analysis of our RNA-seq profiling data identified the top 3 most significantly enhanced genes (FUT1, heat shock protein family A member 6 [HSPA6], and fosB proto-oncogene, AP-1 transcription factor subunit [FOSB]) when cells were cultured under glucose restriction. Two of these harbor ATF4 binding sites on their promoter regions (Figure 2A). With ATF4 being an important downstream transcription factor of PERK-mediated UPR signaling, we hypothesized that PERK-UPR-ATF4 induction under glucose restriction would enhance the liver TIC phenotype through HSPA6, FUT1, or FOSB. Analysis of FUT1 and ATF4 mRNA levels in The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC) data set revealed that ATF4 expression was positively correlated with FUT1 (Figure 2B) but not FOSB expression (Supplemental Figure 4). Notably, HSPA6, which was also overexpressed when HCC cells were cultured under low glucose, also showed a positive correlation with FUT1 expression in HCC (Supplemental Figure 4), although it did not harbor an ATF4 binding site and thus was not further analyzed. In addition to the TCGA LIHC data set, we further confirmed our observations by qPCR, where we found FUT1 to be significantly enriched by approximately 8-fold when the HCC cell lines Huh7 and CLC13 and HCC patient–derived spheroids were cultured under glucose restriction (Figure 2C). Likewise, in vivo HCC tumors formed by injecting HCC cells treated with low glucose also showed elevated FUT1 expression compared with that of HCC cells treated with high glucose (Supplemental Figure 2). Notably, the expression of FUT2, a close family member of FUT1, was not altered when cells were cultured under the same conditions, suggesting the importance of FUT1 alone in this process (Supplemental Figure 5).

(A) A volcano plot revealed the top 3 most significantly enhanced genes when HCC cells were cultured in low glucose. (B) Analysis of FUT1 and ATF4 expression in The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC) database (unpaired Student’s t test and Pearson’s correlation test). (C) FUT1 expression in low-glucose culturing conditions was validated by qPCR in Huh7 and CLC13 cells and HCC patient–derived spheroids (unpaired Student’s t test). The data shown are representative of 3 independent experiments. (D) Confirmation of ATF4 binding to both predicted sites on FUT1 by ChIP-qPCR using anti-ATF4 and control IgG antibodies (unpaired Student’s t test and 1-way ANOVA). The data shown are representative of at least 3 independent experiments. (E) Luciferase reporter assays found that only ATF4 binding site 2 (Δ3′) was critical in modulating FUT1 transcriptional activity and FUT1 transcriptional activity was upregulated under glucose-deprived conditions in Huh7 and CLC13 HCC cells. ATF4 expression was induced by low-glucose treatment (unpaired Student’s t test and 1-way ANOVA). The data shown are representative of 3 independent experiments. HG, high glucose; LG, low glucose; FL, full-length. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To determine whether ATF4 directly regulates FUT1 expression, we assessed the binding of ATF4 to the FUT1 promoter, which is predicted to contain 2 consensus binding sites (at –74 to –102 and –533 to –591 relative to the transcription start site) (Supplemental Figure 6A). Chromatin immunoprecipitation (ChIP) of ATF4 followed by qPCR with 2 different sets of primers spanning the 2 putative ATF4 binding sites in the FUT1 promoter showed a 4- to 6-fold enrichment in the FUT1 signal over ChIP with nonspecific IgG in both Huh7 and CLC13 HCC cells (Figure 2D). To confirm the ability of ATF4 to control FUT1 expression, we tested the effects of introducing full-length ATF4 with one or both putative ATF4 binding sites deleted on luciferase activity (with luciferase expression driven by the FUT1 promoter). Although ATF4 bound to both predicted binding sites, we observed only a significant reduction in luciferase activity when the predicted ATF4 binding site close to the 3′ end (i.e., site 2) was deleted, suggesting that only 1 of the 2 predicted ATF4 binding sites is critical in controlling FUT1 transcription (Supplemental Figure 6B and Figure 2E). We also tested the level of FUT1 promoter activity under glucose-deprived conditions, and consistently, an increase in luciferase activity was observed in both Huh7 and CLC13 cells treated with glucose-restricted medium (Figure 2E). ATF4 suppression in HCC cells cultured under low glucose consistently led to a reduction in FUT1 expression in both Huh7 and CLC13 cells (Supplemental Figure 7). Notably, GRP78, p-PERK, p-eIF2α, ATF4, and FUT1 showed a stepwise increase in expression when going from high (hyperglycemic; 4.5 g/L) to medium (physiological; 1 g/L) to low (hypoglycemic; 0.45 g/L) glucose concentrations (Supplemental Figure 8A), while the addition of the PERKi reversed the change in FUT1 expression (Figure 1D). Culturing the immortalized normal liver cell line MIHA in high or low glucose did not alter ATF4 or FUT1 expression, suggesting that the observed phenomenon was HCC specific (Supplemental Figure 8B). Overall, our data identified PERK-mediated UPR-driven ATF4 as a key regulator of FUT1 transcriptional induction in HCC under glucose-restriction stress. Notably, we also cultured Huh7 and CLC13 HCC cells under normoxic and hypoxic conditions, but ATF4 and FUT1 expression was not altered, suggesting that PERK-mediated UPR activation of FUT1 promoter activity via ATF4 induction is not relevant to the hypoxic tumor microenvironment (Supplemental Figure 9).
FUT1 overexpression is tightly associated with aggressive clinical features. FUT1 encodes the FUT1 enzyme, which is responsible for the addition of fucose to glycans via the α-(1,2) linkage, resulting in the formation of Lewis and H antigens. To confirm the clinical relevance of FUT1 expression, we interrogated publicly available HCC data sets in which we could compare FUT1 expression in specimens with different tumor grades and survival oucomes (GSE14520, TCGA LIHC; ref. 10). FUT1 mRNA expression was significantly elevated in HCC tumor tissues compared with nontumor liver or normal liver tissues in both data sets (Figure 3A). High FUT1 expression was also correlated with the presence of multinodular tumors, advanced tumor stage (nontumor liver vs. stage III), and worse overall survival (Figure 3, B–D). Immunohistochemical analysis of 61 paired primary HCC and corresponding nontumor liver tissues also showed that the FUT1 protein was overexpressed in most HCC samples examined (Figure 3E and Supplemental Table 2). Analysis of the GSE109211 data set also found that HCC patients with low FUT1 and ALDH1L1 expression were more responsive to sorafenib treatment, suggesting that FUT1 together with the cancer stem cell (CSC) marker ALDH1L1 may be a predictive marker for the response to sorafenib in the clinic (Supplemental Figure 10).
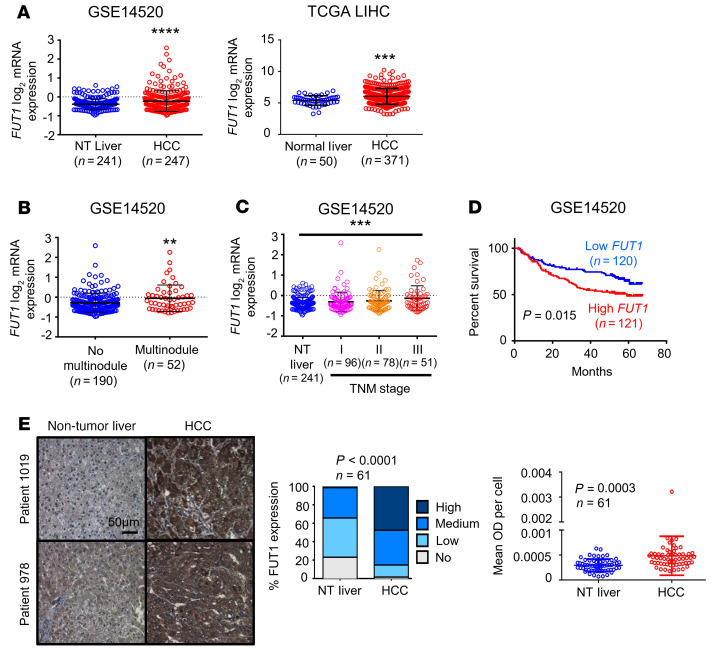
(A) Box-and-whisker plot analysis of FUT1 mRNA levels in nontumor (NT) liver or normal liver and HCC tissues using information gathered from the GSE14520 data set from the NCBI Gene Expression Omnibus (GEO) (left) or The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC) database (right) revealed FUT1 to be frequently overexpressed in HCC (unpaired Student’s t test). (B–D) FUT1 overexpression in HCC correlated significantly with multinodular tumors (B), advanced TNM stage (C), and worse overall survival (D) (unpaired Student’s t test, 1-way ANOVA, and Kaplan-Meier survival curve using the log-rank test, respectively). (E) FUT1 immunostaining on a tissue microarray comprising 61 paired NT liver and HCC tissue samples found FUT1 to be frequently overexpressed in HCC. Representative images of immunostaining in 2 patients are shown. Scale bar: 50 μm. The bar graph shows the percentage of cases displaying no, low, medium, and high staining intensity of FUT1 in paired NT and HCC samples (Fisher’s exact test). The dot plot illustrates the quantification of FUT1 by ImageJ (NIH), with both the intensity (mean OD) and number of cells according to the number of nuclei taken into consideration (paired Student’s t test). Mean OD per cell indicates mean OD relative to the number of nuclei. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Knockdown of endogenous Fut1 expression in the liver of an immunocompetent HCC mouse model attenuates hepatocarcinogenesis.
To investigate the causative relationship between FUT1 overexpression and the functional role of FUT1 in HCC, we performed endogenous knockdown of Fut1 expression in an immunocompetent mouse model. Using the hydrodynamic tail vein injection (HTVI) delivery approach, we induced HCC tumors in C57BL/6 mice using a combination of the activated forms of the myristylated AKT (myr-AKT) and N-RasV12 proto-oncogenes (NRAS+AKT) and sleeping beauty (SB) transposase (Figure 4A). A stepwise increase in Fut1 mRNA expression was noted from harvested livers of the mice injected with the empty vector (EV) control alone from early (1 to 2 weeks after injection) to advanced HCC (3 to 5 weeks after injection) to the endpoint, at which time the mice died naturally (6 to 8 weeks after injection) (Figure 4B). Knockdown of endogenous Fut1 expression delivered via tail vein injection of lentiviral particles significantly attenuated the liver to body weight ratio as well as the number and size of tumor nodules compared with those of the nontargeting shRNA control (shNTC) (Figure 4, C and D, and Supplemental Figure 11). Notably, the liver-free body weight was not found to be significantly different between the 2 groups, excluding data bias due to tumor burden–induced cachexia (Supplemental Figure 12). Immunohistochemical analysis of resected tumors showed a marked decrease in the expression of FUT1, the liver CSC markers CD133 and ALDH1L1, and the proliferation marker PCNA in the Fut1-knockdown group (Supplemental Figure 13). The apoptotic marker cleaved caspase 3 did not exhibit any expression in either the control or shFut1 groups and TUNEL staining was negative, suggesting that FUT1 does not affect spontaneous HCC cell death (Supplemental Figure 13). Subsequent ex vivo limiting-dilution assays using cells isolated from the liver of this HCC mouse model demonstrated that the frequency of TICs capable of forming spheres also decreased upon Fut1 knockdown (Figure 4E). These findings support a key role of FUT1 in promoting HCC growth and self-renewal.
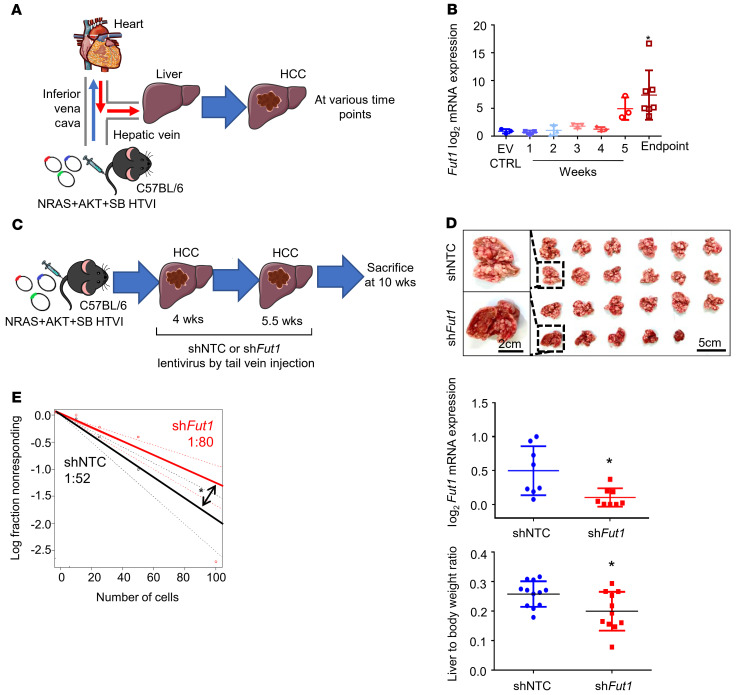
(A) Schematic representation of the hydrodynamic tail vein injection (HTVI) model in C57BL/6 mice. (B) qPCR analysis of Fut1 expression in mice that received HTVI of either empty vector control (EV CTRL) or NRAS, AKT, and sleeping beauty (SB) transposase for HCC induction with samples collected at various time points. n = 22 in total (EV, n = 3; 1 week, n = 3; 2 weeks, n =3; 3 weeks, n = 3; 4 weeks, n = 3; 5 weeks, n = 3; endpoint, n = 7) (1-way ANOVA). (C) Strategy for testing the functional significance of Fut1 in hepatocarcinogenesis. NRAS, AKT, and SB transposase were delivered by HTVI for HCC induction. Lentiviral particles with nontargeting control shRNA (shNTC) or shFut1 were administered twice at 4 and 5.5 weeks. Mice were sacrificed at 10 weeks after plasmid injection. (D) Representative images of dissected livers at the end of the experiment. Scale bars: 5 cm and 2 cm (enlarged images). Successful Fut1 knockdown confirmed by qPCR. n = 11–12 per group. (E) An ex vivo limiting-dilution assay of HCC tumor cells harvested from HTVI mouse models found that the frequency of tumor-initiating cells decreased in the mice administered shFut1 lentiviral particles. The data shown are representative of 3 independent experiments (pairwise tests for differences in stem cell frequencies). *P < 0.05.
FUT1 overexpression is important in mediating growth, self-renewal, and drug resistance in HCC cells.
To functionally demonstrate the importance of FUT1 in TIC maintenance, we used a lentivirus-based approach to stably overexpress FUT1 in Huh7 and CLC13 HCC cells. Efficient FUT1 overexpression was confirmed by Western blot analysis (Figure 5A). Note that manipulation of FUT1 expression did not affect FUT2 expression, suggesting a specific effect of FUT1 on HCC cells (Supplemental Figure 14). FUT1 overexpression resulted in a 2.5- to 7-fold increase in the frequency of TICs, as demonstrated by an in vitro limiting-dilution assay (Figure 5B). Overexpression of FUT1 also resulted in an enhanced ability of the cells to resist the molecularly targeted therapeutic sorafenib, as demonstrated by a decrease in the number of dead cells measured by annexin V/propidium iodide (PI) flow cytometry (Figure 5C). As a complementary approach, we also used lentivirus-based targeting to express shRNAs to reduce FUT1 in Huh7 cells (shFUT1 clones 544 and 565) and observed the opposite functional phenomena (Figure 5, B and C). Of note, FUT1 suppression completely reversed the enhanced self-renewal ability induced by culturing cells in low glucose (Figure 5, B and C). To further determine whether FUT1 overexpression resulted in enhanced tumor propagation in vivo, we subcutaneously implanted HCC cells with or without FUT1 overexpression into immunocompromised NOD/SCID mice. Overexpression of FUT1 resulted in a profound increase in the ability of cells to initiate tumor growth. FUT1 overexpression in HCC cells also resulted in increased tumor incidence, expedited tumor latency, and a higher frequency of TICs. HCC cells with high FUT1 also exhibited significantly worse tumor-free survival than HCC cells without FUT1 overexpression (Figure 5D and Supplemental Table 3).
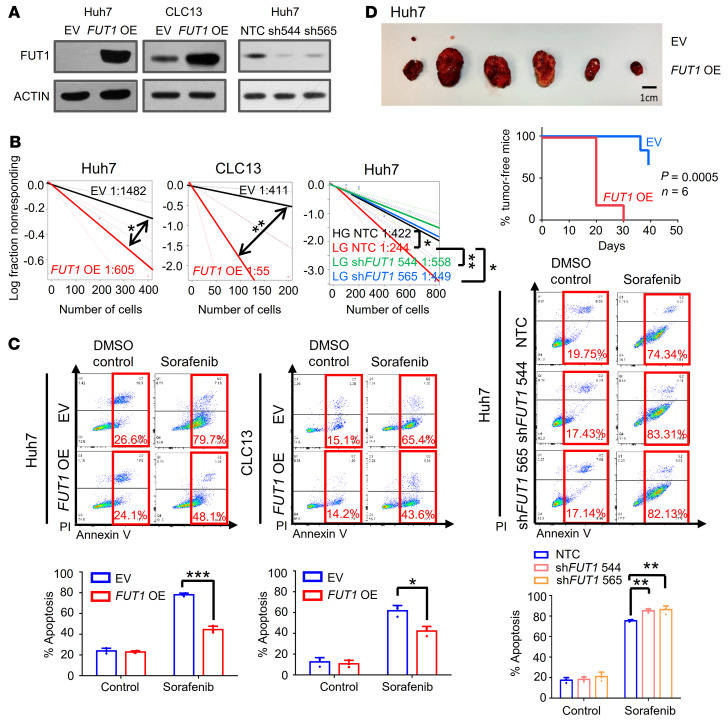
(A) Validation of FUT1 overexpression or knockdown in Huh7 and CLC13 HCC cells by Western blotting. (B) An in vitro limiting-dilution assay revealed that FUT1 overexpression and knockdown promoted and attenuated the frequency of tumor-initiating cells, respectively (pairwise tests for differences in stem cell frequencies). (C) Annexin V/PI analysis found that FUT1 overexpression augmented apoptosis in Huh7 cells treated with 4 μM sorafenib (72 hours) and CLC13 cells treated with 4 μM sorafenib (48 hours). DMSO (2%) was used as a control (1-way ANOVA). The reverse phenotype was observed in a Huh7 FUT1-knockdown model (4 μM sorafenib, 72 hours). (D) FUT1 overexpression promoted HCC tumor growth in an in vivo subcutaneous implantation model. Representative image of tumors harvested from mice injected with 5000 cells. Scale bar: 1 cm. Kaplan-Meier curve showing a significant decrease in the percentage of tumor-free survival upon FUT1 overexpression (n = 6). In vivo limiting-dilution assays found that FUT1 overexpression enhanced tumor incidence, expedited tumor latency, and increased the frequency of tumor-initiating cells (pairwise tests for differences in stem cell frequencies and Kaplan-Meier survival curve using the log-rank test). The data shown in A–C are representative of 3 independent experiments. NTC, nontargeting control; OE, overexpression. *P < 0.05; **P < 0.01; ***P < 0.001.
Inhibition of α-(1,2) fucosylation by 2DGal increases the efficacy of sorafenib and eradicates TICs.
In light of the functional and clinical significance of FUT1 in mediating the TIC phenotype, we then speculated whether targeting FUT1 would be beneficial in the treatment of HCC. As a proof of concept, we tested the effects of inhibiting α-(1,2) fucosylation with 2DGal on the ability of sorafenib to suppress HCC proliferation and self-renewal (11). HCC cells stably overexpressing FUT1 were subjected to 2DGal and sorafenib treatment. While FUT1 overexpression enhanced the ability of the cells to resist sorafenib, combination treatment with 2DGal reversed this protective effect and sensitized cells to sorafenib (Figure 6, A and B). Treatment of HCC cells cultured under low glucose with 2DGal also reversed the TIC frequency, as demonstrated by an in vitro limiting-dilution spheroid assay (Figure 6C). We also extended our study to a more physiological setting, utilizing organotypic ex vivo culture of HCC tumor tissues where the pathophysiology of the original tumor is better preserved than that of cell lines. Note that all 3 HCC patient–derived organoids (HCC-HK P1, HCC-HK P2, and HCC10) have been thoroughly characterized, either in-house or by our collaborator, at both the molecular and phenotypic levels, with comparisons made against the original tissue samples (12). HCC-HK P2 and HCC10 both tested positive for FUT1 expression, while HCC-HK P1 did not express FUT1 (Figure 6D). The treatment efficacy of sorafenib and 2DGal was evaluated in ex vivo cultures of HCC patient–derived organoids treated with either sorafenib or 2DGal alone or the combination of both. In the FUT1-expressing HCC-HK P2 and HCC10 cells, combination treatment resulted in the most significant reduction in tumor growth, where 2DGal treatment was found to sensitize HCC cells to sorafenib (Figure 6E). Conversely, in the FUT1-deficient HCC-HK P1 cells, combination treatment did not further enhance the sorafenib response (Figure 6E). Additional analyses were performed to discern the in vivo benefit of their combinatorial effect in HCC in the NRAS+AKT HTVI immunocompetent HCC mouse model, where we found Fut1 to be overexpressed (Figure 4B). Sorafenib and/or 2DGal were administered following HCC formation at 5–7 weeks. At the endpoint, HCC tumor cells were harvested for flow cytometric cell sorting to capture live cells for the ex vivo limiting-dilution assay (Figure 6F). Although sorafenib treatment alone led to a decrease in the frequency of TICs, combination treatment led to an even more significant decrease in the self-renewal ability of the cells compared with the control (Figure 6G), suggesting that 2DGal sensitizes HCC cells to sorafenib and preferentially eradicates the TIC subset of the tumor. FUT1 is a key rate-limiting enzyme in the synthesis of Lewis Y, a membrane-associated carbohydrate antigen (13, 14). Hence, we used the Lewis Y antigen as a readout of the level of FUT1 modification on its target proteins. Immunohistochemical analysis of Lewis Y expression in the resected livers of the 2DGal and combination treatment groups showed a decrease in expression, suggesting that 2DGal effectively impaired the synthesis of Lewis Y antigen on cell-surface proteins (Supplemental Figure 15). Collectively, these data demonstrated the potential of utilizing 2DGal to enhance the efficiency and efficacy of sorafenib and to preferentially eradicate the TIC subset of HCC for long-term tumor eradication.

(A) Schematic representation of the effects of 2DGal and sorafenib on Huh7 HCC cell proliferation (10 mM 2DGal and 4 μM sorafenib) and HCC patient-derived organoids (HCC-HK P1 and HCC-HK P2, 10 mM 2DGal and 2 μM sorafenib; HCC10, 10 mM 2DGal and 4 μM sorafenib) after 72 hours. (B) CellTiter Glo analysis found that Huh7 cells with FUT1 overexpression (OE) responded to a combination of 2DGal and sorafenib more significantly than either drug alone when compared with the empty vector (EV) control (1-way ANOVA). (C) In vitro limiting-dilution assays of Huh7 cells cultured in low glucose treated with 2DGal (10 mM) (pairwise tests for differences in stem cell frequencies). (D) Western blot analysis of FUT1 expression in HCC-HK P1, HCC-HK P2, and HCC10. (E) CellTiter Glo analysis found that FUT1-expressing HCC organoids (HCC-HK P2 and HCC10) responded to a combination of 2DGal and sorafenib more significantly than either drug alone when compared with HCC-HK-P1 (1-way ANOVA). (F) Strategy for testing the effects of 2DGal and sorafenib in NRAS+AKT+SB HTVI–driven HCC immunocompetent mouse models. (G) Ex vivo limiting-dilution assay of HCC tumor cells harvested from the HTVI mouse models (pairwise tests for differences in stem cell frequencies). The data shown in B–E and G are representative of 3 independent experiments. 2DGal, 2-deoxy-D-galactose; LDA, limiting dilution assay; SB, sleeping beauty; HTVI, hydrodynamic tail vein injection; Combo, combination. *P < 0.05; **P < 0.01; ****P < 0.0001. NS, not significant.
CD147, ICAM-1, EGFR, and EPHA2 are mediators of FUT1 that regulate cancer stemness via the AKT/mTOR/4EBP1 signaling axis.
For determination of the downstream signaling caused by FUT1 deregulation, HCC cells transduced with shFUT1 compared with shNTC or the FUT1 overexpression vector and the EV control were subjected to reversed-phase protein array (RPPA) functional proteomics analysis, measuring the levels of 436 key cancer-related proteins. A heatmap of proteins in enriched pathways with greater than 20% difference in expression between shNTC and FUT1-knockdown cells or between EV-treated and FUT1-overexpressing cells is depicted in Figure 7A. Consistently, analysis of TCGA LIHC data revealed that high-FUT1-expressing HCC tumors were positively correlated with PI3K/AKT/mTOR signaling activation (Figure 7B). We successfully confirmed enhanced AKT/mTOR signaling, as evident by p-AKT (Ser473), p-mTOR, and p-4EBP1 expression, in HCC cells cultured in low glucose or when overexpressing FUT1 (Figure 7C). A decrease in the p-AKT, p-mTOR, and p-4EBP1 levels was also consistently evident when FUT1 expression was stably suppressed in HCC cells (Figure 7C). To further demonstrate the significance of AKT signaling in FUT1-driven HCC, we performed a functional rescue experiment in which the addition of an AKT inhibitor to HCC cells cultured in low glucose reversed the TIC frequency (Supplemental Figure 16) to levels comparable to those in HCC cells cultured under high glucose. Addition of an AKT inhibitor could also reverse FUT1-mediated sorafenib resistance in HCC cells (Supplemental Figure 17). Consistently, we also observed a significant decrease in Lewis Y antigen staining and, most importantly, p-AKT (Ser473) staining in tumors harvested from HTVI-induced HCC with endogenous knockdown of Fut1 compared with shNTC (Supplemental Figure 18) and in vivo HCC tumors formed by injecting HCC cells treated with low glucose (Supplemental Figure 2), further reinforcing our in vitro observations in an in vivo model. Additionally, treatment of the FUT1-overexpressing HCC cells with 2DGal not only decreased FUT1 expression and Ulex europaeus agglutinin 1 (UEA-1) binding but also attenuated AKT/mTOR/4EBP1 signaling activation (Figure 7D).
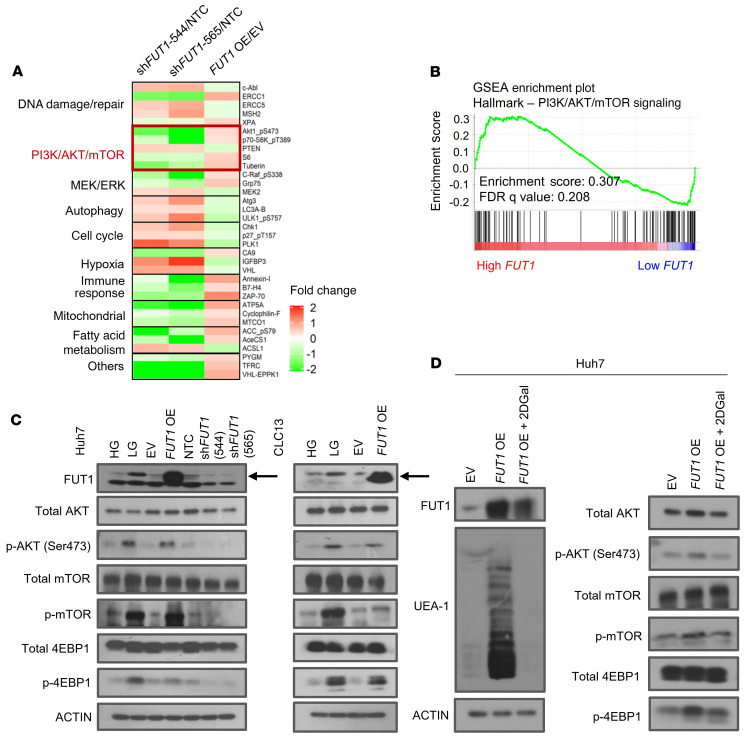
(A) A reversed-phase protein array was performed using lysates of Huh7 HCC cells transfected with shNTC or shFUT1 (clones 544 and 565) or transfected with empty vector control or FUT1 overexpression vector. A heatmap of differentially expressed proteins is shown. (B) GSEA of high-FUT1-expressing HCC (top 50%) and low-FUT1-expressing HCC (bottom 50%) with data extracted from The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC) database showing that high FUT1 expression was significantly correlated with PI3K/AKT/mTOR signaling. (C) Western blot analysis shows enhanced p-AKT, p-mTOR, and p-4EBP1 levels in HCC cells treated with low glucose or when transduced with FUT1-overexpressing plasmid in both Huh7 and CLC13 HCC cells. Decreases in p-AKT, p-mTOR, and p-4EBP1 expression were also observed when Huh7 cells had suppressed FUT1 expression. (D) Western blot analysis shows that treatment of the FUT1-overexpressing cells with 2DGal (10 mM) attenuated FUT1, p-AKT, p-mTOR, and p-4EBP1 expression and reduced UEA-1 expression. The data shown in C and D are representative of 3 independent experiments. EV, empty vector control; HG, high glucose; LG, low glucose; NTC, nontargeting control; OE, overexpression; FDR, false discovery rate; NS, not significant; 2DGal, 2-deoxy-D-galactose.
To identify α-(1,2)-fucosylated proteins that could mediate the effects of FUT1 dysregulation on TIC maintenance in HCC, we then performed fucosylated peptide profiling of α-(1,2)-fucosylated total proteins. We prepared total protein extracts of HCC Huh7 cells with or without FUT1 overexpression and enriched fucosylated proteins using lectin chromatography with Lens culinaris agglutinin (LCA) and UEA-1. LCA recognizes α-(1,6) fucosylation catalyzed by FUT8 and was used as a positive control, while UEA-1 recognizes α-(1,2) fucosylation catalyzed by FUT1 (Figure 8A). Mass spectrometric analysis identified 454 proteins in common between the EV control and FUT1-overexpressing cells. Because our current data do suggest that FUT1 regulates cancer stemness features, we sought to narrow down 30 candidates by matching proteins that are also highlighted in the Cancer Stem Cell database (CSCdb) to identify proteins that may be biologically related to cancer stemness processes (Figure 8B, Supplemental Table 4, and ref. 15). GO analysis performed on the 30 shortlisted proteins revealed enrichment in biological processes relevant to cellular response to stress, programmed cell death, and membrane-associated proteins (Supplemental Figure 19). This finding suggests that these candidates were stress-response proteins and might play a role in counteracting the glucose deprivation stress–induced UPR. By integrating RPPA and fucosylated peptide profiling analysis, we shortlisted 23 PI3K/AKT signaling–related proteins that were also identified as potential targets of FUT1. Stemness- and PI3K/AKT–related proteins that bound to UEA-1 and that are localized on the plasma membrane include ICAM-1, EGFR, CD147, and EPHA2 (Figure 8B and Supplemental Table 5). Of interest, many of these membrane-associated candidate proteins were previously reported to be N-linked glycosylated proteins and functionally regulated by the presence of fucosylation, suggesting that our approach is indeed valid (16–18). To validate our proteomic analysis, we further examined protein targets that showed a minimum of 2 unique hits in the FUT1-overexpressing model (Supplemental Table 5). By UEA-1 lectin pulldown analysis, we successfully validated the α-(1,2)-fucosylated state of CD147, ICAM-1, EGFR, and EPHA2 in Huh7 HCC cells treated with low glucose compared with high glucose, while the input showed no differences in their expression. The same effect was observed in Huh7 HCC cells overexpressing FUT1 compared with EV controls, while the opposite result was obtained when FUT1-overexpressing cells were treated with 2DGal (Figure 8C). Similar observations could also be recapitulated in the CLC13 HCC cell model (Supplemental Figure 20). LAMP2 was used as a positive control (19). Consistently, immunoprecipitation of CD147, ICAM-1, EGFR, and EPHA2 followed by UEA-1 blot analysis showed enhanced UEA-1 binding to the CD147, ICAM-1, EGFR, and EPHA2 proteins in the FUT1-overexpressing HCC cells compared with the EV control cells (Supplemental Figure 21). Notably, EGFR could not be detected in CLC13 HCC cells. Other protein targets, including HDGF, PEBP1, PARP1, HMGB1, CALR, HMGA2, and SRSF3, showed no difference in the UEA-1 pulldown analysis of the EV control and FUT1-overexpressing cells (Supplemental Figure 22). To further demonstrate FUT1-mediated fucosylation of CD147, ICAM-1, EGFR, and EPHA2 and cancer stemness as well as AKT/mTOR/4EBP1 signaling, we performed rescue experiments whereby CD147, ICAM-1, EGFR, and EPHA2 expression levels were individually suppressed by lentivirus-based knockdown in FUT1-overexpressing HCC cells. Knockdown of CD147, ICAM-1, EGFR, and EPHA2 expression consistently resulted in attenuated TIC frequency as well as a concomitant decrease in p-AKT, p-mTOR, and p-4EBP1 expression (Supplemental Figures 23–26). Overall, our results suggest that aberrant α-(1,2) fucosylation of the membrane proteins CD147, ICAM-1, EGFR, and EPHA2 by FUT1 in response to the glucose-deprived microenvironment plays a critical role in driving cancer stemness via AKT/mTOR/4EBP1 signaling.
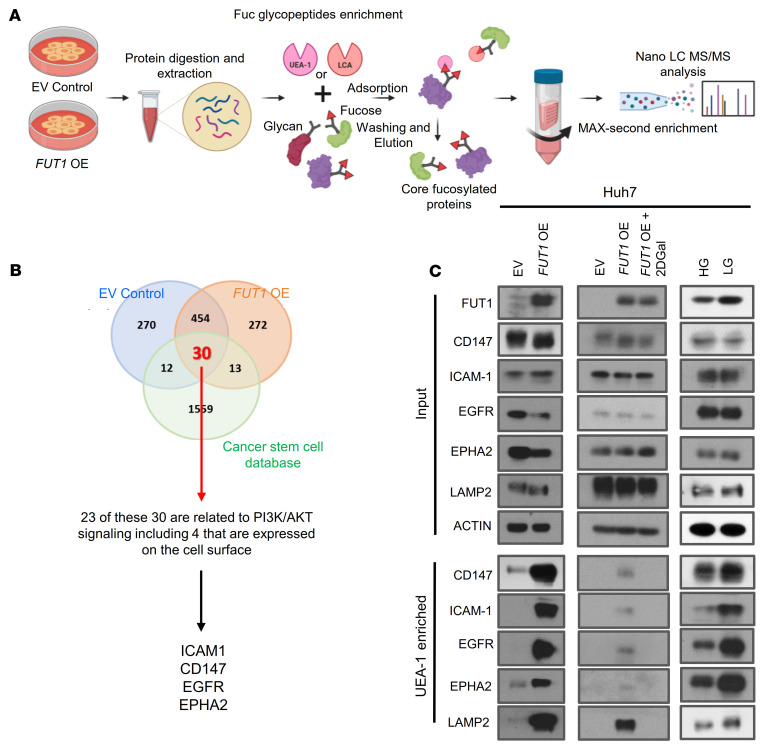
(A) Schematic representation of our experimental approach to fucosylated-peptide profiling. (B) Venn diagram showing the number of proteins identified by nano–LC MS/MS analysis of the UEA-1–enriched fractions of cells treated with empty vector control and cells with FUT1 overexpression, as well as the common proteins that matched the Cancer Stem Cell database (CSCdb; ref. 15). Of the 30 proteins, 24 are related to PI3K/AKT signaling, including 4 cell surface proteins (ICAM1, CD147, EGFR, and EPHA2). (C) UEA-1 affinity chromatography of whole-cell lysates of Huh7 HCC cells transfected with empty vector or with FUT1 overexpression and in the absence or presence of 2DGal or Huh7 HCC cells cultured in high or low glucose, followed by Western blotting with LAMP2, CD147, ICAM-1, EGFR, and EPHA2 antibodies. Input shows no effect of glucose restriction and FUT1 overexpression on protein expression. LAMP2 was used as positive control. The data shown are representative of 3 independent experiments. EV, empty vector control; OE, overexpression; HG, high glucose; LG, low glucose; 2DGal, 2-deoxy-D-galactose.
Discussion
Fucosylation, one of the most important types of glycosylation, is a posttranslational modification of glycans, proteins, and lipids that is responsible for many biological processes. Fucose conjugation via α-(1,2), α-(1,3), α-(1,4), α-(1,6), and O-linkages to glycans and variations in fucosylation linkages have important implications for cancer biology (20). Fucosylation is catalyzed by a family of FUTs (21). In HCC, core fucosylated AFP (AFP-L3) has been shown to be a better marker than AFP alone, with good clinical value, and is currently the only serum biomarker approved by the US FDA for the detection of the disease. Increased fucosylation is also documented in HCC tissue compared with cirrhotic or adjacent nontumor liver tissue. Various FUTs (FUT1, FUT2, FUT6, FUT7, and FUT8) have also been separately implicated in HCC, although with seemingly divergent effects. High expression of FUT1 was found to be significantly associated with advanced stages and poor outcome in HCC, while FUT1 overexpression in HepG2 cells inhibited tumor formation in nude mice (22, 23). Specifically, in the study by Mathieu et al., overexpression of FUT1 in HepG2 cells inhibited tumor formation in nude mice, which is contradictory to the findings of our current study (23). This discrepancy may be explained by the fact that HepG2 is a hepatoblastoma cell line derived from a 15-year-old male adolescent. Hepatoblastoma is a liver tumor type of early childhood and is distinct from HCC. According to information provided by ATCC, HepG2 cells do not form tumors when injected into immunosuppressed mice. The apparently contradictory findings presented in Mathieu et al. and our current work may be a result of cell line differences and perhaps varying effects of FUT1 in hepatoblastoma versus HCC cells. With the exception of these 2 studies, there have been no other reports to our knowledge on the functional role of FUT1 or the molecular mechanisms regulating FUT1 expression. Findings from our current study showed that FUT1 expression was enhanced in HCC and that increased FUT1 expression in HCC was correlated with more aggressive clinical features. Functionally, FUT1 exhibited an oncogenic role in HCC, regulating tumor initiation, self-renewal, and drug resistance, as demonstrated in HCC cell lines as well in an immunocompetent HCC mouse model where endogenous Fut1 expression was suppressed. Notably, in our immunocompetent HCC mouse model, treatment with a combination of sorafenib and 2DGal did not result in any beneficial antitumor effect but only a marked decrease in self-renewal in this group of cells, as demonstrated by their attenuated tumor-initiating potential. Previous studies have indicated terminal α-(1,2) fucosylation to be a hallmark of M1 inflammatory macrophages and to play a major role in their antigen processing and presentation. Treatment with 2DGal has also been shown to effectively suppress type II collagen–induced rheumatoid arthritis (24). M1 macrophages were shown to have proinflammatory and tumor suppressive roles in various tumor models, including HCC (25, 26). Hence, we hypothesize that 2DGal may exert a similar effect on the inflammatory response in our immunocompetent HCC mouse model.
Extensive evidence has shown that tumors are flexible and able to respond to the endogenous environments in which they grow. Here, we demonstrated that glucose restriction in HCC tumor microenvironments results in the clonal selection of a more malignant TIC-like phenotype. By transcriptome profiling, we observed a significant enrichment in PERK/UPR signaling in low-glucose conditions, with subsequent follow-up studies showing that the enhanced PERK/eIF2α/ATF4 signaling axis drives FUT1 transcription via direct binding of ATF4 to the FUT1 promoter. FUT1 and FUT2 share a common role in the catalysis of α-(1,2) fucosylation, and only FUT1 expression is differentially upregulated under glucose-restricted conditions and responsible for driving cancer stemness in HCC. Additionally, because 2DGal can inhibit both FUT1 and FUT2 activity, there is a clear need to identify an FUT1-specific inhibitor that targets the cancer stemness subset in the nutrient-deprived HCC tumor bulk.
We next focused on specific α-(1,2)-fucosylated glycoproteins underlying the FUT1-mediated effects in HCC. Glycoproteomic analysis of HCC cells with or without stable FUT1 overexpression identified 454 common α-(1,2)-fucosylated proteins. This group included known fucosylated proteins, such as ICAM-1, AFP, integrin β1, LAMP2, HMGB1, and EGFR (16, 19, 27, 28). Because our functional studies suggest that FUT1 confers cancer stemness properties, we further narrowed down our list of targets using CSCdb to select for cancer stemness–related proteins, which yielded 30 candidates with enrichment in the cellular stress response, programmed cell death, and membrane-associated cellular components such as ICAM-1, CD147, EPHA2, and EGFR (15), and the results were subsequently validated. This result might explain why such an increased level of α-(1,2) fucosylation is critical to maintain cell survival upon glucose deprivation–induced UPR. ICAM-1 is a cell surface glycoprotein that can be stimulated by ER stress, and glycosylation of ICAM-1 is essential to activate its cellular adhesive interactions in cell metastasis and various inflammatory responses via the PI3K/AKT pathway (29–31). Regulation of CD147 has been reported to function through proteolysis and N-glycosylation (32, 33). The highly glycosylated form of CD147 has been shown to interact more with CD44 and EGFR to drive the Ras/MAPK signaling cascade than the less glycosylated form of CD147 (33). Glucose metabolism regulated by CD147 via an AKT/mTOR-dependent pathway has been reported in non–small cell lung cancer (34). Fucosylation of EGFR mediated by FUT1 has been found to be important in activating the AKT/ERK pathway (18). Together, our current data and findings from past studies suggest that increased levels of α-(1,2) fucosylation on these cell surface proteins contribute to cell survival and cancer stemness in response to UPR in glucose-deprived HCC via AKT/mTOR/4EBP1 signaling.
Our study highlights the therapeutic potential of targeting FUT1 to treat nutrient-deprived HCC and other advanced tumors. Exemplifying the value of glycosylation enzymes as druggable targets are both glycosyltransferase (Zavesca) for Gaucher’s disease and glycosidase inhibitors (aznamivir/Relenza and oseltamivir/Tamiflu) for influenza, all currently in use or in advanced clinical trials (35–37). Our work provides a rationale for the future design of small molecule inhibitors against FUT1 to sensitize tumors to sorafenib and treat tumors by eradicating the CSC subset.
Methods
Cell lines, HCC patient–derived organoids, and culture conditions.
The HCC cell line Huh7 was provided by the JCRB Cell Bank. The HCC cell line CLC13 was a gift from Lijian Hui (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China) (38). The 293T cell line was purchased from ATCC, while 293FT cells were purchased from Invitrogen, Thermo Fisher Scientific. The immortalized normal liver cell line MIHA was provided by J.R. Chowdhury, Albert Einstein College of Medicine, New York, New York, USA. The cell lines used in this study were authenticated by STR profiling and tested for the absence of mycoplasma contamination. Huh7, CLC13, and patient-derived spheroids were cultured in 4.5 g/L glucose and 0.45 g/L glucose in complete DMEM, complete RPMI, or serum-free DMEM/F12 spheroid medium supplemented with growth factors (refer to details below), respectively, to mimic high- and low-glucose conditions. Specifically, 4.5 g/L and 0.45 g/L glucose mimicked hyperglycemic and hypoglycemic conditions, respectively. A hypoxic environment was created by culturing cells in 1% O2/5% CO2 in a modular incubator chamber at 37°C for 48 hours. For patient-derived organoid cultures, cells were isolated and cultured as previously described (12, 39, 40). HCC tissues used for organoid establishment of HK-HCC P1 and HK-HCC P2 were obtained from HCC patients undergoing hepatectomy or liver transplantation at Queen Mary Hospital, Hong Kong. Samples were collected from patients who had not received any previous local or systemic treatment prior to operation. HCC patient–derived organoid-labeled HCC10 was a gift from Meritxell Huch (The Gurdon Institute at the University of Cambridge, Cambridge, United Kingdom).
HTVI NRAS+AKT mouse model of HCC.
Six- to 8-week-old male wild-type C57BL/6 mice (Jackson Laboratory) were used, and the procedure was performed as previously described (41, 42). In brief, 20 μg of plasmid encoding human AKT1 (myr-AKT1) and human neuroblastoma Ras viral oncogene homolog (N-RasV12) along with sleeping beauty (SB) transposase at a ratio of 25:1 was diluted in 2 mL of saline (0.9% NaCl), filtered through a 0.22 μm filter, and injected into the lateral tail vein of C57BL/6 mice in 5 to 7 seconds. The constructs used in this study showed long-term expression of genes via hydrodynamic injection (42). Four weeks after HTVI of proto-oncogenes and SB transposase, the mice were separated into 2 groups and administered 5 × 107 transducing units of lentiviruses encoding either shNTC or shFut1 in 100 μL of PBS via tail vein injection. At 5.5 weeks, mice were given another dose of the shNTC or shFut1 lentiviruses. Mouse shNTC (CGTTCTCCGAACGTGTCACGT) and mouse shFut1 target (GTTCTCCGAACGTGTCACGT) sequences were cloned into the LV3 vector.
Tumor-initiating and self-renewal animal studies.
Tumor-initiating and self-renewal abilities were investigated by limiting-dilution and serial transplantation assays. Four- to 6-week-old male NOD/SCID mice (Jackson Laboratory) were injected subcutaneously with 2500, 5000, 10,000, or 50,000 cells. For the high- and low-glucose study, HCC cells were cultured under low- or high-glucose conditions for 6 days prior to primary implantation. For secondary implantation, only 1 tumor was dissociated from each of the low- and high-glucose treatment groups for passage into an additional 5 mice. Tumor incidence and tumor latency were recorded. Tumor-initiating frequency was calculated using extreme-limiting-dilution analysis. Tumor sizes were measured every 3 days by calipers, and tumor volumes were calculated as volume (cm3) = L × W2 × 0.5, with L and W representing the largest and smallest diameters, respectively. Tumor initiation and incidence were determined to be positive when the tumor volume reached 14 mm3. Tumors formed were harvested for histological analysis. Only tumors with volumes less than 200 mm3 were harvested and dissociated for subsequent passage to secondary mouse recipients or for in vitro limiting-dilution analysis to minimize the number of necrotic cells within the tumor bulk. Animals that were injected with tumor cells but showed no sign of tumor burden were generally euthanized 6 months after tumor cell inoculation, and animals were opened up at the injection sites to confirm that there was no tumor development.
RNA-seq.
See Supplemental Methods. Gene expression data were deposited in the Gene Expression Omnibus, and the accession number is GSE152927.
Statistics.
Statistical analyses were performed using GraphPad Prism 5.0 or SPSS 21.0. Unpaired and paired Student’s t tests were used to compare the mean values of 2 groups. One-way ANOVA with Tukey’s honestly significant difference (HSD) post hoc test was used to compare the mean values of more than 2 groups. Clinicopathological significance in clinical samples was evaluated by Fisher’s exact test and independent Student’s t test for categorical data and continuous data, respectively. The differences in Kaplan-Meier survival curves were calculated using the log-rank test. The differences in survival analysis in the mouse model were determined by the log-rank test. Data are shown as the mean ± standard deviation. Statistical significance was defined as P < 0.05. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Study approval.
HCC tissues used for organoid establishment were obtained from HCC patients undergoing hepatectomy or liver transplantation at Queen Mary Hospital, Hong Kong, with written informed consent obtained from all patients and a protocol approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster. Formalin-fixed, paraffin-embedded primary human HCC and adjacent nontumor liver tissue samples were obtained from HCC patients undergoing hepatectomy at the Sun Yat-Sen University Cancer Centre in Guangzhou, China, with informed consent obtained from all patients and a protocol approved by the Institutional Review Board of the University Cancer Centre. All animal study protocols were approved by and performed in accordance with the Committee of the Use of Live Animals in Teaching and Research at The University of Hong Kong and the Animals (Control of Experiments) Ordinance of Hong Kong.
Author contributions
JHCL and SKYM conceived the project and designed the studies. JHCL performed the research and analyzed and interpreted the data, with the help of TLW, MT, LZ, KYN, and CHL. HJY performed hydrodynamic tail vein injection for HCC mouse models. RS performed the research and analyzed and interpreted the data relating to the fucosylated peptide profiling studies. JPY, CML, and KM obtained patient consent and provided clinical samples for analysis. XYG provided reagents. TKL provided reagents and critical scientific input. JHCL and SKYM wrote the manuscript. SKYM supervised the project and provided funding for the study.
Acknowledgments
This project is supported in part by grants from the Research Grants Council of Hong Kong – Theme Based Research Scheme (T12-704/16-R and T12-710/16-R), Collaborative Research Fund (C7026-18G), and the Health and Medical Research Fund from Food and Health Bureau of the Hong Kong Government (07183206). We also acknowledge funding support from “Laboratory for Synthetic Chemistry and Chemical Biology” under the Health@InnoHK Program launched by the Innovation and Technology Commission, The Government of Hong Kong Special Administrative Region of the People’s Republic of China. We thank the Centre for PanorOmic Sciences (The University of Hong Kong) for providing and maintaining the equipment and technical support needed for transcriptome sequencing, mass spectrometry, flow cytometry, animal imaging, and confocal microscopy studies. We thank the Centre for Comparative Medicine Research (The University of Hong Kong) for supporting our animal work studies. We also sincerely thank Meritxell Huch of The Gurdon Institute at the University of Cambridge for providing the patient-derived HCC organoid line HCC10; Lijian Hui of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, for providing CLC13 HCC cells; and Phillip Chiu and Keith Lee of The University of Hong Kong for sharing antibodies.
Version Changes
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(11):e143377.https://doi.org/10.1172/JCI143377.
References
Articles from The Journal of Clinical Investigation are provided here courtesy of American Society for Clinical Investigation
Full text links
Read article at publisher's site: https://doi.org/10.1172/jci143377
Read article for free, from open access legal sources, via Unpaywall:
http://www.jci.org/articles/view/143377/files/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/104473514
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1172/jci143377
Article citations
H Antigen expression modulates epidermal Keratinocyte Integrity and differentiation.
Biol Res, 57(1):72, 18 Oct 2024
Cited by: 0 articles | PMID: 39420441 | PMCID: PMC11487879
New insights into allergic rhinitis treatment: MSC nanovesicles targeting dendritic cells.
J Nanobiotechnology, 22(1):575, 19 Sep 2024
Cited by: 0 articles | PMID: 39294599 | PMCID: PMC11411834
Targeting Hepatic Cancer Stem Cells (CSCs) and Related Drug Resistance by Small Interfering RNA (siRNA).
Cell Biochem Biophys, 82(4):3031-3051, 26 Jul 2024
Cited by: 1 article | PMID: 39060914
Review
Activating transcription factor 4: a regulator of stress response in human cancers.
Front Cell Dev Biol, 12:1370012, 27 Mar 2024
Cited by: 1 article | PMID: 38601083 | PMCID: PMC11004295
Review Free full text in Europe PMC
Fut2 Deficiency Promotes Intestinal Stem Cell Aging by Damaging Mitochondrial Functions via Down-Regulating α1,2-Fucosylation of Asah2 and Npc1.
Research (Wash D C), 7:0343, 27 Mar 2024
Cited by: 1 article | PMID: 38550777 | PMCID: PMC10976588
Go to all (37) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE152927
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
microRNA-448 inhibits stemness maintenance and self-renewal of hepatocellular carcinoma stem cells through the MAGEA6-mediated AMPK signaling pathway.
J Cell Physiol, 234(12):23461-23474, 24 Jun 2019
Cited by: 21 articles | PMID: 31232474
Targeting AMP-activated kinase impacts hepatocellular cancer stem cells induced by long-term treatment with sorafenib.
Mol Oncol, 13(5):1311-1331, 15 Apr 2019
Cited by: 21 articles | PMID: 30959553 | PMCID: PMC6487713
IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway.
BMC Cancer, 18(1):609, 30 May 2018
Cited by: 38 articles | PMID: 29848298 | PMCID: PMC5977745
Fucosylation in Urological Cancers.
Int J Mol Sci, 22(24):13333, 11 Dec 2021
Cited by: 7 articles | PMID: 34948129 | PMCID: PMC8708646
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Health and Medical Research Fund from Food and Health Bureau of the Hong Kong Government (1)
Grant ID: 07183206
Innovation and Technology Commission (2)
Grant ID: Chemical Biology” under the Health@InnoHK Program
0 publications
Grant ID: “Laboratory for Synthetic Chemistry
0 publications
Research Grants Council of Hong Kong – Collaborative Research Fund (1)
Grant ID: C7026-18G
Research Grants Council of Hong Kong – Theme Based Research Scheme (2)
Grant ID: T12-710/16-R
Grant ID: T12-704/16-R




