Abstract
Key points
Impairment of muscle biogenesis contributes to the progression of Duchenne muscular dystrophy (DMD). As a muscle enriched microRNA that has been implicated in muscle biogenesis, the role of miR-133b in DMD remains unknown. To assess miR-133b function in DMD-affected skeletal muscles, we genetically ablated miR-133b in the mdx mouse model of DMD. We show that deletion of miR-133b exacerbates the dystrophic phenotype of DMD-afflicted skeletal muscle by dysregulating muscle stem cells involved in muscle biogenesis, in addition to affecting signalling pathways related to inflammation and fibrosis. Our results provide evidence that miR-133b may underlie DMD pathology by affecting the proliferation and differentiation of muscle stem cells.Abstract
Duchenne muscular dystrophy (DMD) is characterized by progressive skeletal muscle degeneration. No treatments are currently available to prevent the disease. While the muscle enriched microRNA miR-133b has been implicated in muscle biogenesis, its role in DMD remains unknown. To assess miR-133b function in DMD-affected skeletal muscles, we genetically ablated miR-133b in the mdx mouse model of DMD. In the absence of miR-133b, the tibialis anterior muscle of P30 mdx mice is smaller in size and exhibits a thickened interstitial space containing more mononucleated cells. Additional analysis revealed that miR-133b deletion influences muscle fibre regeneration, satellite cell proliferation and differentiation, and induces widespread transcriptomic changes in mdx muscle. These include known miR-133b targets as well as genes involved in cell proliferation and fibrosis. Altogether, our data demonstrate that skeletal muscles utilize miR-133b to mitigate the deleterious effects of DMD.Free full text

The microRNA, miR-133b, functions to slow Duchenne muscular dystrophy pathogenesis
Abstract
Duchenne muscular dystrophy (DMD) is characterized by progressive skeletal muscle degeneration. No treatments are currently available to prevent the disease. While the muscle enriched microRNA, miR-133b, has been implicated in muscle biogenesis, its role in DMD remains unknown. To assess miR-133b function in DMD-affected skeletal muscles, we genetically ablated miR-133b in the mdx mouse model of DMD. In the absence of miR-133b, the tibialis anterior muscle of P30 mdx mice is smaller in size and exhibits a thickened interstitial space containing more mononucleated cells. Additional analysis revealed that miR-133b deletion influences muscle fiber regeneration, satellite cell proliferation and differentiation, and induces widespread transcriptomic changes in mdx muscle. These include known miR-133b targets as well as genes involved in cell proliferation and fibrosis. Altogether, our data demonstrate that skeletal muscles utilize miR-133b to mitigate the deleterious effects of DMD.
Introduction
Duchenne muscular dystrophy (DMD) is the most prevalent of the muscular dystrophies, primarily affecting boys, with symptoms arising between 3-5 years of age. While advances in medical care have allowed DMD patients to live into adulthood, there is still no cure for the disease (Guiraud et al., 2015). DMD is caused by loss of function of the dystrophin gene, an X-linked gene important for maintaining the integrity of the muscle fiber sarcolemma (Hoffman et al., 1987; Rybakova et al., 2000; McNally & Pytel, 2007; Guiraud et al., 2015) through its association with dystroglycan and sarcoglycans (Ibraghimov-Beskrovnaya et al., 1992; Lapidos et al., 2004; McNally & Pytel, 2007). While the initial formation of skeletal muscles proceeds mostly unimpeded during development, muscle fibers rapidly degenerate as individuals mature due to damages on the peripheral membrane caused by mechanical stress associated with muscle contraction (McNally & Pytel, 2007; Guiraud et al., 2015). As the disease becomes more severe, skeletal muscle progenitor cells, known as satellite cells, fail to adequately proliferate and differentiate in order to replace damaged muscle fibers (Almada & Wagers, 2016). DMD-pathogenesis is further exacerbated by chronic immune cell infiltration and fibrosis (Kharraz et al., 2014) that results from deposition of collagens and fat emanating from fibroblasts and adipocytes (Yin et al., 2013b; Kharraz et al., 2014; Almada & Wagers, 2016). These pathological changes invariably impair the function and stability of remaining muscle fibers, compromising voluntary movements and ultimately causing death of affected individuals (Guiraud et al., 2015). Thus, it is important to continue to explore the molecular basis for DMD progression.
The progressive nature of DMD (Guiraud et al., 2015) suggests that endogenous mechanisms could be recruited to mitigate damages caused by DMD and to maintain the regenerative capacity of skeletal muscles. Arguably, the most attractive endogenous molecular mechanisms would be those that bolster the regenerative capacity of skeletal muscles and endow muscle fibers with the ability to overcome fibrosis and inflammation resulting from the loss of dystrophin (Chang et al., 2016). In this regard, microRNAs have received attention as candidate molecules for modifying DMD-pathology because of their functions in normal and disease-affected skeletal muscles and the relative ease with which they can be packaged into vectors (Williams et al., 2009a; Sharma et al., 2014; Horak et al., 2016; Ballarino et al., 2016). These ubiquitous small non-coding RNAs are approximately 22 nucleotides in length and impact a wide range of cellular processes by inhibiting translation of target mRNAs (Meister, 2013; Ha & Kim, 2014).
There is ample evidence suggesting that the muscle-enriched microRNA, miR-133b, may be a candidate for modifying DMD-pathogenesis. In vitro studies have shown that targeting miR-133b and its close relative, miR-133a, affect different stages of myogenesis, including the proliferation of satellite cells (Chen et al., 2006; Cui et al., 2019) and myoblasts (Feng et al., 2013), the differentiation of satellite cells into either myoblasts or brown adipose tissue (Yin et al., 2013a), and the formation of muscle fibers (Hak et al., 2006; Feng et al., 2013; Cui et al., 2019). In vivo, published data indicate that skeletal muscles upregulate miR-133b during regeneration following injury (Liu et al., 2012) and may utilize miR-133b to maintain homeostasis during cellular, functional and biochemical changes induced by exercise (Russell et al., 2013) and denervation (Valdez et al., 2014) as well as in sarcopenia (Iannone et al., 2020). Additionally, miR-133b was shown to accelerate muscle regeneration following injury in young adult rats alongside miR-1 and miR-206 (Nakasa et al., 2010). Importantly, there is evidence indicating that miR-133b is elevated in the serum and muscles of dystrophin-null (mdx) mice (Coenen-Stass et al., 2018) and in the serum of DMD patients, however the latter study did not distinguish miR-133b from miR-133a (Cacchiarelli et al., 2011). Despite these findings, the role of miR-133b in DMD remains unclear. This is because miR-133b has yet to be examined independently of other non-coding RNAs in DMD (Cesana et al., 2011; Boettger et al., 2014). In one published study, miR-133b was proposed to collaborate with a long noncoding RNA, linc-MD1, to affect the timing and rate of muscle fiber formation in DMD (Cesana et al., 2011). In another study, it was shown that simultaneously deleting miR-133b and miR-206, another muscle enriched microRNA found in the same long non-coding RNA as miR-133b, has no effect on DMD pathogenesis (Boettger et al., 2014). Therefore, it remains unknown if and how miR-133b plays a role in DMD.
To directly test the role of miR-133b in DMD-pathogenesis, we generated mdx mice lacking miR-133b. Analysis of these mice during various stages of disease severity revealed that miR-133b does in fact play important roles in DMD. Loss of miR-133b decreased the size of muscle fibers while increasing the area of the interstitial space and the presence of nuclei within it in P30 mdx hindlimb muscles. These changes are partly due to alterations in satellite cell abundance and changes in expression of myriad genes, including both known targets of miR-133b and genes shown to be involved in cell cycle regulation and promotion of fibrosis. Altogether, these findings strongly suggest that skeletal muscles increase miR-133b levels to mitigate damages caused by DMD.
Methods
Ethical Approval and Animal Information
With the exception of the Pax7 antibody, a kind donation from Dr. Julia von Maltzahn, all reagents were obtained from commercial vendors as described below. Dystrophin-null DMD mice (mdx, Jackson Labs 001801, RRID: IMSR_JAX:001801 (Bulfield et al., 1984)) were purchased from Jackson Laboratories (Bar Harbor, Maine) and miR-133b−/− mice were obtained from the Feng lab (Heyer et al., 2012). All mice were maintained on a mixed genetic background. miR-133b−/− mice were crossed with mdx mice to generate mdx;miR-133b+/− mice which were subsequently bred to generate mdx;miR-133b+/+ and mdx;miR-133b−/− mice for experimental use. miR-133b−/− mice were also crossed with WT mice to generate miR-133b+/− mice which were subsequently bred to generate miR-133b+/+ and miR-133b−/− littermates for experimental use. Wild-type control mice used for qPCR were age and sex-matched to mdx mice. Male and female mice were used for all in vivo studies and for primary cell culture experiments and the number of male and female mice were balanced across genotypes for each individual experiment, with the exception of body weight measurements. For comparisons between mdx;miR-133b+/+ and mdx;miR-133b−/− muscle, mice were aged 30, 60, or 90 days as described in the results section. For comparisons between miR-133b+/+ and miR-133b−/− muscle, mice were aged 60 days. Mice aged 15-21 days were used for primary cell culture. Prior to cardiotoxin administration, mice were anesthetized with a solution of ketamine (50 mg/kg IP) and xylazine (7.5 mg/kg) injected into the peritoneum. Sufficient anesthesia was checked with a hind foot pinch and ophthalmic ointment was applied to the animal’s eyes. Respiration and arousal were monitored throughout the procedure. All mice were sacrificed with a lethal dose of isoflurane prior to tissue collection or tissue dissociation for primary cell culture. Animals were housed in a 12 hour light/dark cycle with access to food and water ad libitum. No animals died before the conclusion of experiments. Breeding, housing, and experimental use of animals were performed in a pathogen free environment in accordance with The Journal of Physiology animal ethics policy as well as the National Institutes of Health and Virginia Tech Institutional Animal Care and Use Committee (Protocol# 18-148 and 18-176) guidelines.
Immunohistochemistry (IHC): Whole mounted muscle
Mice were anesthetized with isoflurane and perfused transcardially with phosphate buffered saline followed by 4% paraformaldehyde. Following dissection, the extensor digitorum longus (EDL) muscle was incubated with Alexafluor 555 conjugated alpha-bungarotoxin (#B35451, Invitrogen, Carlsbad, CA) diluted 1:1000 in PBS for 2 h at room temperature, washed 3 times with PBS for 5 minutes and mounted to a glass slide in Vectashield (Vector Laboratories, Burlingame, CA).
IHC: Muscle Cross Sections
The tibialis anterior (TA) muscle was dissected from paraformaldehyde perfused mice, post-fixed in 30% sucrose in PBS for 48 h at 4C, cross-sectioned at 16 μm in Tissue Freezing Medium (#TFM-5, General Data, Cincinnati, OH) with a cryostat, and mounted to gelatin coated glass slides. For laminin IHC, TA sections were incubated in blocking buffer (5% BSA, 3% goat serum, 0.1% Triton X-100 in PBS) for 1 h at room temperature, incubated in primary antibody (Rabbit anti-laminin, #L9393, RRID: AB_477163, Sigma-Aldrich, St. Louis, MO) diluted 1:250 in blocking buffer overnight at 4C, washed 3 times in PBS, incubated in Alexafluor 568 conjugated polyclonal goat anti-rabbit antibody (#A11036, Invitrogen, Carlsbad, CA) diluted 1:1000 in blocking buffer for 1 h at room temperature, washed twice in PBS, incubated in DAPI (#D1306, Fisher Scientific, Waltham, MA) diluted 1:1000 in PBS for 20 minutes at room temperature, and washed twice in PBS before Vectashield (Vector Laboratories) mounting medium and coverslip application. Pax7 IHC was performed according to (Mayerl et al., 2018), briefly, sections were incubated in M.O.M. blocking buffer (#MKB-2213, Vector Laboratories, Burlingame, CA) for 1 h at room temperature, incubated in mouse anti-Pax7 antibody (undiluted, RRID: AB_528428) and rabbit anti-laminin (1:250, #L9393, RRID: AB_477163, Sigma-Aldrich, St. Louis, MO) overnight at 4°C, washed in PBS 3 times for 5 minutes, incubated in Alexafluor 546 conjugated polyclonal goat anti-mouse IgG antibody (#A-11010, RRID: AB_2534077, Invitrogen, Carlsbad, CA) and Alexafluor 488 conjugated polyclonal goat anti-rabbit IgG (#A-110008, RRID: AB_143165, Invitrogen, Carlsbad, CA) diluted 1:1000 in 5% horse serum for 1 h at room temperature, washed twice in PBS, incubated in DAPI (1:1000 in PBS) for 20 minutes at room temperature, and washed twice in PBS before Vectashield mounting medium and coverslip application. Specificity of the Pax7 antibody in adult mouse skeletal muscle has been described previously (Von Maltzahn et al., 2013). The Pax7 antibody was a generous donation from Dr. Julia von Maltzahn.
Immunocytochemistry (ICC)
Cell cultures were fixed in 4% paraformaldehyde for 30 minutes at room temperature and washed 3 times in PBS for 5 minutes. Pax7 immunocytochemistry was performed according to Pax7 IHC protocol above. Myosin ICC was performed with mouse anti-myosin (1:100, #MF20, DSHB, Iowa City, IA, RRID: 2147781) according to laminin IHC protocol above.
Imaging
Imaging was performed with a Zeiss LSM 710 laser scanning confocal microscope (Carl Zeiss Microscopy, Berlin, Germany) using a 20× (0.8 numerical aperture) objective. Zeiss Zen software (RRID: SCR_013672) was used for maximum intensity projections and stitching of tile scans.
Muscle fiber cross sectional area and central nuclei analyses
Muscle fiber analysis was performed on four tile scan images of laminin/DAPI stained TA cross sections. Cross sections of 16 μm thickness were selected from the medial TA at 0.3 mm intervals. Muscle fibers were identified by laminin positive rings within muscle tissue. Muscle fibers with central nuclei were defined by the presence of a DAPI-positive nucleus within laminin positive rings that are not immediately adjacent to the laminin positive staining. The fractionator method (Mouton, 2011) was used to randomly sample an average of 290 muscle fibers per muscle. To achieve this, a grid was superimposed on the images with ImageJ software (RRID:SCR_003070) and all muscle fibers located on a grid point were selected for measurement. Muscle fiber CSA measurements were performed with ImageJ.
Necrotic Muscle Fiber Analysis
Analysis of necrotic muscle fibers was performed on laminin/Pax7/DAPI stained TA cross sections as previously described (Bencze et al., 2019). Necrotic muscle fibers were identified by intense labeling of immunoglobulin G (IgG) throughout the sarcolemma with Alexafluor 546 conjugated polyclonal goat anti-mouse IgG antibody (#A-11010, RRID: AB_2534077, Invitrogen, Carlsbad, CA), indicating muscle fiber permeability to this serum protein. Additional morphological analysis of fragmented sarcoplasm and increased presence of nuclei (Radley & Grounds, 2006) was used to confirm necrosis in IgG+ muscle fibers. The fractionator method (Mouton, 2011) was used to randomly sample an average of 195 muscle fibers per muscle. To achieve this, a grid was superimposed on the images with ImageJ software and all muscle fibers located on a grid point were selected for analysis. Counting and sampling were performed on blinded images with ImageJ software.
Interstitial Nuclei Analysis
Interstitial nuclei counts were performed on laminin/DAPI stained 16 μm TA cross sections using the fractionator sampling method (Mouton, 2011). On average, 30 regions measuring 7800 μm2 were randomly sampled across two cross sections collected from the medial TA. Interstitial nuclei were identified as DAPI positive nuclei located in areas of thickened laminin deposition in the interstitial space and not immediately adjacent to muscle fibers. Counting and sampling were performed on blinded images with ImageJ software.
Interstitial Space Analysis
Analysis of the area of the interstitial space was performed on laminin/DAPI stained 16 μm thick TA cross sections using the fractionator sampling method (Mouton, 2011). On average, 22 regions measuring 20,000 μm2 were randomly sampled across two cross sections collected from the medial TA. Interstitial area was identified by the presence of thick laminin staining occupied by nuclei in the interstices of muscle fibers. Sampling and measurements were obtained from blinded images with ImageJ software and reported as a percent of the total cross-sectional area sampled.
In Vivo Satellite Cell Counts
Counting of Pax7+ satellite cells was performed on blinded images obtained from Pax7/laminin/DAPI stained TA cross sections of 16 μm thickness using the fractionator sampling method. Four cross sections were selected from the medial TA at 0.3 mm intervals. Pax7+ cells were identified by the presence of a Pax7+/DAPI+ nucleus located immediately adjacent to a laminin+ muscle fiber.
Satellite Cell Proliferation Analysis
Satellite cell proliferation analysis was performed on blinded images of satellite cell enriched cultures using the fractionator sampling method. Following muscle dissociation from a single mouse, cells were seeded to two chamber slides. The first slide was fixed immediately after the cells attached to the slide (4-16 h post-seed) and the second slide was fixed at 72 h post-seed. The cell cultures were stained with Pax7/DAPI, imaged, and satellite cells were identified by the presence of a Pax7+/DAPI+ nucleus. One replicate represents 3 technical replicates derived from the dissociation of skeletal muscles of a single miR-133b+/+ mouse or its miR-133b−/− littermate. Data are presented as the average fold change in the number of Pax7+ cells at 72 h post-seed relative to the number of Pax7+ cells immediately after seeding for a given replicate.
Myotube Size
Myotube size analysis was performed on blinded images of primary muscle cultures derived from miR-133b+/+ and miR-133b−/− littermates that were fixed at 2 or 5 d post-differentiation and stained with myosin/DAPI. Myotubes were identified by the presence of myosin and at least two nuclei. Area measurements were obtained by outlining the myosin+ area of a myotube using ImageJ software.
Cardiotoxin Injury
Cardiotoxin from Naja melanoleuca (#v9000, Sigma-Aldrich, St. Louis, MO) was diluted in sterile PBS to a final concentration of 70 μg/mL. Following anesthesia of P60 miR-133b+/+ and miR-133b−/− littermates with ketamine (50 mg/kg IP), 50 μL of cardiotoxin solution was delivered to the belly of the TA muscle with a 31 gauge needle.
Primary Myoblast Culture
Hindlimb muscles were collected from p15-p21 miR-133b+/+ and miR-133b−/− littermates. After removal of connective tissue and fat, muscles were cut into 5 mm2 pieces with forceps and digested in 2 mg/mL collagenase II (Worthington Chemicals, Lakewood, NJ) for 1h at 37°C. Muscles were further dissociated by mechanical trituration in high glucose Dulbecco’s modified eagle medium (DMEM, #11965118, Life Technologies, Carlsbad, CA) containing 10% horse serum (Life Technologies #16050122) and passed through a 40 μm filter to generate a single cell suspension. Excess debris was removed from the suspension by centrifugation in 4% BSA followed by a second centrifugation in 40% Optiprep solution (Sigma-Aldrich, St. Louis, MO) from which the interphase was collected. Cells were washed in 10% horse serum in DMEM and seeded to laminin (#23017015, Life Technologies, Carlsbad, CA) coated chamber slides (NUNC Permanox, #160005, NUNC, Rochester, NY) in growth media consisting of 20% fetal bovine serum (#FP-0500-A, Atlas Biologicals, Fort Collins, CO), 0.1% penicillin/streptomycin (Life Technologies, Carlsbad, CA), 2 mm glutamine (Life Technologies, Carlsbad, CA), and 0.1% Fungizone (Life Technologies, Carlsbad, CA) in high glucose DMEM. For analysis of satellite cell proliferation, cells were seeded at a density of 20,000 cells per well. For analysis of myotube formation, cells were seeded at a density of 100,000 cells per well. Once cells were 100% confluent, growth medium was replaced with differentiation medium consisting of 2% horse serum (Life Technologies, Carlsbad, CA) and 0.1% penicillin/streptomycin in high glucose DMEM to induce myotube formation. Myotube cultures were fixed with 4% PFA at 2 and 5 d after addition of differentiation media. Cell cultures were maintained at 37°C and 5% CO2.
mRNA Isolation and qPCR
RNA was extracted from the TA muscle using Trizol reagent (Life Technologies, Carlsbad, CA) and the Aurum Total RNA Mini Kit (Bio-Rad, Hercules, CA) and treated with DNase. Reverse transcription of RNA was performed with iScript™ Reverse Transcription Supermix (Bio-Rad, Hercules, CA). qPCR was performed with iTAQ SYBR Green Supermix (Bio-Rad, Hercules, CA) using the CFX Connect Real Time PCR System (Bio-Rad, Hercules, CA). Primers were used at a concentration of 300 nM, primer sequences used in this study are listed in Table 1. To distinguish expression of miR-133a from miR-133b and miR-1 from miR-206, primers were designed to amplify the unique sequences of the precursor miRNAs. Primer specificity and genomic contamination were evaluated by comparisons of melt curves between samples and no reverse transcriptase and no template controls. A sample was considered to be not detected if it did not produce a cycle threshold (Ct) value after 40 PCR cycles or if it produced a Ct value and/or a melt peak temperature that resembled either the NTC or the NRT control samples. Expression values are normalized to β2 Microglobulin using the 2−ΔΔCT method.
Table 1.
qPCR primers sequences and range of cycle threshold (Ct) values for samples.
| Gene | Forward Primer (5’-3’) | Reverse Primer (5’-3’) | Ct Range |
|---|---|---|---|
| pre miR-1a | AGCACATACTTCTTTATGTACCCA | ACTTCTTTACATTCCATAGCACTGA | 33-37 |
| pre miR-133a | GGTAAAATGGAACCAAATCGCCT | GGTTGAAGGGGACCAAATCCA | 30-32 |
| pre miR-133b | GCTGGTCAAACGGAACCAAG | ATATTGAGCTTTGCCAGCCCT | 28.5-31 |
| pre miR-206 | GGCCACATGCTTCTTTATATCC | AAACCACACACTTCCTTACATTCC | 30-38 |
| β2 Microglobulin | TGCTATCCAGAAAACCCCTCAA | GGATTTCAATGTGAGGCGGG | 21-25 |
RNA-seq
RNA sequencing was performed by the Genomics Sequencing Center at Virginia Tech using an mRNA-stranded Seq library prep (Illumina, San Diego, CA) and NextSeq High Output 75SR sequencing (Illumina, San Diego, CA). Following sequencing, data were trimmed for both adaptor and quality using a combination of ea-utils and Btrim (Kong, 2011; Aronesty, 2013). Sequencing reads were aligned to the genome using HiSat2 (Kim et al., 2015) and counted via HTSeq (Anders et al., 2015). QC summary statistics were examined to identify any problematic samples (e.g. total read counts, quality and base composition profiles (+/− trimming), raw fastq formatted data files, aligned files (bam and text file containing sample alignment statistics), and count files (HTSeq text files). Following successful alignment, mRNA differential expression was determined using contrasts of and tested for significance using the Benjamini-Hochberg corrected Wald Test in the R-package DESeq (Love et al., 2014). Signaling pathway analysis was generated through the use of IPA (QIAGEN Inc., https://www.qiagenbio-informatics.com/products/ingenuity-pathway-analysis). RNA seq data are deposited at NCBI GEO, (https://www.ncbi.nlm.nih.gov/geo/, accession # GSE156267).
Statistical analysis
Unpaired Student’s T-tests and unpaired Student’s T-test with Welch’s correction were used to make comparisons between 2 means. Two-way ANOVA with Bonferroni post-hoc analysis was used to make comparisons between 4 means. Kolmogorov–Smirnov test was used to compare muscle fiber CSA distributions. Statistical analyses performed with the Microsoft Excel (RRID:SCR_016137) Data Analysis plugin and GraphPad Prism (RRID: SCR_002798) software. For in vivo experiments, an n is defined as one animal. For in vitro experiments, an n is defined as a cell culture generated from dissociation of skeletal muscles from one animal. Data are expressed as mean ± standard deviation.
Results
miR-133b is induced in dystrophin-deficient mice
Although miR-133 is elevated in the serum of DMD patients (Cacchiarelli et al., 2011) and adult mdx mice (Coenen-Stass et al., 2018), its expression in DMD-afflicted muscle is less clear. Previous studies have shown that miR-133 (without distinction between miR-133a and miR-133b) or miR-133b is upregulated, downregulated, or unchanged in various hindlimb skeletal muscles of young adult mdx mice (Greco et al., 2009; Coenen-Stass et al., 2018). Additionally, the temporal relationship between miR-133b levels and severity of DMD in skeletal muscles remains unknown. To answer these questions, we examined miR-133b expression in the tibialis anterior (TA) muscle at postnatal day 30 (P30), an age at which muscle fiber degeneration is typically pronounced in mdx hindlimb muscles, and at P60, an age at which subsequent pathology remission is typical (McGeachie et al., 1993; Willmann et al., 2009; Duddy et al., 2015). Since one nucleotide distinguishes mature miR-133b from miR-133a, we designed primers against the unique sequences of the precursor miRNAs (pre-miRNAs) to evaluate expression using qPCR (Table 1). Our analysis revealed that miR-133b is increased at P30 and P60 in mdx mice compared to control mice (Fig. 1A). While miR-133b was not detected in the TA of adult control mice (Fig. 1A), it is possible that it is present at levels that are too low to be detected by our assay. In mdx mice, miR-133b expression is significantly higher in the severe period of dystrophy (P30) as compared to the subsequent period of remission (P60) in the TA. This expression pattern was specific to miR-133b, as compared to miR-133a, which was expressed at similar or lower levels in mdx TA muscle (Fig. 1B). It also differed from other muscle-enriched microRNAs in mdx muscle. While miR-206 was significantly higher at both ages in mdx versus control muscle, it was expressed at higher levels at P60 compared to P30 in mdx TA (Fig. 1C). Levels of miR-1 were similar at P30 and somewhat decreased at P60 in the TA muscle of mdx mice compared to control mice (Fig. 1D), confirming previous reports (Roberts et al., 2012).
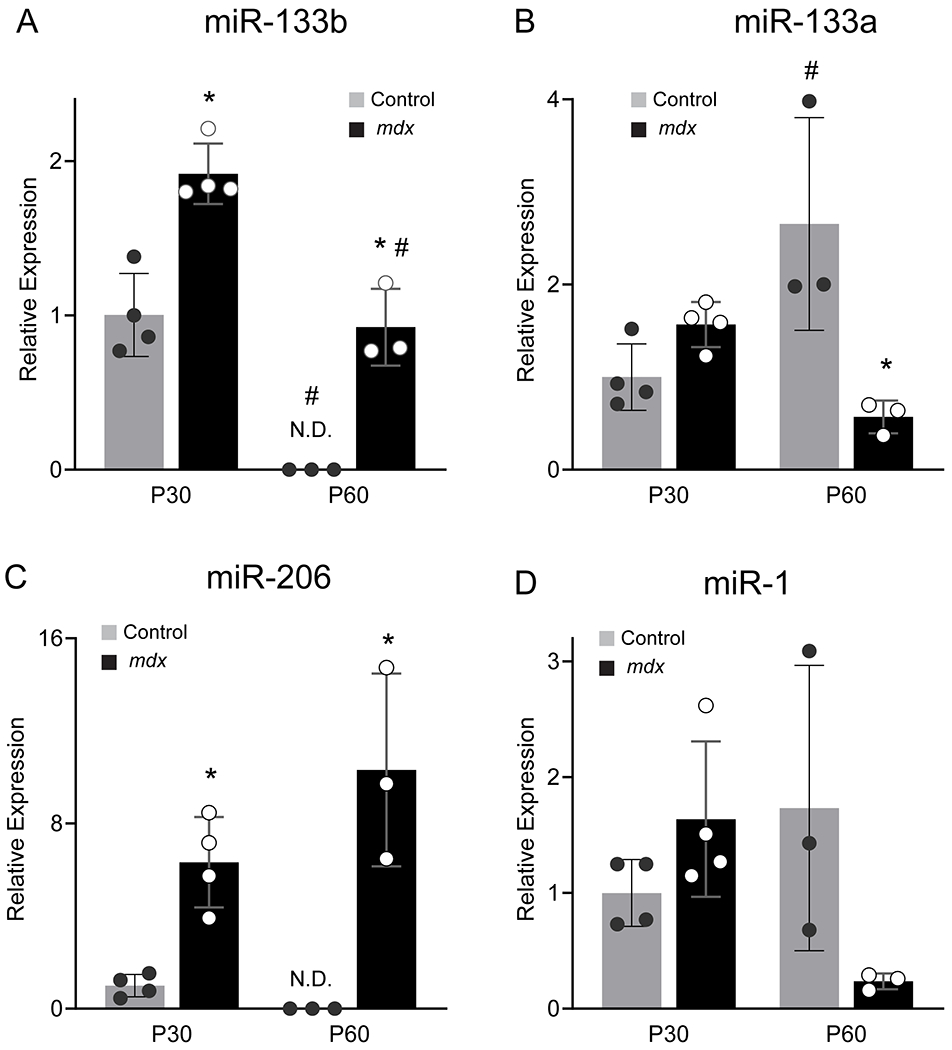
Expression analysis of miR-133b and related miRNAs in mdx muscle. Levels of precursor miRNAs in the TA muscle of P30 and P60 control and mdx mice were assessed using qPCR. (A) miR-133b (genotype effect, p < 0.0001; age effect, p < 0.0001; genotype X age interaction, p = 0.9719; two-way ANOVA). (B) miR-133a (genotype effect, p = 0.0339; age effect, p = 0.3130; genotype X age interaction, p = 0.0016; two-way ANOVA). (C) miR-206 (genotype effect, p < 0.0001; age effect, p = 0.2293; genotype X age interaction, p = 0.0586; two-way ANOVA). (D) miR-1 (genotype effect, p = 0.2708; age effect, p = 0.3864; genotype X age interaction, p = 0.0160; two-way ANOVA). All values normalized to β2 Microglobulin using the 2−ΔΔCT method and reported as mean ± standard deviation; P30, n = 4; P60 n = 3. * p < 0.05 versus age-matched control, # p < 0.05 versus genotype-matched group at P30, Bonferroni’s multiple comparisons. N.D. not detected.
Deletion of miR-133b exacerbates DMD-pathogenesis in skeletal muscles of mdx mice
To date, the role of miR-133b in DMD pathogenesis remains debated mainly because it has yet to be examined independently of the precursor long-noncoding RNA in which both miR-133b and miR-206 are encoded (LINCMD1) (Eisenberg et al., 2009; Williams et al., 2009a). Previous work has demonstrated directly opposing myogenic functions of miR-206, miR-133b and other regions of the LINCMD1 gene (Cesana et al., 2011). While deletion of miR-206 alone exacerbates DMD-related pathology (Liu et al., 2012), the simultaneous deletion of mir-133b and miR-206 has no effect on it (Boettger et al., 2014). To examine the function of miR-133b independently of miR-206 in DMD, we analyzed mice lacking only miR-133b (miR-133b−/−). First, we examined muscles lacking miR-133b (Heyer et al., 2012) in otherwise healthy young adult mice (Fig. 2A–B). Analysis of pre-miR-133b levels in miR-133b−/− TA muscle with qPCR confirmed deletion of miR-133b in this mouse line (Fig. 2C). We found that deletion of miR-133b did not cause obvious changes in body weight (Fig. 2D) or skeletal muscles (Fig. 2E–I). There were no differences in TA muscle mass (Fig. 2E), average muscle fiber cross-sectional area (CSA, Fig 2A, ,B,B, ,F),F), distribution of muscle fiber size (Fig. 2G), numbers of mononucleated cells occupying the interstitial space (Fig. 2H), or numbers of Pax7+ satellite cells (Fig. 2I) between miR-133b+/+ and miR-133b−/− mice.
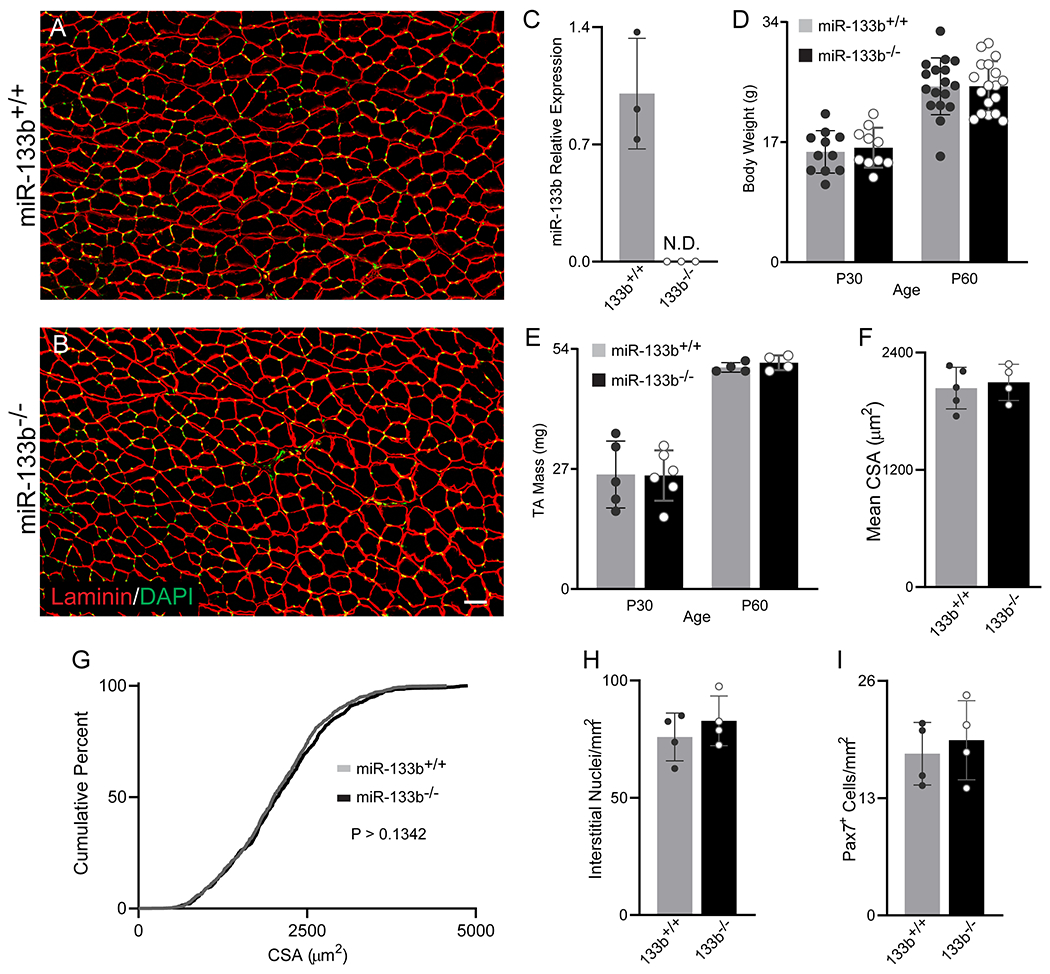
Analysis of healthy control mice lacking miR-133b. (A and B) Representative images of laminin (red) and DAPI (green) IHC of P60 miR-133b+/+ and miR-133b−/− TA cross sections. (C) qPCR analysis of pre-miR-133b in miR-133b+/+ and miR-133b−/− TA of P60 mice (n = 3, values normalized to β2 Microglobulin using the 2−ΔΔCT method). (D) Mouse body weight of P30 and P60 mice (n = 10-17, genotype effect, p = 0.7514, age effect, p < 0.0001, genotype X age interaction, p = 0.7693, two-way ANOVA). (E) Overall TA muscle mass at P30 and P60 (n = 4-6, genotype effect, p = 0.8687, age effect, p < 0.0001, genotype X age interaction, p = 0.7911, two-way ANOVA). (F-I) Analysis of healthy P60 miR-133b+/+ and miR-133b−/− TA cross sections, including (F) mean muscle fiber CSA (p = 0.6765, unpaired T-test), (G) muscle fiber CSA distribution (p = 0.1342, Kolmogorov-Smirnov test), (H) numbers of interstitial nuclei (p = 0.3860, unpaired T-test), and (I) numbers of Pax7+ satellite cells (p = 0.6613, unpaired T-test). All values except (G) reported as mean ± standard deviation. For (F-I), n = 5, miR-133b+/+; n = 4, miR-133b−/−. Scale bar = 50 μm. N.D. not detected.
We next examined skeletal muscles from mdx mice lacking miR-133b, following crossing of the miR-133b−/− and mdx mouse lines (herein referred to as mdx;miR-133b−/−). To start, we examined the histopathology of TA muscle cross sections at P30, an age at which the hindlimb muscles of mdx mice typically experience severe DMD pathology, characterized by elevated levels of muscle fiber damage and regeneration (McGeachie et al., 1993; Willmann et al., 2009; Duddy et al., 2015). This analysis revealed that the CSA of the entire TA muscle (Fig. 3C) as well as the CSA of the muscle fibers within the TA (Fig. 3A, ,B,B, ,D)D) were decreased in mdx;miR-133b−/− mice compared to mdx;miR-133b+/+ mice. Further analysis of muscle fiber CSA distribution revealed that this reduction in muscle fiber CSA is due to fewer muscle fibers with large CSAs and more muscle fibers with small CSAs (Fig. 3E). Interestingly, the number of muscle fibers with centralized nuclei, a characteristic of regenerating juvenile and adult muscle fibers, was somewhat decreased in mdx;miR-133b−/− muscle, however this difference was not statistically significant (Fig. 3F, p = 0.0808). Despite alterations in indices of muscle fiber regeneration, the abundance of necrotic muscle fibers was roughly similar in mdx TA with and without miR-133b (Fig. 3G). Analysis of the interstitial space revealed a larger laminin-positive area (Fig. 3H), indicative of increased ECM deposition. This was accompanied by elevated numbers of mononucleated cells within the interstitial space (Fig. 3I), suggestive of an increased presence of fibroblasts, immune cells, or adipocytes (Uezumi et al., 2010; Kharraz et al., 2014; Almada & Wagers, 2016).
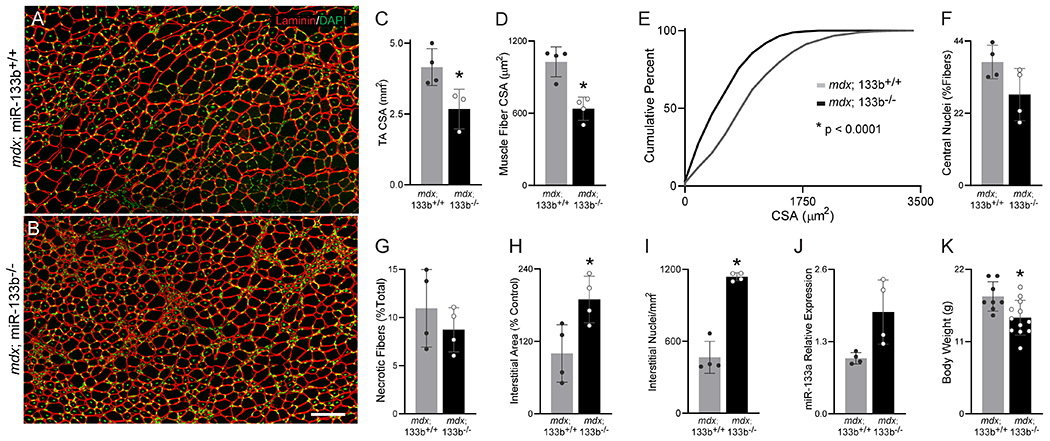
Deletion of miR-133b exacerbates DMD pathology in the TA of P30 mdx mice. (A-B) Representative images of laminin (red) and DAPI (green) staining in (A) P30 mdx;miR-133b+/+ and (B) mdx;miR-133b−/− TA muscle cross-sections. (C) Overall TA muscle CSA (p = 0.0344, n = 4, mdx;miR-133b+/+, n = 3, mdx;miR-133b−/−, unpaired T-test). (D) Average muscle fiber CSA (p = 0.0025, unpaired T-test). (E) Cumulative frequency distribution curve of muscle fiber CSAs (p < 0.0001, Kolmogorov-Smirnov test). (F) Analysis of muscle fibers with central nuclei (p = 0.0808, unpaired T-test). (G) Analysis of necrotic muscle fibers (p = 0.3741, unpaired T-test). (H & I) Analysis of the interstitial space, including (H) interstitial area (p = .0265, unpaired T-test) and (I) the number of nuclei within the interstitial area (p = 0.0014, unpaired T-test with Welch’s correction). (J) Expression analysis of pre-miR-133a levels in the TA using qPCR (p = 0.0612, unpaired T-test with Welch’s correction). (K) Mouse body weight (p = 0.0106, n = 8, mdx;miR-133b+/+; n = 12, mdx;miR-133b−/−, unpaired T-test). n = 4 unless otherwise noted. All values, except E, reported as mean ± standard deviation. miR-133a expression normalized to β2 Microglobulin using the 2−ΔΔCT method. * p < 0.05, scale bar = 100 μm.
In consideration of the high degree of sequence similarity between miR-133b and miR-133a, we analyzed pre-miR-133a levels in P30 mdx;miR-133b−/− TA using qPCR. We observed a trend towards increased pre-miR-133a levels in P30 mdx muscle lacking miR-133b (Fig. 3J, p = 0.0612), suggestive of compensatory expression. In addition to the observed differences in the TA muscle, P30 mdx;miR-133b−/− mice had reduced body weight (Fig. 3K) but did not display other differences in outward appearance when compared to mdx;miR-133b+/+ mice. Altogether, these data suggest that miR-133b deletion impacts the severity of muscular dystrophy by altering both muscle fiber maturation and interstitial space composition but not the rate of muscle fiber degeneration in mdx mice.
We next assessed the effect of miR-133b deletion in mdx TA muscle at P60 and P90, ages at which remitting DMD pathology is typical in the hindlimb muscles of mdx mice (McGeachie et al., 1993; Willmann et al., 2009; Duddy et al., 2015). At P60, TA muscle mass (Fig. 4C) and whole muscle CSA (Fig. 4D) were similar between mdx;miR-133b+/+ and mdx;miR-133b−/− mice. Likewise, the average TA muscle fiber CSA was similar between mdx littermates with and without miR-133b (Fig. 4A, ,B,B, ,E).E). However, a muscle fiber CSA distribution analysis revealed a slight, but statistically significant, shift towards increased abundance of muscle fibers with smaller CSAs in mdx;miR-133b−/− versus mdx;miR-133b+/+ mice (Fig. 4F). We also observed an uptick in muscle fibers with centralized nuclei (Fig. 4G) alongside a trend towards reduced presence of necrotic muscle fibers in mdx;miR-133b−/− mice (Fig. 4H, p = 0.1208). Other histopathological indices were similar in the TA muscle at P60 between mdx;miR-133b+/+ and mdx;miR-133b−/− mice, including the laminin-positive areas within the interstitial space (Fig. 4I) and the number of interstitial nuclei (Fig. 4J). Additionally, miR-133a levels were similar between mdx;miR-133b+/+ and mdx;miR-133b−/− mice (Fig. 4K). Deletion of miR-133b also did not affect the body weight (Fig. 4L) or overall outward appearance of P60 mdx mice. At P90, all histopathological differences between mdx;miR-133b−/− and mdx;miR-133b+/+ muscle were absent in our observations. We found no difference in the overall TA muscle CSA (Fig. 5C), muscle fiber CSA (Fig. 5A, ,B,B, ,D,D, ,E)E) and numbers of muscle fibers with central nuclei (Fig. 5F). The interstitial area (Fig. 5G), numbers of interstitial nuclei (Fig. 5H), and presence of necrotic muscle fibers (Fig. 5I) were also similar in the TA muscle of P90 mdx;miR-133b−/− compared to mdx;miR-133b+/+ mice. These results suggest that the effects of miR-133b deletion become less pronounced with the continued remission of DMD pathology that is typical of skeletal muscles in P60 and P90 mdx mice (Willmann et al., 2009).
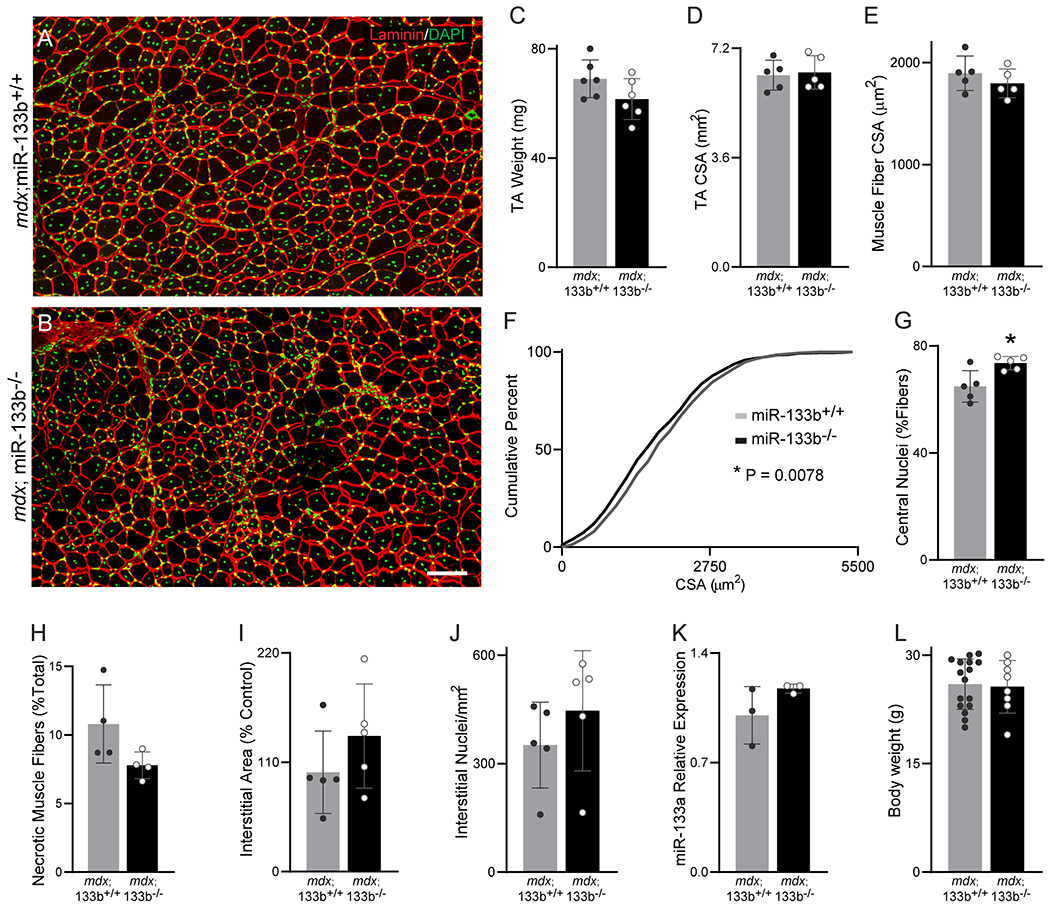
Assessment of miR-133b deletion on DMD pathogenesis in P60 mdx TA. (A-B) Representative images of laminin (red) and DAPI (green) staining of P60 (A) mdx;miR-133b+/+ and (B) mdx;miR-133b−/− TA cross sections. (C) TA mass (n = 6, p = 0.1035, unpaired T-test). (D) Overall CSA of the TA muscle (p = 0.7879, unpaired T-test). (E) Mean muscle fiber CSA (p = 0.3459, unpaired T-test). (F) Cumulative frequency distribution curve of muscle fiber CSAs (p = 0.0078, Kolmogorov-Smirnov test). (G) Numbers of muscle fibers with centrally located nuclei (p = 0.0163, unpaired T-test). (H) Numbers of necrotic muscle fibers (n = 4, p = 0.1208, unpaired T-test with Welch’s correction). (I) Area of the interstitial space (p = 0.2574, unpaired T-test). (J) Levels of interstitial nuclei (p = 0.3291, unpaired T-test). (K) Expression analysis of pre-miR-133a levels in the TA using qPCR (n = 3, p = 0.1856, unpaired T-test). (L) Mouse body weight (p = 0.8147, n = 8-16, unpaired T-test). n = 5 unless otherwise noted. All values, except F, reported as mean ± standard deviation. miR-133a expression normalized to β2 Microglobulin using the 2−ΔΔCT method. * p < 0.05, scale bar = 100 μm.
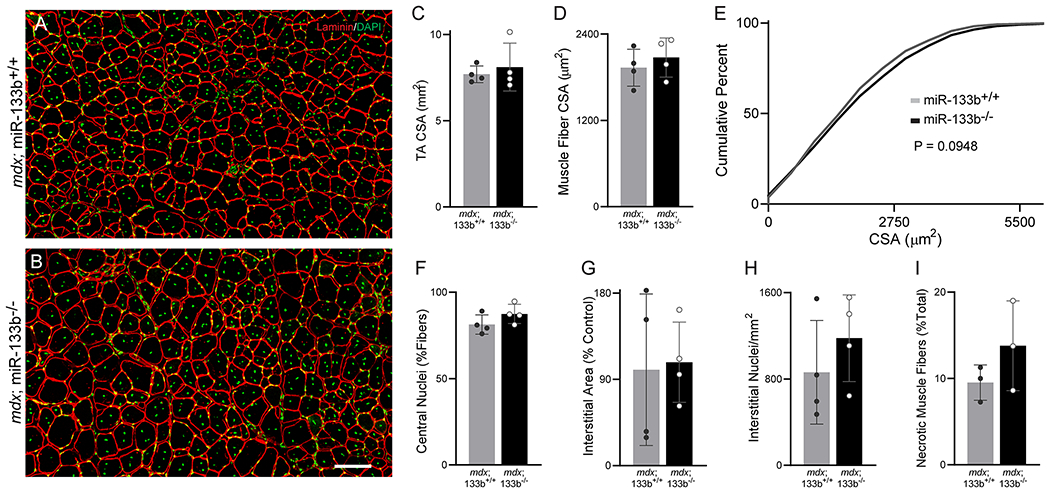
Assessment of miR-133b deletion on DMD pathogenesis in P90 mdx muscle. (A-B) Representative images of laminin (red) and DAPI (green) staining of P60 (A) mdx;miR-133b+/+ and (B) mdx;miR-133b−/− TA cross sections. (C) Measurement of overall CSA of the TA muscle (p = 0.5936, unpaired T-test). (D) Mean muscle fiber CSA (p = 0.4773, unpaired T-test). (E) Cumulative frequency distribution curve of muscle fiber CSAs (p = 0.0948, Kolmogorov-Smirnov test). (F) Number of muscle fibers with centrally located nuclei (p = 0.1715, unpaired T-test). (G) Interstitial area (p = 0.8660, unpaired T-test). (H) Numbers of interstitial nuclei (p = 0.3513, unpaired T-test). (I) Abundance of necrotic muscle fibers (n = 3, p = 0.2563, unpaired T-test). n = 4 unless otherwise noted. All values, except E, reported as mean ± standard deviation. Scale bar = 100 μm.
Loss of miR-133b does not directly affect NMJs in mdx mice
Although we previously showed that neuromuscular junctions (NMJs) are unaffected in miR-133b null mice (Valdez et al., 2014), it is possible that NMJs afflicted with DMD require miR-133b, due to the high rate of NMJ denervation and reinnervation associated with muscle fiber turnover (Pratt et al., 2015). To address this possibility, we examined NMJs in the extensor digitorum longus (EDL) muscle of mdx;miR-133b−/− mice following labeling of nicotinic acetylcholine receptors (nAChRs) with fluorescently-tagged alpha-bungarotoxin (fBTX). As a hindlimb muscle located adjacent to the TA with similar muscle fiber type composition, the EDL was selected for analysis due to the relative ease with which NMJs can be imaged as compared to the TA. This analysis showed that loss of miR-133b resulted in diminished endplate area (Fig. 6A, ,B,B, ,C)C) and postsynaptic area, marked by nAChRs, (Fig. 6D). These differences in P30 mdx;miR-133b−/− mice may be due to a reduction in muscle size, as shown for the TA (Fig. 3), and not from direct changes in the number of NMJs or their architecture. In support of this hypothesis, we found no difference in post-synaptic fragmentation of NMJs between P30 mdx;miR-133b−/− and mdx;miR-133b+/+ mice (Fig. 6E). Furthermore, NMJs were indistinguishable at P60 between mdx;miR-133b−/− mice and mdx;miR-133b+/+ mice (Fig. 6F–J). These findings are also in line with the muted effect of miR-133b deletion on skeletal muscles of mdx mice at P60.
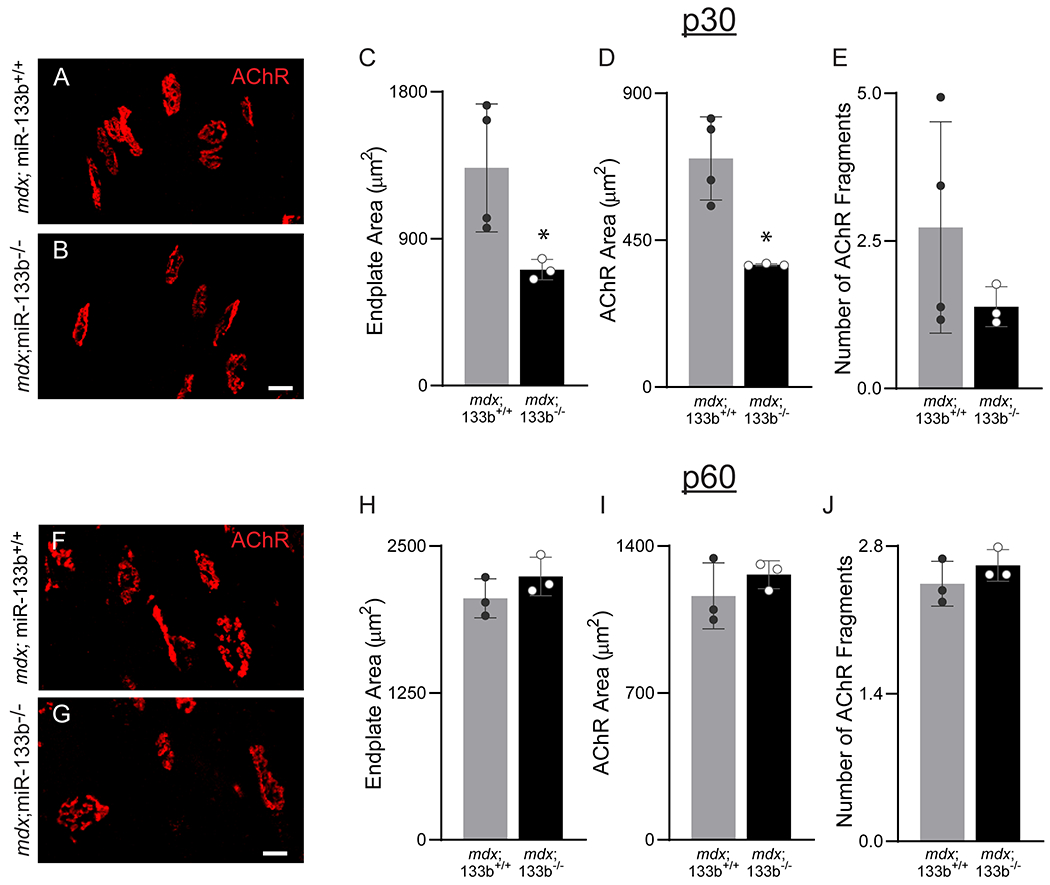
Assessment of miR-133b deletion on the NMJ in P30 and P60 mdx muscle. (A-B) Representative images of nAChRs at the NMJ using fBTX in P30 (A) mdx;miR-133b+/+ and (B) mdx;miR-133b−/− EDL muscle. Analysis of (C) endplate area (p = 0.0479, unpaired T-test with Welch’s correction), (D) nAChR Area (p = 0.0143, unpaired T-test with Welch’s correction), and (E) post-synaptic fragmentation (p = 0.2318, unpaired T-test with Welch’s correction) in P30 mdx;miR-133b+/+ and mdx;miR-133b−/− EDL. (F-G) Representative images of nAChRs at the NMJ using fBTX in (F) P60 mdx;miR-133b+/+ and (G) mdx;miR-133b−/− EDL muscle. Analysis of (H) endplate area (p = 0.2376, unpaired T-test), (I) nAChR Area (p = 0.3633, unpaired T-test with Welch’s correction), and (J) post-synaptic fragmentation (p = 0.3124, unpaired T-test) in P60 mdx;miR-133b+/+ and mdx;miR-133b−/− EDL. n = 3, all groups except P30 mdx;miR-133b+/+ where n = 4. All values reported as mean ± standard deviation. * p < 0.05, scale bar = 100 μm.
Impact of miR-133b deletion on satellite cells in mdx mice
In vitro studies have uncovered important roles for miR-133b in satellite cell proliferation and differentiation (Horak et al., 2016; Mok et al., 2017; Cui et al., 2019), raising the possibility that loss of miR-133b exacerbates DMD-pathogenesis by affecting satellite cells. To address this possibility, we examined the number of Pax7+ satellite cells in TA muscle cross sections of P30, P60, and P90 mdx;miR-133b+/+ and mdx;miR-133b−/− mice. We did not observe a statistically significant difference in numbers of Pax7+ satellite cells in the TA of mdx;miR-133b−/− as compared to mdx;miR-133b+/+ mice during the initial stage of muscle regeneration at P30 (Fig. 7A, ,B,B, ,G,G, p = 0.1474). However, fewer Pax7+ satellite cells were observed in mdx;miR-133b−/− compared to mdx;miR-133b+/+ muscle with increased age, at both P60 (Fig. 7C, ,D,D, ,H)H) and P90 (Fig. 7E, ,F,F, ,I).I). These results suggest that miR-133b deletion affects the proliferative response of satellite cells in DMD-afflicted muscle.
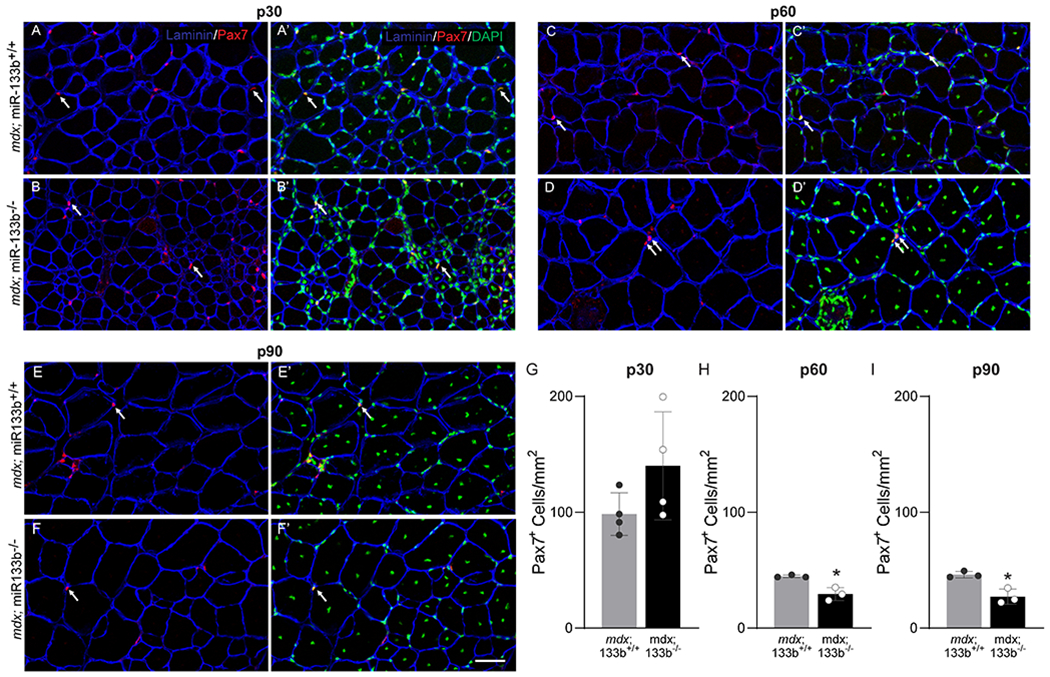
The number of satellite cells is altered in mdx;miR-133b−/− muscle. (A-F) Representative images of laminin (blue), Pax7 (red), and DAPI (green) IHC performed on TA cross sections collected from P30 (A-B), P60 (C-D), and P90 (E-F) mdx;miR-133b+/+ and mdx;miR-133b−/− mice. White arrows indicate examples of Pax7/DAPI double positive nuclei used to identify satellite cells. (G-I) Quantification of satellite cell density based on the number of Pax7+ satellite cells at (G) P30 (n = 4, p = 0.1474, unpaired T-test), (H) P60 (n = 3, p = 0.0097, unpaired T-test) and (I) P90 (n = 3, p = 0.0101, unpaired T-test) in mdx;miR-133b−/− as compared to mdx;miR-133b+/+ muscle. All values reported as mean ± standard deviation. * p < 0.05, scale bar = 50 μm.
Deletion of miR-133b moderately affects muscle regeneration in acutely stressed skeletal muscles in healthy young adult mice
The results above raised the prospect that miR-133b also plays a critical role in the repair of skeletal muscles affected by other stressors. We thus asked if miR-133b must be present for muscle fibers to regenerate properly and in a timely fashion following an acute injury. For this, we administered cardiotoxin (CTX), which causes muscle fibers to rapidly degenerate (Duchen et al., 1973, 1974), to the TA muscle of healthy miR-133b+/+ and miR-133b−/− littermates. We found that as muscle fibers begin to reform, at 7 days (7d) post-CTX injection, the average muscle fiber CSA was lower (Fig. 8A, ,B,B, ,G)G) and there were fewer muscle fibers with larger CSAs (Fig. 8E) in miR-133b−/− as compared to miR-133b+/+ mice. This trend towards impaired muscle regeneration continued at 14d post-CTX injection (Fig. 8C, ,D).D). We observed a downward shift in muscle fiber CSA distribution (Fig. 8F) and an overall decrease in average muscle fiber CSA (Fig. 8G) in miR-133b−/− TA, suggesting that regenerating muscle fibers mature at a slower rate in the absence of miR-133b. These differences do not appear to result from changes associated with the interstitial space since the number of interstitial nuclei was similar in miR-133b−/− versus miR-133b+/+ muscle at both 7d and 14d post-CTX (Fig. 8H). The slower rate of muscle fiber regeneration also does not seem to be caused by a lower abundance of Pax7+ satellite cells (Fig. 8I). Thus, miR-133b does not have an obvious impact on satellite cells or mononucleated cells in the interstitial space in transiently stressed skeletal muscles. This is in stark contrast to chronically stressed skeletal muscles in the mdx TA, where miR-133b must be present to minimize the accumulation of pathological features.
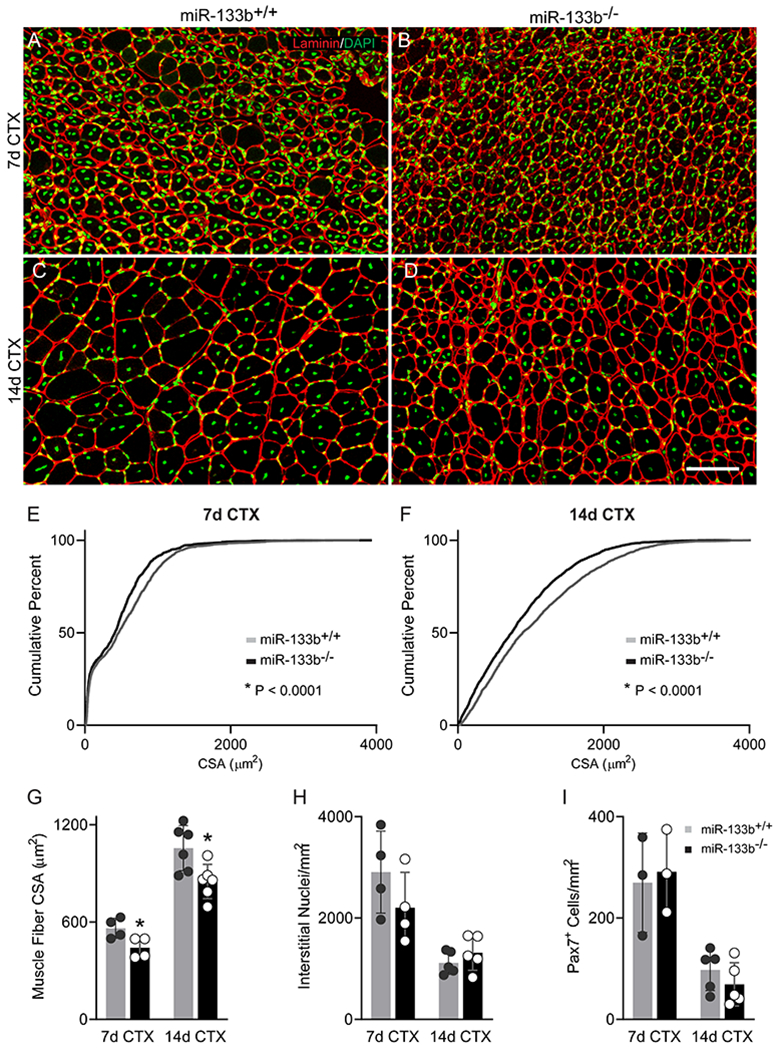
Deletion of miR-133b from control mice delays muscle fiber regeneration following an acute injury. Cardiotoxin (CTX) was administered to the TA muscle of healthy P60 miR-133b+/+ and miR-133b−/− littermates to induce muscle regeneration. (A-D) Representative images of laminin (red) and DAPI (green) IHC in TA muscle collected at 7d (A-B) and 14d (C-D) post-CTX from miR-133b+/+ and miR-133b−/− mice. (E-F) Cumulative frequency distribution curve of muscle fiber CSAs in miR-133b+/+ versus miR-133b−/− TA at (E) 7d post-CTX (p < 0.0001, Kolmogorov–Smirnov test) and (F) 14d post-CTX (p < 0.0001, Kolmogorov-Smirnov test). (G) Average muscle fiber CSA at 7 and 14d post-CTX (7d post-CTX, p = 0.0352, 14d post-CTX, p = 0.0081, unpaired T-test). (H) Analysis of numbers of nuclei in the interstitial space (7d post-CTX, n = 4, p = 0.2370; 14d post-CTX, n = 5, p = 0.3105, unpaired T-test). (I) Quantification of Pax7+ satellite cells (7d post-CTX, n = 3, p = 0.7845; 14d post-CTX, n = 5, p = 0.3129, unpaired T-test). All values, except E and F, reported as mean ± standard deviation. Unless otherwise noted, 7d CTX, n = 4; 14d CTX, n = 6. * p < 0.05, scale bar = 50 μm.
Deleting miR-133b accelerates satellite cell proliferation but slows the formation of myotubes
To determine if loss of miR-133b directly impacts satellite cell proliferation and their ability to generate new myofibers, we isolated and cultured satellite cells from p15-p20 miR-133b+/+ and miR-133b−/− littermates. In order to assess rates of proliferation, satellite cells were seeded at a low density so that proliferation could be observed before the culture approached confluency. After 72 h in vitro, Pax7+ satellite cells lacking miR-133b proliferated faster compared to miR-133b+/+ controls (Fig. 9A–C). To determine whether miR-133b impacts satellite cell differentiation and fusion into myotubes, we seeded satellite cells at a high density and treated with differentiation media. At both 2 (Fig. 9D, ,E)E) and 5 (Fig. 9F,,G)G) days post-differentiation we observed smaller myosin+ myotubes in miR-133b−/− versus miR-133b+/+ cultures (Fig. 9H), suggesting an impairment of myotube formation in the absence of miR-133b. Taken together, these results show that miR-133b functions to control satellite cell proliferation and formation of myofibers, which are two cellular processes altered in in mdx;miR-133b−/− muscle.
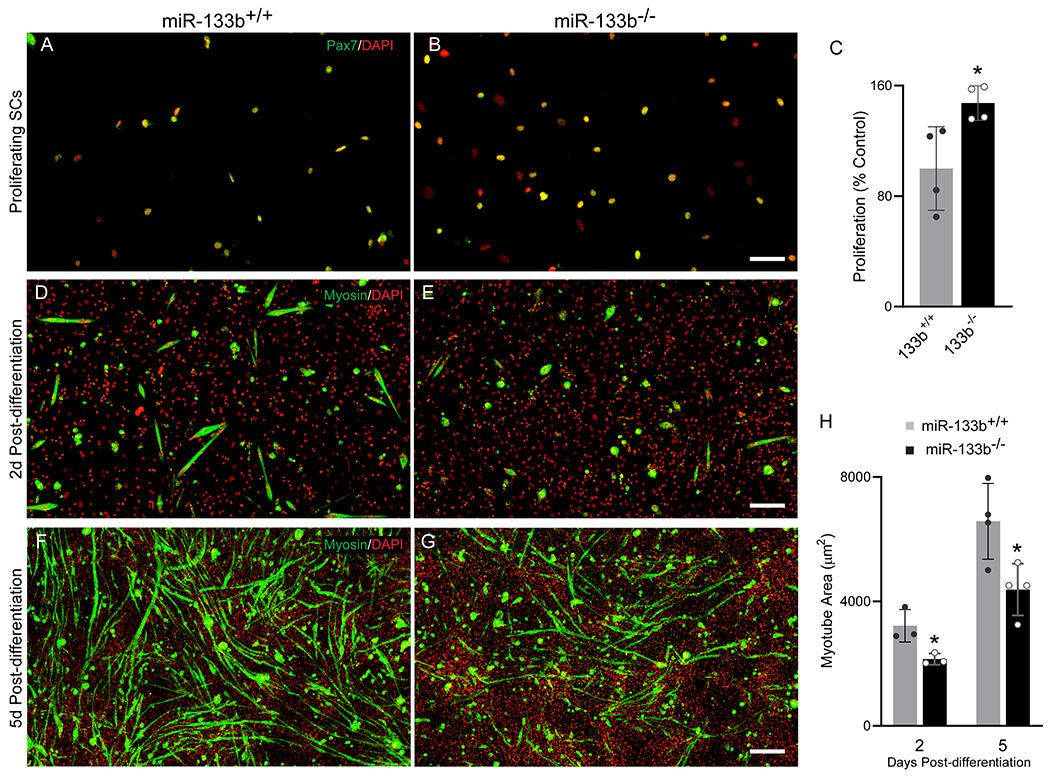
Deletion of miR-133b enhances satellite cell proliferation but inhibits myotube formation. (A & B) Representative images of Pax7 (green) and DAPI (red) immunocytochemistry of miR-133b+/+ and miR-133b−/− primary satellite cell-enriched cultures at 72 h in vitro. (C) Quantification of satellite cell proliferation in miR-133b+/+ and miR-133b−/− primary satellite cell-enriched cultures, determined by measuring the ratio of Pax7+ satellite cells at 72 h versus <16 h in vitro (n = 4, p = 0.0275, unpaired T-test). (D-E) Representative images of myosin (green) and DAPI (red) immunocytochemistry at 2d post-differentiation (PD) in miR-133b+/+ and miR-133b−/− primary satellite cell-enriched cultures. (F-G) Myosin/DAPI immunocytochemistry at 5d PD. (H) Analysis of average myotube area in miR-133b+/+ and miR-133b−/− primary satellite cell-enriched cultures at 2d and 5d PD. (2d PD, n = 3, p = 0.0284; 5d PD, n = 4, p = 0.0248, unpaired T-test). All values reported as mean ± standard deviation. * p < 0.05, scale bar = 200 μm.
Molecular signatures associated with loss of miR-133b in mdx mice
The increased DMD-pathogenesis in mdx;miR-133b−/− mice indicates that miR-133b impacts expression of protein-coding genes with important functions in DMD-afflicted skeletal muscles. To identify genes altered in the TA muscle lacking miR-133b, we compared the transcriptome between wild type (miR-133b+/+), miR-133b−/−, mdx;miR-133b+/+, and mdx;miR-133b−/− mice using RNA seq. Genes with an absolute log2 fold change greater than 1 and adjusted p-value less than 0.01 were deemed differentially expressed between genotypes. This analysis revealed the following: 1) very few genes were differentially expressed between healthy miR-133b+/+ and miR-133b−/− mice (Fig. 10A), partly corroborating cellular analysis showing that loss of miR-133b has no obvious effect on otherwise healthy skeletal muscles (Fig. 2) and NMJs (Valdez et al., 2014); 2) myriad genes were significantly altered in the TA muscle of mdx compared to wild type control mice (Fig. 10B); 3) deletion of miR-133b further altered the transcriptome of the TA muscle in mdx mice (Fig. 10C).
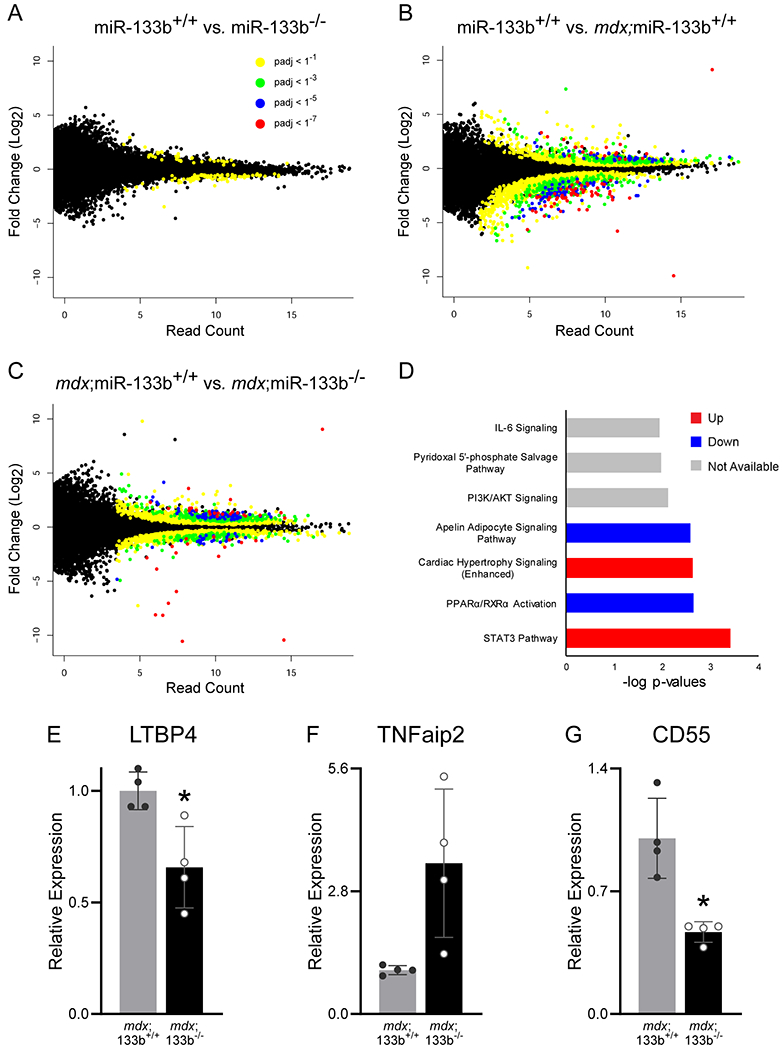
Transcriptomic changes resulting from miR-133b deletion in P30 healthy and mdx TA muscle. (A) Scatter plot comparison of differentially expressed genes in miR-133b+/+ versus miR-133b−/− TA muscle, as identified by RNA seq. Read count in miR-133b+/+ muscle is represented on the x-axis and log2 fold change versus miR-133b−/− is represented on the y-axis. Adjusted p-value for each gene expression comparison is represented by dot coloration. (B) Scatter plot comparison of genes expressed in healthy (miR-133b+/+) versus DMD-afflicted (mdx;miR-133b+/+) muscle. Read count in miR-133b+/+ muscle is represented on the x-axis and log2 fold change versus mdx;miR-133b+/+ is represented on the y-axis, p-values color coded according to legend in panel A. (C) Scatter plot comparison of genes expressed in mdx;miR-133b+/+ versus mdx;miR-133b−/− muscle. Read count in mdx;miR-133b+/+ muscle is represented on the x-axis and log2 fold change versus mdx;miR-133b−/− muscle is represented on the y-axis, p-values color coded according to legend in panel A. (D) Top signaling pathways impacted by deletion of miR-133b in mdx muscle as identified by Ingenuity Pathway Analysis of genes with a fold change absolute value of greater than 2 and adjusted p value of less than 0.01 in P30 mdx;miR-133b+/+ versus mdx;miR-133b−/− TA. Pathways identified as being differentially regulated where the directionality could not be determined are labeled as unknown. (E-G) qPCR validation of differentially regulated genes related to fibrosis and inflammation, including (E) LTBP4 (p = 0.0160, unpaired T-test), (F) TNFaip2 (p = 0.0629, unpaired T-test with Welch’s correction), and (G) CD44 (p = 0.0152, unpaired T-test with Welch’s correction). n = 4. Values in E-G normalized to β2 Microglobulin using the 2−ΔΔCT method and reported as mean ± standard deviation. * p < 0.05.
To gain insights into the signaling pathways that miR-133b alters to exacerbate DMD-pathology in mdx mice, we used Ingenuity Pathway Analysis (IPA). This revealed several pathways with particular relevance to DMD pathogenesis that were closely associated with deletion of miR-133b in mdx mice (Fig. 10D) including STAT3, IL-6, and Apelin. STAT3 has been shown to be expressed by satellite cells where it plays a pivotal role in myogenesis by promoting satellite cell proliferation and inhibiting the formation of myotubes (Guadagnin et al., 2018). Recent studies have demonstrated that IL-6 is closely associated with infiltrating cells of the interstitial space and is downregulated in response to glucocorticoid treatment in DMD patients (Guadagnin et al., 2018). Apelin has been characterized as an exercised-induced signaling molecule that prevents sarcopenia, improves muscle regeneration, and decreases inflammation in aging muscle (Vinel et al., 2018). Modulation of these pathways by miR-133b may serve to protect muscle from DMD pathogenesis.
Genes involved in inflammation, TGF-β signaling, and other aspects of DMD pathology were prominent among those upregulated in mdx;miR-133b−/− muscle. This list of genes included a number of known miR-133b targets that were identified using miRTarBase (Hsu et al., 2011) (Table 2). These included RhoA, a well characterized target of miR-133b (Qin et al., 2012) that is associated with myoblast fusion, muscle regeneration, fatty infiltration and effectiveness of glucocorticoid treatment in mdx mice (Hosoyama et al., 2009; Doherty et al., 2011; Hindi et al., 2013; Mu et al., 2013, 2017). It also included members of the TGF-β signaling pathway (Table 3), which promotes fibrosis in DMD-afflicted muscle (Juban et al., 2018). Specifically, the TGF-β receptor, TGF-β-induced factor homeobox 1 (TGIF1), SMAD3, and SMAD5 were found upregulated in mdx;miR-133b−/− muscle (Table 3) (Wotton et al., 1999; Burks & Cohn, 2011). We confirmed trends in expression patterns that are suggestive of heightened inflammation using qPCR. In particular, we observed decreased expression of latent TGF-β binding protein 4 (LTBP4) in mdx;miR-133b−/− muscle (Fig. 10E). LTBP4 inhibits TGF-β by sequestering it in the extracellular matrix and it is one of a handful of genes known to affect disease progression in DMD patients (Flanigan et al., 2013; Juban et al., 2018). TNFα induced protein 2 (TNFAIP2) is induced in response to TNFα signaling (Jia et al., 2018) and has been shown to influence epithelial to mesenchymal transition (EMT) (Niwa et al., 2019). Given the role of TNFα in inflammation, increased levels of TNFAIP2 in mdx;miR-133b−/− mice may be indicative of a heightened M1 inflammatory response (Jia et al., 2018). This difference was statistically significant in the RNA seq data, however it was not statistically significant in the qPCR results (Fig. 10F, p = 0.0629) despite a similar trend. Similarly, the decreased levels of CD55, a complement inhibitor that is associated with anti-inflammatory responses in immune cells (Sutavani et al., 2013), in mdx;miR-133b−/− muscle (Fig. 10G) further suggest a heightened and dysregulated inflammatory response in the absence of miR-133b. Overall, this expression dataset supports important roles for miR-133b in mitigating DMD pathogenesis. It also provides promising candidate genes and signaling modules that could be directly targeted to attenuate the various pathological features associated with DMD.
Table 2.
miR-133b targets upregulated in mdx; miR-133b−/− versus mdx; miR-133b+/+ TA muscle. Gene upregulation determined from RNA seq. Targets of miR-133b identified by the miRTarBase database. Data are available at NCBI GEO, (https://www.ncbi.nlm.nih.gov/geo/, accession # GSE156267).
| Gene | Function | Implications in DMD | Validation Methods | References |
|---|---|---|---|---|
| RhoA | GTPase | Impairs myoblast fusion and promotes adipogenesis | 4 | (Hosoyama et al., 2009; Doherty et al., 2011; Hindi et al., 2013; Mu et al., 2013, 2017) |
| SP1 | Transcription factor | Regulates myogenesis | 2 | (Viñals et al., 1997) |
| Tnrc6b | RNA-induced silencing | Myo-miR related myogenesis | 1 | (Meister, 2013; Horak et al., 2016) |
| PIAS2 | E3 ubiquitin ligase | Inhibits the satellite cell differentiation molecule STAT2 | 1 | (Wang et al., 2008) |
| PTBP2 | miRNA biogenesis | Myo-miR related myogenesis | 3 | (Xu & Hecht, 2011; Horak et al., 2016) |
Table 3.
TGF-β pathway components differentially regulated in mdx; miR-133b−/− as compared to mdx; miR-133b+/+ TA muscle, as identified by RNA seq. Data are available at NCBI GEO, (https://www.ncbi.nlm.nih.gov/geo/, accession # GSE156267).
| Gene | Name | Function | Up or down in absence of miR-133b? | References |
|---|---|---|---|---|
| LTBP4 | Latent TGF-β binding protein 4 | Sequesters TGF-β in ECM | Down | (Burks & Cohn, 2011) |
| SMAD3 | SMAD3 | Transcription factor | Up | (Burks & Cohn, 2011) |
| SMAD5 | SMAD5 | Transcription factor | Up | (Burks & Cohn, 2011) |
| TAB3 | TAK3/MAP3K7 binding protein 3 | Activate TGFB activated kinase | Up | (Broglie et al., 2010) |
| TGFβR | TGFβ Receptor | Transmembrane receptor | Up | (Burks & Cohn, 2011) |
| TGIF1 | TGFβ-induced factor homeobox 1 | Transcriptional co-repressor | Up | (Wotton et al., 1999) |
Discussion
This is the first study to directly assess the role of miR-133b on DMD pathogenesis and the regenerative capacity of skeletal muscle in vivo. We show that deletion of miR-133b reduces the size of muscle fibers without profoundly impacting muscle fiber degeneration in the severe stage of muscle dystrophy in the TA muscle of mdx mice. Importantly, we provide cellular and molecular mechanistic insights on how miR-133b impacts DMD pathogenesis. We demonstrate that deletion of miR-133b impacts the proliferation of satellite cell and their ability to form myofibers. We also uncover genes and signaling modules that are altered when miR-133b is deleted in mdx mice. Altogether, our findings provide evidence that miR-133b and its downstream targets affect the regenerative capacity of skeletal muscles, possibly through satellite cells.
The impact of miR-133b on satellite cells is a significant finding in this study because of the role these cells play in the progression of DMD pathogenesis. Specifically, it is clear that satellite cell dysfunction contributes to the loss of skeletal muscle mass and increased presence of fibrotic and adipose tissue that accompanies the progression of DMD (Yin et al., 2013b; Chang et al., 2016). We show that miR-133b plays an important role in modulating the capacity of satellite cells to generate myofibers. In culture, we found that miR-133b deletion in satellite cells inhibited myotube formation. This suggests that miR-133b may be upregulated in satellite cells to promote differentiation into myotubes, a finding that has been demonstrated by others (Hak et al., 2006; Feng et al., 2013; Cui et al., 2019). Corroborating these findings, deletion of miR-133b increased the number of small muscle fibers in mdx mice at P30, an age at which skeletal muscle fiber degeneration is severe. Loss of miR-133b from wild type mice also slowed regeneration of muscle fibers following CTX-induced injury. Providing further evidence that miR-133b deletion impacts the regenerative response of skeletal muscle precursor cells, STAT3 signaling was foremost among upregulated pathways in mdx;miR-133b−/− muscle. STAT3 signaling has a well described role in inhibiting satellite cell differentiation. Together, these findings suggest that deletion of miR-133b impairs muscle fiber regeneration, possibly by disrupting the differentiation of satellite cells.
Along with promoting differentiation, miR-133b may be upregulated in DMD to promote quiescence of activated satellite cells, a process that is necessary to replenish and maintain the pool of muscle precursor cells (Zammit et al., 2006). In support of this, we observed elevated rates of proliferation in satellite cells lacking miR-133b in culture, suggestive of a preference towards activation versus quiescence. Following acute muscle damage, either in the initial stages of degeneration in mdx muscle at P30 or following CTX-induced injury, we observed similar or slightly higher numbers of satellite cells in the absence of miR-133b. Following an extended period of skeletal muscle fiber degeneration and regeneration in P60 and P90 mdx mice, however, we observed fewer satellite cells in miR-133b−/− muscle. Thus, miR-133b may play a role in regulating continuous proliferation of satellite cells during chronic, asynchronous muscle regeneration in dystrophic muscle. An examination of molecular markers of activated satellite cells, such as MyoD (Zammit et al., 2006), in future studies would shed light on this possibility. Regardless, these data add to published studies indicating that miR-133b is an important regulator of the myogenic program of satellite cells (Hak et al., 2006; Chen et al., 2006; Yin et al., 2013a; Feng et al., 2013; Cui et al., 2019). Thus, the varying levels of miR-133b at specific stages of DMD severity in mdx muscle (i.e. P30 and P60) may be necessary to both control the proliferation and differentiation of satellite cells to promote regeneration of muscle fibers.
In addition to orchestrating muscle regeneration, this study demonstrates an additional and previously unknown role for miR-133b in influencing the expression of genes related to inflammation and fibrosis in mdx muscle. Of particular therapeutic relevance is miR-133b regulation of IL-6 signaling, reductions of which are associated with improved outcomes following steroid treatment (Guadagnin et al., 2018), and LTBP4, which has been identified as a genetic factor of disease progression in DMD patients (Flanigan et al., 2013; Juban et al., 2018). Evidence of increased inflammation is supported by an elevated presence of mononucleated cells in the interstitial space. While the identity of these cells was not determined in this study, it is possible that miR-133b may directly influence levels of fibroblasts, adipocytes, and other cells residing in skeletal muscles. Along these lines, previous work has established a role of miR-133 in regulating levels of adipocytes in skeletal muscle tissue (Yin et al., 2013a).
In contrast to mdx mice, deletion of miR-133b from healthy control mice had a limited effect on skeletal muscles, suggesting that miR-133b is utilized in responses to damage rather than prevention of degeneration. Muscle fiber size, satellite cell number, ECM thickness, and levels of infiltrating cells were normal in healthy muscle lacking miR-133b. Similarly, RNA seq analysis identified only two differentially regulated genes related to deletion of miR-133b in healthy muscle while over 600 genes were differentially regulated in mdx muscle lacking miR-133b. These findings are in line with previous studies demonstrating that miRNAs are primarily utilized by cells in mature tissues to coordinate responses under stress (Williams et al., 2009b; Russell et al., 2013; Emde & Hornstein, 2014; Horak et al., 2016). Along these lines, deletion of miR-133b greatly influenced skeletal muscle fiber regeneration but had a limited impact on muscle fiber degeneration in mdx muscle, as levels of necrotic fibers were roughly similar between mdx muscle with and without miR-133b at all ages examined. Taken together, these results support roles for miR-133b in promoting regeneration but not degeneration of muscle fibers.
Impairments in myogenesis, both on the level of satellite cell proliferation and muscle fiber maturation, and increases in cell signaling associated with fibrosis and inflammation are hallmarks of DMD pathology. These cellular and molecular features inevitably contribute to progressive loss of muscle fibers, muscle weakness, and death. Hence, it is critical to continue to identify targets that could slow these features of DMD. Our results provide evidence that miR-133b, a short oligonucleotide, could be used to mitigate DMD pathology by impacting the proliferation and differentiation of satellite cells while reducing expression of genes associated with inflammation and fibrosis. Thus, future experiments will assess the impact of manipulating miR-133b levels and the action of downstream targets at specific stages of DMD pathology.
Acknowledgements
The authors wish to thank Dr. Julia von Maltzahn for donating the Pax7 antibody used in this study and for assistance with establishing a Pax7 IHC protocol for the laboratory.
Funding
This work was funded through grants from the National Institute on Aging (R01AG055545 and R56AG051501) and the National Institute of Neurological Disorders and Stroke (R21NS106313) awarded to GV.
Author Profile:
Tom Taetzsch has been a member of Gregorio Valdez’s laboratory since 2015. During this time, he has researched molecules that impact the health of skeletal muscles and the neuromuscular junction, particularly in disease in aging. Tom aspires to make discoveries in the laboratory that will make people’s lives better.
Footnotes
Data Availability
RNA seq data are deposited at NCBI GEO, (https://www.ncbi.nlm.nih.gov/geo/, accession # GSE156267).
Competing Interests
The authors have no competing interests to declare.
References
- Almada AE & Wagers AJ (2016). Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat Rev Mol Cell Biol 17, 267–279. [Europe PMC free article] [Abstract] [Google Scholar]
- Anders S, Pyl PT & Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [Europe PMC free article] [Abstract] [Google Scholar]
- Aronesty E (2013). Comparison of Sequencing Utility Programs. Open Bioinforma J 7, 1–8. [Google Scholar]
- Ballarino M, Morlando M, Fatica A & Bozzoni I (2016). Non-coding RNAs in muscle differentiation and musculoskeletal disease. J Clin Invest 126, 2021–2030. [Europe PMC free article] [Abstract] [Google Scholar]
- Bencze M, Periou B, Baba-Amer Y & Authier FJ (2019). Immunolabelling myofiber degeneration in muscle biopsies. J Vis Exp 2019, 59754. [Abstract] [Google Scholar]
- Boettger T, Wüst S, Nolte H & Braun T (2014). The miR-206/133b cluster is dispensable for development, survival and regeneration of skeletal muscle. Skelet Muscle 4, 1–13. [Europe PMC free article] [Abstract] [Google Scholar]
- Broglie P, Matsumoto K, Akira S, Brautigan DL & Ninomiya-Tsuji J (2010). Transforming growth factor β-activated kinase 1 (TAK1) Kinase Adaptor, TAK1-binding protein 2, plays dual roles in TAK1 signaling by recruiting both an activator and an inhibitor of TAK1 kinase in tumor necrosis factor signaling pathway. J Biol Chem 285, 2333–2339. [Europe PMC free article] [Abstract] [Google Scholar]
- Bulfield G, Siller WG, Wight PA & Moore KJ (1984). X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A 81, 1189–1192. [Europe PMC free article] [Abstract] [Google Scholar]
- Burks TN & Cohn RD (2011). Role of TGF-β signaling in inherited and acquired myopathies. Skelet Muscle 1, 19. [Europe PMC free article] [Abstract] [Google Scholar]
- Cacchiarelli D, Legnini I, Martone J, Cazzella V, D’Amico A, Bertini E & Bozzoni I (2011). miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol Med 3, 258–265. [Europe PMC free article] [Abstract] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A & Bozzoni I (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. [Europe PMC free article] [Abstract] [Google Scholar]
- Chang NC, Chevalier FP & Rudnicki MA (2016). Satellite Cells in Muscular Dystrophy - Lost in Polarity. Trends Mol Med 22, 479–496. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen J-F, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL & Wang D-Z (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38, 228–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Coenen-Stass AML, Sork H, Gatto S, Godfrey C, Bhomra A, Krjutškov K, Hart JR, Westholm JO, O’Donovan L, Roos A, Lochmüller H, Puri PL, EL Andaloussi S, Wood MJA & Roberts TC (2018). Comprehensive RNA-Sequencing Analysis in Serum and Muscle Reveals Novel Small RNA Signatures with Biomarker Potential for DMD. Mol Ther - Nucleic Acids 13, 1–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Cui S, Li L, Mubarokah SN & Meech R (2019). Wnt/β-catenin signaling induces the myomiRs miR-133b and miR-206 to suppress Pax7 and induce the myogenic differentiation program. J Cell Biochem 120, 12740–12751. [Abstract] [Google Scholar]
- Doherty JT, Lenhart KC, Cameron M V, Mack CP, Conlon FL & Taylor JM (2011). Skeletal muscle differentiation and fusion are regulated by the BAR-containing Rho-GTPase-activating protein (Rho-GAP), GRAF1. J Biol Chem 286, 25903–25921. [Europe PMC free article] [Abstract] [Google Scholar]
- Duchen LW, Excell BJ, Patel R & Smith B (1973). Proceedings: Light and electron microscopic changes in mouse muscle fibres and motor end-plates caused by the depolarizing fraction (cardiotoxin) of the of the venom of Dendroaspis jamesoni. J Physiol 234, 1P–2P. [Abstract] [Google Scholar]
- Duchen LW, Excell BJ, Patel R & Smith B (1974). Changes in motor end-plates resulting from muscle fibre necrosis and regeneration. A light and electron microscopic study of the effects of the depolarizing fraction (cardiotoxin) of Dendroaspis jamesoni venom. J Neurol Sci 21, 391–417. [Abstract] [Google Scholar]
- Duddy W, Duguez S, Johnston H, Cohen TV, Phadke A, Gordish-Dressman H, Nagaraju K, Gnocchi V, Low SH & Partridge T (2015). Muscular dystrophy in the mdx mouse is a severe myopathy compounded by hypotrophy, hypertrophy and hyperplasia. Skelet Muscle 5, 16. [Europe PMC free article] [Abstract] [Google Scholar]
- Eisenberg I, Alexander MS & Kunkel LM (2009). miRNAS in normal and diseased skeletal muscle. J Cell Mol Med 13, 2–11. [Europe PMC free article] [Abstract] [Google Scholar]
- Emde A & Hornstein E (2014). miRNAs at the interface of cellular stress and disease. EMBO J 33, 1428–1437. [Europe PMC free article] [Abstract] [Google Scholar]
- Feng Y, Niu LL, Wei W, Zhang WY, Li XY, Cao JH & Zhao SH (2013). A feedback circuit between miR-133 and the ERK1/2 pathway involving an exquisite mechanism for regulating myoblast proliferation and differentiation. Cell Death Dis 4, e934–e934. [Europe PMC free article] [Abstract] [Google Scholar]
- Flanigan KM, Ceco E, Lamar K-M, Kaminoh Y, Dunn DM, Mendell JR, King WM, Pestronk A, Florence JM, Mathews KD, Finkel RS, Swoboda KJ, Gappmaier E, Howard MT, Day JW, McDonald C, McNally EM, Weiss RB & United Dystrophinopathy Project (2013). LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann Neurol 73, 481–488. [Europe PMC free article] [Abstract] [Google Scholar]
- Greco S, De Simone M, Colussi C, Zaccagnini G, Fasanaro P, Pescatori M, Cardani R, Perbellini R, Isaia E, Sale P, Meola G, Capogrossi MC, Gaetano C & Martelli F (2009). Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J 23, 3335–3346. [Abstract] [Google Scholar]
- Guadagnin E, Mázala D & Chen YW (2018). STAT3 in skeletal muscle function and disorders. Int J Mol Sci; DOI: 10.3390/ijms19082265. Available at: http://www.ncbi.nlm.nih.gov/pubmed/30072615 [Accessed February 12, 2020]. [Europe PMC free article] [Abstract] [CrossRef]
- Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, van Ommen G-JB & Kunkel LM (2015). The Pathogenesis and Therapy of Muscular Dystrophies. Annu Rev Genomics Hum Genet 16, 281–308. [Abstract] [Google Scholar]
- Ha M & Kim VN (2014). Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524. [Abstract] [Google Scholar]
- Hak KK, Yong SL, Sivaprasad U, Malhotra A & Dutta A (2006). Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174, 677–687. [Europe PMC free article] [Abstract] [Google Scholar]
- Heyer MP, Pani AK, Smeyne RJ, Kenny PJ & Feng G (2012). Normal Midbrain Dopaminergic Neuron Development and Function in miR-133b Mutant Mice. J Neurosci 32, 10887–10894. [Europe PMC free article] [Abstract] [Google Scholar]
- Hindi SM, Tajrishi MM & Kumar A (2013). Signaling mechanisms in mammalian myoblast fusion. Sci Signal 6, re2. [Europe PMC free article] [Abstract] [Google Scholar]
- Hoffman EP, Brown RH & Kunkel LM (1987). Dystrophin: The protein product of the duchenne muscular dystrophy locus. Cell 51, 919–928. [Abstract] [Google Scholar]
- Horak M, Novak J & Bienertova-Vasku J (2016). Muscle-specific microRNAs in skeletal muscle development. Dev Biol 410, 1–13. [Abstract] [Google Scholar]
- Hosoyama T, Ishiguro N, Yamanouchi K & Nishihara M (2009). Degenerative muscle fiber accelerates adipogenesis of intramuscular cells via RhoA signaling pathway. Differentiation 77, 350–359. [Abstract] [Google Scholar]
- Hsu Da, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP & Huang H Da (2011). MiRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res; DOI: 10.1093/nar/gkq1107. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Iannone F, Montesanto A, Cione E, Crocco P, Caroleo MC, Dato S, Rose G & Passarino G (2020). Expression Patterns of Muscle - Specific miR - 133b and miR - 206 Correlate with Nutritional Status and Sarcopenia. Nutrients 12, 297. [Europe PMC free article] [Abstract] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW & Campbell KP (1992). Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 355, 696–702. [Abstract] [Google Scholar]
- Jia L, Shi Y, Wen Y, Li W, Feng J & Chen C (2018). The roles of TNFAIP2 in cancers and infectious diseases. J Cell Mol Med 22, 5188–5195. Available at: /pmc/articles/PMC6201362/?report=abstract [Accessed July 30, 2020]. [Europe PMC free article] [Abstract] [Google Scholar]
- Juban G, Saclier M, Yacoub-Youssef H, Kernou A, Arnold L, Boisson C, Ben Larbi S, Magnan M, Cuvellier S, Théret M, Petrof BJ, Desguerre I, Gondin J, Mounier R & Chazaud B (2018). AMPK Activation Regulates LTBP4-Dependent TGF-β1 Secretion by Pro-inflammatory Macrophages and Controls Fibrosis in Duchenne Muscular Dystrophy. Cell Rep 25, 2163–2176.e6. [Abstract] [Google Scholar]
- Kharraz Y, Guerra J, Pessina P, Serrano AL & Mu??oz-C??noves P (2014). Understanding the process of fibrosis in duchenne muscular dystrophy. Biomed Res Int; DOI: 10.1155/2014/965631. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kim D, Langmead B & Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [Europe PMC free article] [Abstract] [Google Scholar]
- Kong Y (2011). Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98, 152–153. [Abstract] [Google Scholar]
- Lapidos KA, Kakkar R & McNally EM (2004). The Dystrophin Glycoprotein Complex. Circ Res. Available at: http://circres.ahajournals.org/content/94/8/1023.long [Accessed June 5, 2017]. [Abstract]
- Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R & Olson EN (2012). MicroRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122, 2054–2065. [Europe PMC free article] [Abstract] [Google Scholar]
- Love MI, Huber W & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [Europe PMC free article] [Abstract] [Google Scholar]
- Von Maltzahn J, Jones AE, Parks RJ & Rudnicki MA (2013). Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A 110, 16474–16479. [Europe PMC free article] [Abstract] [Google Scholar]
- Mayerl S, Schmidt M, Doycheva D, Darras VM, Hüttner SS, Boelen A, Visser TJ, Kaether C, Heuer H & von Maltzahn J (2018). Thyroid Hormone Transporters MCT8 and OATP1C1 Control Skeletal Muscle Regeneration. Stem Cell Reports 10, 1959–1974. [Europe PMC free article] [Abstract] [Google Scholar]
- McGeachie JK, Grounds MD, Partridge TA & Morgan JE (1993). Age-related changes in replication of myogenic cells in mdx mice: Quantitative autoradiographic studies. J Neurol Sci 119, 169–179. [Abstract] [Google Scholar]
- McNally EM & Pytel P (2007). Muscle Diseases: The Muscular Dystrophies. Annu Rev Pathol Mech Dis 2, 87–109. [Abstract] [Google Scholar]
- Meister G (2013). Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 14, 447–459. [Abstract] [Google Scholar]
- Mok GF, Lozano-Velasco E & Münsterberg A (2017). microRNAs in skeletal muscle development. Semin Cell Dev Biol 72, 67–76. [Abstract] [Google Scholar]
- Mouton PR (2011). Unbiased stereology : a concise guide /. Johns Hopkins University Press, Baltimore: : [Google Scholar]
- Mu X, Tang Y, Takayama K, Chen W, Lu A, Wang B, Weiss K & Huard J (2017). RhoA/ROCK inhibition improves the beneficial effects of glucocorticoid treatment in dystrophic muscle: implications for stem cell depletion. Hum Mol Genet 26, 2813–2824. [Europe PMC free article] [Abstract] [Google Scholar]
- Mu X, Usas A, Tang Y, Lu A, Wang B, Weiss K & Huard J (2013). RhoA mediates defective stem cell function and heterotopic ossification in dystrophic muscle of mice. FASEB J 27, 3619–3631. [Europe PMC free article] [Abstract] [Google Scholar]
- Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N & Ochi M (2010). Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med 14, 2495–2505. [Europe PMC free article] [Abstract] [Google Scholar]
- Niwa N, Tanaka N, Hongo H, Miyazaki Y, Takamatsu K, Mizuno R, Kikuchi E, Mikami S, Kosaka T & Oya M (2019). TNFAIP2 expression induces epithelial-to-mesenchymal transition and confers platinum resistance in urothelial cancer cells. Lab Investig 99, 1702–1713. [Abstract] [Google Scholar]
- Pratt SJP, Valencia AP, Le GK, Shah SB & Lovering RM (2015). Pre- and postsynaptic changes in the neuromuscular junction in dystrophic mice. Front Physiol 6, 252. [Europe PMC free article] [Abstract] [Google Scholar]
- Qin W, Dong P, Ma C, Mitchelson K, Deng T, Zhang L, Sun Y, Feng X, Ding Y, Lu X, He J, Wen H & Cheng J (2012). MicroRNA-133b is a key promoter of cervical carcinoma development through the activation of the ERK and AKT1 pathways. Oncogene 31, 4067–4075. [Abstract] [Google Scholar]
- Radley HG & Grounds MD (2006). Cromolyn administration (to block mast cell degranulation) reduces necrosis of dystrophic muscle in mdx mice. Neurobiol Dis 23, 387–397. [Abstract] [Google Scholar]
- Roberts TC, Blomberg KEM, McClorey G, Andaloussi S EL, Godfrey C, Betts C, Coursindel T, Gait MJ, Edvard Smith C & Wood MJ (2012). Expression Analysis in Multiple Muscle Groups and Serum Reveals Complexity in the MicroRNA Transcriptome of the mdx Mouse with Implications for Therapy. Mol Ther - Nucleic Acids 1, e39. [Europe PMC free article] [Abstract] [Google Scholar]
- Russell AP, Lamon S, Boon H, Wada S, Güller I, Brown EL, Chibalin A V, Zierath JR, Snow RJ, Stepto N, Wadley GD & Akimoto T (2013). Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J Physiol 591, 4637–4653. [Abstract] [Google Scholar]
- Rybakova IN, Patel JR & Ervasti JM (2000). The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol 150, 1209–1214. [Europe PMC free article] [Abstract] [Google Scholar]
- Sharma M, Juvvuna PK, Kukreti H & McFarlane C (2014). Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front Physiol 5, 239. [Europe PMC free article] [Abstract] [Google Scholar]
- Sutavani RV, Bradley RG, Ramage JM, Jackson AM, Durrant LG & Spendlove I (2013). CD55 Costimulation Induces Differentiation of a Discrete T Regulatory Type 1 Cell Population with a Stable Phenotype. J Immunol 191, 5895–5903. [Abstract] [Google Scholar]
- Uezumi A, Fukada SI, Yamamoto N, Takeda S & Tsuchida K (2010). Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol 12, 143–152. [Abstract] [Google Scholar]
- Valdez G, Heyer MP, Feng G & Sanes JR (2014). The role of muscle microRNAs in repairing the neuromuscular junction. PLoS One 9, e93140. [Europe PMC free article] [Abstract] [Google Scholar]
- Viñals F, Fandos C, Santalucia T, Ferré J, Testar X, Palacín M & Zorzano A (1997). Myogenesis and MyoD down-regulate Sp1. A mechanism for the repression of GLUT1 during muscle cell differentiation. J Biol Chem 272, 12913–12921. [Abstract] [Google Scholar]
- Vinel C et al. (2018). The exerkine apelin reverses age-associated sarcopenia. Nat Med 24, 1360–1371. [Abstract] [Google Scholar]
- Wang K, Wang C, Xiao F, Wang H & Wu Z (2008). JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol Chem 283, 34029–34036. [Europe PMC free article] [Abstract] [Google Scholar]
- Williams AH, Liu N, van Rooij E & Olson EN (2009a). MicroRNA control of muscle development and disease. Curr Opin Cell Biol 21, 461–469. [Europe PMC free article] [Abstract] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR & Olson EN (2009b). MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554. [Europe PMC free article] [Abstract] [Google Scholar]
- Willmann R, Possekel S, Dubach-Powell J, Meier T & Ruegg MA (2009). Mammalian animal models for Duchenne muscular dystrophy. Neuromuscul Disord 19, 241–249. [Abstract] [Google Scholar]
- Wotton D, Lo RS, Lee S & Massagué J (1999). A Smad transcriptional corepressor. Cell 97, 29–39. [Abstract] [Google Scholar]
- Xu M & Hecht NB (2011). Polypyrimidine Tract-Binding Protein 2 Binds to Selective, Intronic Messenger RNA and MicroRNA Targets in the Mouse Testis. Biol Reprod 84, 435–439. [Abstract] [Google Scholar]
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van IJcken W, Grosveld F, Dekemp RA, Boushel R, Harper ME & Rudnicki MA (2013a). MicroRNA-133 Controls Brown Adipose Determination in Skeletal Muscle Satellite Cells by Targeting Prdm16. Cell Metab 17, 210–224. [Europe PMC free article] [Abstract] [Google Scholar]
- Yin H, Price F & Rudnicki M a (2013b). Satellite cells and the muscle stem cell niche. Physiol Rev 93, 23–67. [Europe PMC free article] [Abstract] [Google Scholar]
- Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA & Beauchamp JR (2006). Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 119, 1824–1832. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1113/jp280405
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1113/JP280405
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1113/jp280405
Article citations
The Role of MicroRNA in the Pathogenesis of Duchenne Muscular Dystrophy.
Int J Mol Sci, 25(11):6108, 01 Jun 2024
Cited by: 2 articles | PMID: 38892293 | PMCID: PMC11172814
Review Free full text in Europe PMC
Stem Cell-Based Therapeutic Approaches in Genetic Diseases.
Adv Exp Med Biol, 1436:19-53, 01 Jan 2023
Cited by: 0 articles | PMID: 36735185
Postnatal Protein Intake as a Determinant of Skeletal Muscle Structure and Function in Mice-A Pilot Study.
Int J Mol Sci, 23(15):8815, 08 Aug 2022
Cited by: 2 articles | PMID: 35955948 | PMCID: PMC9369224
MicroRNAs as serum biomarkers in Becker muscular dystrophy.
J Cell Mol Med, 26(17):4678-4685, 26 Jul 2022
Cited by: 2 articles | PMID: 35880500 | PMCID: PMC9443944
Smad8 Is Increased in Duchenne Muscular Dystrophy and Suppresses miR-1, miR-133a, and miR-133b.
Int J Mol Sci, 23(14):7515, 07 Jul 2022
Cited by: 5 articles | PMID: 35886863 | PMCID: PMC9323105
Go to all (10) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (4 citations) GEO - GSE156267
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy.
Exp Physiol, 99(4):675-687, 17 Jan 2014
Cited by: 20 articles | PMID: 24443351
miR-146a deficiency does not aggravate muscular dystrophy in mdx mice.
Skelet Muscle, 9(1):22, 14 Aug 2019
Cited by: 16 articles | PMID: 31412923 | PMCID: PMC6693262
Pannexin 1 dysregulation in Duchenne muscular dystrophy and its exacerbation of dystrophic features in mdx mice.
Skelet Muscle, 14(1):8, 26 Apr 2024
Cited by: 0 articles | PMID: 38671506 | PMCID: PMC11046831
Humanization of the mdx Mouse Phenotype for Duchenne Muscular Dystrophy Modeling: A Metabolic Perspective.
J Neuromuscul Dis, 10(6):1003-1012, 01 Jan 2023
Cited by: 1 article | PMID: 37574742 | PMCID: PMC10657711
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIA NIH HHS (4)
Grant ID: T32 AG041688
Grant ID: R56 AG051501
Grant ID: R01 AG055545
Grant ID: R56AG051501
NINDS NIH HHS (1)
Grant ID: R21 NS106313
National Institute of Neurological Disorders and Stroke (1)
Grant ID: R21NS106313
National Institute on Aging (1)
Grant ID: R56AG051501





