Abstract
Background
Hospitalization of patients infected with the severe acute respiratory syndrome virus 2 (SARS-CoV-2) have remained considerable worldwide. Patients often develop severe complications and have high mortality rates. The cycle threshold (Ct) value derived from nasopharyngeal swab samples using real time polymerase chain reaction (RT-PCR) may be a useful prognostic marker in hospitalized patients with SARS-CoV-2 infection, however, its role in predicting the course of the pandemic has not been evaluated thus far.Methods
We conducted a retrospective cohort study which included all patients who had a nasopharyngeal sample positive for SARS-CoV-2 between April 4 -June 5, 2020. The Ct value was used to estimate the number of viral particles in a patient sample. The trend in initial viral load on admission on a population level was evaluated. Moreover, patient characteristics and outcomes stratified by viral load categories were compared and initial viral load was assessed as an independent predictor of intubation and in-hospital mortality.Results
A total of 461 hospitalized patients met the inclusion criteria. This study consisted predominantly of acutely infected patients with a median of 4 days since symptom onset to PCR. As the severity of the pandemic eased, there was an increase in the percentage of samples in the low initial viral load category, coinciding with a decrease in deaths. Compared to an initial low viral load, a high initial viral load was an independent predictor of in-hospital mortality (OR 5.5, CI 3.1-9.7, p < 0.001) and intubation (OR 1.82 CI 1.07-3.11, p = 0.03), while an initial intermediate viral load was associated with increased risk of inpatient mortality (OR 1.9, CI 1.14-3.21, p = 0.015) but not with increased risk for intubation.Conclusion
The Ct value obtained from nasopharyngeal samples of hospitalized patients on admission may serve as a prognostic marker at an individual level and may help predict the course of the pandemic when evaluated at a population level.Free full text

SARS-CoV-2 infection: Initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic
Abstract
Background
Hospitalization of patients infected with the severe acute respiratory syndrome virus 2 (SARS-CoV-2) have remained considerable worldwide. Patients often develop severe complications and have high mortality rates. The cycle threshold (Ct) value derived from nasopharyngeal swab samples using real time polymerase chain reaction (RT-PCR) may be a useful prognostic marker in hospitalized patients with SARS-CoV-2 infection, however, its role in predicting the course of the pandemic has not been evaluated thus far.
Methods
We conducted a retrospective cohort study which included all patients who had a nasopharyngeal sample positive for SARS-CoV-2 between April 4 –June 5, 2020. The Ct value was used to estimate the number of viral particles in a patient sample. The trend in initial viral load on admission on a population level was evaluated. Moreover, patient characteristics and outcomes stratified by viral load categories were compared and initial viral load was assessed as an independent predictor of intubation and in-hospital mortality.
Results
A total of 461 hospitalized patients met the inclusion criteria. This study consisted predominantly of acutely infected patients with a median of 4 days since symptom onset to PCR. As the severity of the pandemic eased, there was an increase in the percentage of samples in the low initial viral load category, coinciding with a decrease in deaths. Compared to an initial low viral load, a high initial viral load was an independent predictor of in-hospital mortality (OR 5.5, CI 3.1–9.7, p < 0.001) and intubation (OR 1.82 CI 1.07–3.11, p = 0.03), while an initial intermediate viral load was associated with increased risk of inpatient mortality (OR 1.9, CI 1.14–3.21, p = 0.015) but not with increased risk for intubation.
Conclusion
The Ct value obtained from nasopharyngeal samples of hospitalized patients on admission may serve as a prognostic marker at an individual level and may help predict the course of the pandemic when evaluated at a population level.
Introduction
As of March 11, 2021, more than 110 million cases of the severe acute respiratory syndrome virus 2 (SARS-CoV-2) have been reported worldwide. The majority of cases have been reported from the United States with more than 500,000 deaths so far in the U.S alone [1]. Several studies evaluated the association between SARS-CoV-2 viral load (VL) and patient outcomes. Zheng S et al. demonstrated that patients with severe disease had late shedding peaks compared to patients with mild disease. The authors also reported that the virus could be isolated up to 29 days after symptom onset with declining VLs over time [2]. More recently, several studies demonstrated that a high SARS-CoV-2 viral load was an independent predictor of mortality in large hospital cohorts [3–6] however, trends in the initial viral load over time at a population level have not been evaluated thus far. Also, similar data from a predominantly African American population have not been reported.
We describe a steady downtrend in the level of initial SARS-CoV-2 VL detected in nasopharyngeal samples of hospitalized patients in Detroit, Michigan as the pandemic evolved coinciding with a decrease in the percent of deaths. Moreover, a higher initial viral load on presentation in our patient population was an independent predictor of in-hospital mortality and risk for intubation, supporting the results of previously published reports.
Methods
Data collection
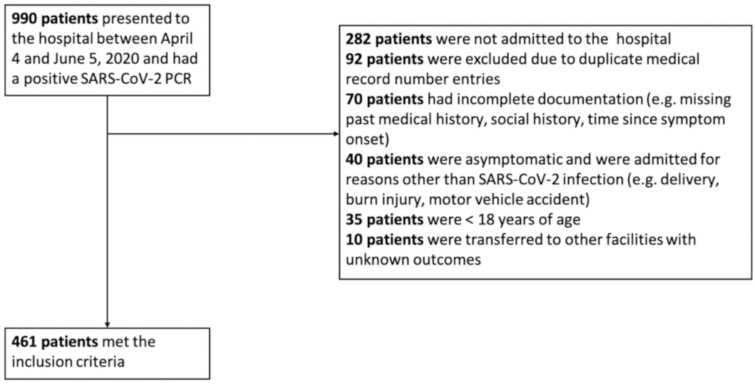
We conducted a retrospective study which included all patients who had a nasopharyngeal swab sample positive for SARS-CoV-2 infection by the Cepheid GenExpert instrument system–Rapid Real Time Polymerase Chain Reaction (RT-PCR) at the Detroit Medical Center during April 4, 2020-June 5, 2020. Data on 990 nasopharyngeal swabs were available for analysis, however, only 461 symptomatic patients met the inclusion criteria for our study (Fig 1). All sample containers, swabs, transport and storage, nucleic acid extraction and PCR protocols were similar for all included patients. We included results of the initial PCR sample obtained on presentation and omitted any subsequent results from the same patient. Medical records through July 5, 2020 were reviewed and all-cause in-hospital mortality was recorded. Patients who were discharged in stable condition were presumed alive at 30-day follow-up. Weeks were divided into intervals of seven days starting from April 4, 2020. In Michigan, the first peak in newly diagnosed COVID-19 cases was observed on April 7, 2020. By the end of week 2 of our study, the number of newly recorded cases had decreased by half and continued to decline steadily thereafter [7]. After May 9, 2020, we noticed a significant drop in the number of hospitalized SARS-CoV-2 cases, therefore, all cases between May 9, 2020 and June 5, 2020 were included in the 5+ week category.
Viral load assessment
Samples were considered positive for SARS-CoV-2 when the N gene (nucleocapsid) was detected. Although the RT-PCR test is qualitative and results are reported as positive and negative, the Exprt Xpress SARS-CoV-2 assay reports the cycle threshold (Ct) when the test is positive [8, 9]. The Ct value can be used to estimate the number of viral particles in a patient sample. The assay used in this study does not have an internal control, however, it contains a Sample Processing Control (SPC) utilized by the GeneXpert Xpress System instrument to confirm adequate processing of the sample and to ensure that the RT-PCR reaction conditions are appropriate. In an ideal setting where the PCR efficiency is 100% and the R-squared value equals 1, a reduction of the Ct value by 3.3 is expected when the target concentration increases by one log10 [9]. Using a commercial quantified standard, our data showed a linear relationship between viral load and Ct values over four “1 to 10” dilutions of standard with an R-squared value of 0.99 (S1 Fig). At 2x102, 2x103 and 2x104 viral particles, the Ct values were 43, 39.4 and 36.7 respectively. We designated high, intermediate, and low VL samples to have a Ct value of ≤ 25, 26–36, and ≥ 37 consecutively. Assuming a linear relationship between the Ct value and target concentration, samples with a Ct value of 26 should have a VL of 2x107 while samples with a Ct value of 36 should have a VL of 2x 104 approximately. The lower limit of detection of our assay is around 250 genome copies/mL (95% confidence).
Statistical analysis
Baseline characteristics and patient outcomes were stratified by viral load category. Medians and interquartile range (IQR) were used to describe continuous variables, while categorical variables were represented as proportions. The Kruskal-Wallis ANOVA test was used to compare medians, and the Fisher’s exact test was used to compare categorical variables. A 2-sided p value ≤ 0.05 was used to designate statistical significance. A time-based analysis using Cox proportional hazards was used to compare the cumulative risk of in-patient mortality and intubation among patients in the different viral load categories. Baseline factors that were associated with an increased risk for in-hospital mortality and intubation were identified on univariate analysis. All variables that were associated with each outcome having a P value of ≤ 0.1 were included in a multivariate logistic regression model (S1 Table) and adjusted odds ratios (OR) were calculated for these variables. Finally, we performed a multivariate logistic regression model using age and Ct values as continuous variables. Statistical analysis was completed using IBM SPSS Statistics version 26 (Armonk, NY: IBM Corp).
Ethics statement
The Institutional Review Board at Wayne State University School of Medicine reviewed the study protocol and ethical approval for the conduction of this study was granted (IRB No. RR19393). The study was conducted under a waiver of informed consent. Data were analyzed anonymously.
Results
Declining trend in initial viral load over time
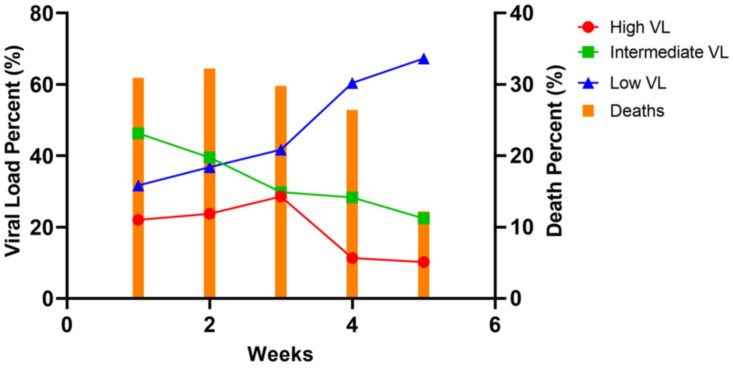
A total of 461 nasopharyngeal swabs were analyzed from hospitalized patients. During the first week of the study (week of April 4, 2020), the initial VL in the PCR-positive respiratory samples was predominantly in the intermediate group (46.3%, n = 57). As the pandemic evolved, there was a steady decline in the percent of positive samples in the high and intermediate VL categories with a concomitant rise in the percent of positive samples in the low VL category. By week five, 67% of the samples were in low VL category. This trend coincided with a decrease in the percentage of deaths in hospitalized patients (Fig 2).
Patient characteristics stratified by viral load
Table 1 represents the baseline characteristics and outcomes of hospitalized patients with SARS-CoV-2 stratified by initial viral load category. The median age of patients with initial high, intermediate, and low viral load was 66, 69 and 71 years respectively (p = 0.002). Patients with a higher initial viral load were more likely to be nursing home residents (45%, n = 44, p = 0.004). There was no significant association between initial viral load on presentation and gender, race, coronary artery disease, hypertension, obstructive lung disease, smoking history, heart failure, diabetes mellitus or body mass index (BMI). In this study, patients with high and intermediate initial viral loads had a median of 3 days from symptom onset to PCR testing compared to 5 days for patients with a low initial viral load. Patients with a higher initial viral load were more likely to require oxygen therapy by nasal canula, non-rebreather mask, or high-flow nasal canula as well as intubation and mechanical ventilation during their hospitalization. They were also more likely to develop shock requiring pressor support and had lower mean arterial pressures on admission.
Table 1
| Variable | Total | Low Viral Load | Intermediate Viral Load | High Viral Load | P Value |
|---|---|---|---|---|---|
| n = 461 | (Ct ≥ 37) | (Ct 26–36) | (Ct ≤ 25) | ||
| n = 195 | n = 168 | n = 98 | |||
 I. Demographic Characteristics I. Demographic Characteristics | |||||
| Age (years), median (IQR) | 68 (60–77) | 66 (55–75) | 69 (63–76) | 71 (61–81) | 0.002 |
| Age (by categories, years), No. (%) | |||||
| 18–39 | 22 (4.8) | 16 (8.2) | 1 (0.6) | 5 (5.1) | 0.001 |
| 40–65 | 166 (36) | 80 (41) | 53 (31.5) | 33 (33.7) | |
| > 65 | 273 (59.2) | 99 (50.8) | 114 (67.9) | 60 (61.2) | |
| Gender—No. (%) | |||||
| Men | 243 (52.7) | 110 (56.4) | 82 (48.8) | 51 (52.0) | 0.35 |
| Women | 218 (47.3) | 85 (43.6) | 86 (51.2) | 47 (48.0) | |
| Race—No. (%) | |||||
| African American | 362 (78.5) | 159 (81.5) | 126 (75) | 77 (78.6) | 0.41 |
| White | 38 (8.2) | 11 (5.6) | 17 (10.1) | 10 (10.2) | |
| Other | 61 (13.2) | 25 (12.8) | 25 (14.9) | 11 (11.2) | |
| Nursing Home Resident | 168 (36.4) | 59 (30.3) | 65 (38.7) | 44 (44.9) | 0.03 |
 II. Comorbidities—No. (%) II. Comorbidities—No. (%) | |||||
| Hypertension | 355 (77) | 146 (74.9) | 129 (76.8) | 80 (81.6) | 0.42 |
| Diabetes Mellitus | 203 (44.0) | 89 (45.6) | 70 (41.7) | 44 (44.9) | 0.73 |
| Chronic Kidney Disease | 128 (27.8) | 65 (33.3) | 46 (27.4) | 17 (17.3) | 0.013 |
| End Stage Renal Disease | 54 (11.7) | 27 (13.8) | 17 (10.1) | 27 (13.8) | 0.47 |
| Coronary Artery Disease | 99 (21.5) | 42 (21.5) | 32 (19) | 25 (25.5) | 0.46 |
| Heart Failure | 45 (9.8) | 17 (8.7) | 16 (9.5) | 12 (12.2) | 0.67 |
| Asthma | 31 (6.7) | 12 (6.2) | 11 (6.5) | 8 (8.2) | 0.80 |
| COPD | 83 (18) | 34 (17.4) | 29 (17.3) | 20 (20.4) | 0.87 |
| Smoking History | 168 (36.4) | 59 (30.3) | 65 (38.7) | 44 (44.9) | 0.57 |
| Autoimmune Disease | 24 (5.2) | 11 (5.6) | 9 (5.4) | 4 (4.1) | 0.84 |
| HIV | 7 (1.5) | 5 (2.6) | 1 (0.6) | 1 (1.0) | 0.28 |
| Hematologic malignancy | 8 (1.7) | 4 (2.1) | 2 (1.2) | 2 (2.0) | 0.82 |
| Solid Malignancy | 39 (8.5) | 17 (8.7) | 11 (6.5) | 11 (11.2) | 0.41 |
| Transplant Recipient | 15 (3.3) | 6 (3.1) | 6 (3.6) | 3 (3.1) | 0.95 |
| Immunosuppressive therapy | 19 (4.1) | 10 (5.1) | 7 (4.2) | 2 (2.0) | 0.08 |
| Body Mass Index (kg/m2) | |||||
| < 18 | 11 (2.4) | 4 (2.1) | 6 (3.6) | 1 (1.0) | 0.13 |
| 18–25 | 123 (26.7) | 53 (27.2) | 40 (23.8) | 30 (30.6) | |
| 25–30 | 135 (29.3) | 53 (27.2) | 45 (26.8) | 37 (37.8) | |
| >30 | 192 (41.6) | 85 (43.6) | 77 (45.8) | 30 (30.6) | |
 III. Symptoms on presentation—No. (%) III. Symptoms on presentation—No. (%) | |||||
| Gastrointestinal | 115 (24.9) | 53 (27.2) | 42 (25) | 20 (20.4) | 0.45 |
| Respiratory | 314 (69) | 117 (60.6) | 126 (75.9) | 71 (74) | 0.004 |
| Neurological | 162 (35.1) | 75 (38.5) | 50 (29.8) | 37 (37.8) | 0.18 |
| Systemic | 267 (57.9) | 107 (54.9) | 102 (60.7) | 58 (59.2) | 0.51 |
| Days since symptom onset-to-PCR, median (IQR) | 4 (2–8) | 5 (2–12) | 3 (2–7) | 3 (1–5) | <.001 |
 IV. Laboratory variables (within 3 days of hospital admission), Median (IQR) IV. Laboratory variables (within 3 days of hospital admission), Median (IQR) | |||||
| Total White Blood Cell Count (x 103 cells/mm3) | 9.5 (6.3–11.6) | 8.7 (5.8–12.4) | 8.3 (5.5–12.3) | 7.9 (5.4–11.5) | 0.64 |
| Absolute Lymphocyte Count (x 10 3 cells/mm 3 ) | 0.9 (0.5–1.2) | 0.95 (0.55–1.3) | 0.7 (0.4–1.0) | 0.75 (0.47–1.2) | <.001 |
| Absolute Neutrophil Count (x 103 cells/mm3) | 7.4 (4.3–9.8) | 6.8 (2.2–9.9) | 7.1 (5.1–10.5) | 8.2 (6.0–10.6) | 0.87 |
| Platelets (x 10 3 cells/mm 3 ) | 226 (167–294) | 235 (177–300) | 234 (174–294) | 202 (155–268) | 0.02 |
| Creatinine (mg/dL) | 1.9 (1.0–4.3) | 1.5 (0.96–4.4) | 2.2 (0.97–6.0) | 2.0 (1.1–4.6) | 0.46 |
| Alanine Aminotransferase (units/L) | 20 (12–33) | 23 (11–31) | 19 (12.2–36.5) | 33.5 (16.2–57.5) | 0.31 |
| Aspartate Aminotransferase (units/L) | 35 (22–59) | 35 (22.25–72.5) | 40.5 (19–65.25) | 44.5 (32.7–75.2) | 0.04 |
| Alkaline Phosphatase (units/L) | 71 (57–95) | 76 (67–89) | 69 (52–97) | 69 (53–95) | 0.75 |
| C-Reactive Protein (mg/L) | 120 (60–161) | 100 (40–150) | 122 (74–167) | 148 (67–209) | 0.001 |
| Ferritin (ng/mL) | 507 (209–1277) | 408 (172–1189) | 512 (247–1097) | 697 (261–1710) | 0.01 |
| D-dimer (mg/L) | 2.07 (1.0–6.96) | 2.2 (1.0–6.7) | 1.83 (0.9–7.22) | 2.22 (1.0–6.99) | 0.69 |
| LDH (units/L) | 364 (252–542) | 311 (230–424) | 372 (260–609) | 417 (244–688) | <.001 |
| Troponin (ng/L) | 28 (11–80) | 28 (9–97) | 29 (26–32) | 28 (24–35) | 0.71 |
 V. Vital Signs on Admission, median (IQR) V. Vital Signs on Admission, median (IQR) | |||||
| Heart rate (beats per minute) | 93 (82–107) | 89 (82–102) | 98 (84–116) | 104 (90–115) | 0.09 |
| Mean Arterial Pressure (mmHg) | 90 (79–102) | 92 (80–104) | 89 (77–101) | 87 (75–101) | 0.04 |
| Temperature (°C) | 37 (36.6–37.7) | 36.9 (36.6–37.5) | 37.1 (36.6–38) | 36.9 (36.6–38) | 0.12 |
 VI. Chest X-ray Findings VI. Chest X-ray Findings | |||||
| No Abnormal Findings | 84 (18.2) | 44 (26.2) | 22 (13.8) | 18 (19.4) | 0.07 |
| Unilateral Infiltrates | 93 (20.2) | 36 (21.4) | 35 (21.9) | 22 (23.7) | |
| Bilateral Infiltrates | 244 (52.9) | 88 (52.4) | 103 (64.4) | 53 (57) | |
 VII. Medications VII. Medications | |||||
| Corticosteroids | 302 (65.5) | 102 (52.3) | 123 (73.2) | 77 (78.6) | < 0.01 |
 VIII. Clinical Outcomes VIII. Clinical Outcomes | |||||
| Highest Oxygen Delivery during hospitalization | |||||
| No supplemental Oxygen Required | 73 (15.8) | 43 (22.1) | 23 (13.7) | 7 (7.1) | 0.01 |
| Non-Invasive Oxygen Delivery ^ | 267 (57.9) | 107 (54.9) | 102 (60.7) | 58 (59.2) | 0.01 |
| Intubation and Mechanical Ventilation | 121 (26.2) | 45 (23.1) | 43 (25.6) | 33 (33.7) | 0.13 |
 Days until intubation, Mean (IQR) Days until intubation, Mean (IQR) | 2 (0–5) | 3.6 (2–5) | 2.3 (1–3) | 3.9 (2–5) | |
| Shock Requiring Pressor support during hospitalization (n, %) | 95 (20.6) | 29 (14.9) | 35 (20.8) | 31 (31.6) | 0.004 |
| In-hospital Mortality (n, %) | 132 (28.6) | 31 (15.9) | 50 (29.8) | 51 (52) | <.001 |
 Days until death, Median (IQR) Days until death, Median (IQR) | 9 (4–16) | 14 (9–15) | 9 (6–10) | 8 (2–5) | 0.01 |
Normal laboratory range: Alanine Aminotransferase: 7–52 units/L; Aspartate Aminotransferase: 13–39 units/L; Alkaline Phosphatase: 45–115 units/L; C-reactive protein: <5 mg/L; Ferritin: 23.9–336.2 ng/mL; LDH: 140–271 units/L; High sensitivity Troponin: < 17 ng/L; D-dimer: < 0.5 mg/L.
^ Non-invasive oxygen delivery includes nasal canula, non-rebreather mask, face mask, high flow oxygen delivery.
Patients with a high initial viral load were more likely to have lower absolute lymphocyte counts, higher lactate dehydrogenase (LDH), C-reactive protein (CRP) and ferritin levels on presentation. There were however no differences in the levels of troponin or chest X-ray findings on presentation among patients with high, intermediate, and low viral load respectively.
Patient outcomes
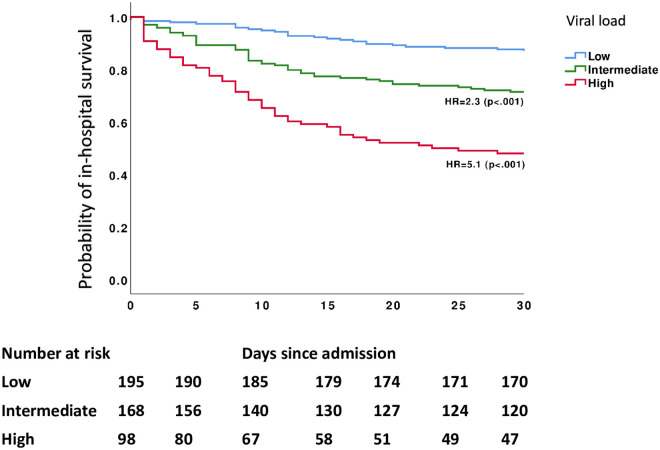
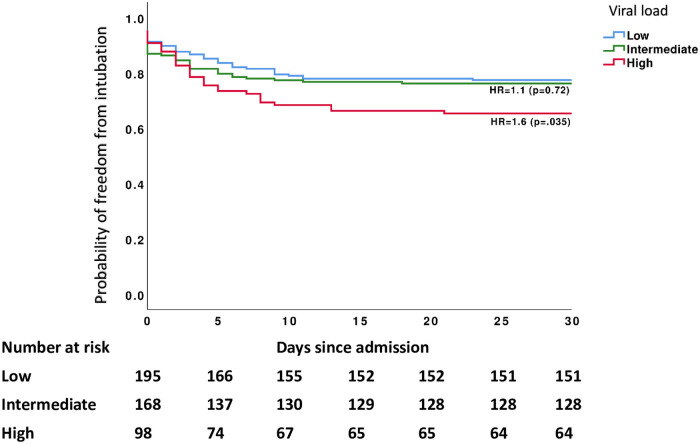
The last day of study follow-up was on July 5, 2020. By that time, 28.6% (n = 132) of the patients had died during their hospital admission while 71% (n = 329) were alive, out of which 58.4% (n = 192) were discharged home and 41.6% (n = 137) were discharged to nursing homes, long-term acute care centers or rehabilitation centers. Patients with high initial viral loads were at increased risk for intubation and death during their hospital admission. Among patients with a high initial viral load, in-hospital mortality was 52% (n = 51), compared to 30% (n = 50) and 16% (n = 31) for patients with intermediate and low initial viral load respectively (P < 0.001). The risk for intubation and mechanical ventilation during hospital admission was 34% (n = 33) in patients with an initial high viral load compared to 26% (n = 43) and 24% (n = 45) in patients with an intermediate and low initial viral load respectively (P = 0.01). In a time-based analysis, an initial high viral load was associated with a hazard ratio (HR) of 5.1 (CI 3.2–8.1 p < 0.001) for in-hospital mortality and a HR of 1.6 (CI 1.03–2.5, p = 0.035) for intubation compared to a low initial viral load, whereas an intermediate initial viral load was associated with an increased risk for in-hospital mortality (HR 2.2, CI 1.4–3.5 p < 0.001) but not with increased risk for intubation (Figs (Figs33 and and44).
Moreover, in a multivariate analysis adjusted to age and acute kidney injury, compared to a low initial viral load, both high and intermediate initial viral load were associated with increased risk of in-hospital mortality (OR 5.5, CI 3.1–9.7, p < 0.001 and OR 1.9, CI 1.14–3.21, p = 0.015) (Table 2). A high initial viral load was found to be associated with an increased risk for intubation and mechanical ventilation OR 1.82 (CI 1.07–3.11, p = 0.03) whereas an initial intermediate viral load was not.
Table 2
| Variables | Mortality | |
|---|---|---|
| Odds Ratio (95% CI) | p-value | |
| Viral Load By Nasopharyngeal Swab | ||
| Low viral load (Ct ≥ 37) | Reference | Reference |
| Intermediate viral load (Ct 26–36) | 1.91 (1.14–3.21) | .015 |
| High viral load (Ct ≤ 25) | 5.50 (3.11–9.73) | <.001 |
| Age (years) | ||
| 18–39 | Reference | Reference |
| 40–65 | 2.24 (0.47–10.70) | 0.312 |
| >65 | 5.42 (1.16–25.2) | 0.031 |
| CKD * | 0.87 (0.52–1.45) | 0.598 |
| Intubation | ||
| Viral Load By Nasopharyngeal Swab | ||
| Low viral load (Ct ≥ 37) | Reference | Reference |
| Intermediate viral load (Ct 26–36) | 1.07 (0.66–1.725) | 0.779 |
| High viral load (Ct ≤ 25) | 1.82 (1.07–3.11) | 0.028 |
*CKD: Chronic Kidney Disease.
Using multiple logistic regression, both age and Ct value were found to be significantly associated with mortality, such as every 1-year increase in age results in 3.5% increase in odds of death and every 1 unit increase in Ct value decreases the odds of death by 8%. The negative association between Ct and mortality reflects the fact that lower Ct values indicate higher viral load (Table 3).
Table 3
| Variables | Mortality | |
|---|---|---|
| Odds Ratio (95% CI) | p-value | |
| Age | 1.035 (1.018–1.053) | <0.001 |
| Ct Value | 0.921 (0.894–0.949) | <0.001 |
Variables entered in the model: Age (years), Ct value and chronic kidney disease (yes/no).
Finally, we explored the use of three drugs: corticosteroids, tocilizumab and remdesivir. At the time of the study, there was not enough evidence to support the use of any of these medications in the management of SARS-CoV-2 infection. A total of 302 patients (65.5%) received corticosteroids, while only 20 (4.3%) and 2 (0.4%) received tocilizumab and remdesivir respectively. Corticosteroids were prescribed to 52.3% (n = 102) of patients with low viral load, 73.2% (n = 123) of patients with intermediate initial viral load and 78.6% (n = 77) of those with high initial viral load (Table 1). Patients who received corticosteroids had higher mortality rates compared to patients who did not (35.8% vs 15.1%, p < 0.011). Since corticosteroids were generally reserved for more severe cases, the increased prevalence of their use in higher viral load categories reflects the relationship between high viral load and increased mortality highlighted in this study.
Discussion
A gradual decline in the number of patients with a high initial viral load over time, coinciding with a declining death rate, corresponded well with the lessening severity of the pandemic. Similar observations were documented by Clementi N et al. from Italy. The authors noted that the mean Ct value of positive SARS-CoV-2 samples collected during the month of April, which represented the peak of the pandemic, was significantly lower than the mean Ct value of positive samples collected in May, a time when the pandemic in Italy became less severe [10].
Mina M et al. suggested that a changing population distribution of SARS-CoV-2 Ct values can be used to predict the course of the pandemic [11]. The authors speculated that when the pandemic is on the rise, the majority of tested patients will be acutely symptomatic, therefore, their SARS-CoV-2 PCR tests will demonstrate a low Ct value compared to high Ct values during pandemic decline as the majority of the patients will be in the convalescent phase [11]. Moreover, rapid implementation of social distancing measures and the widespread use of facemasks may have contributed to “variolation “, which may have helped slow down the spread of the virus [12], although, a recent study by Masia et al. demonstrated that patients with a lower viral load were less likely to seroconvert [13]. A change in the virus may also explain this observation, however, no data are available to support this conclusion. Major limitations for the use of the Ct value in clinical practice stem from the variability of sample processing protocols and the use of different PCR platforms. In our study, all sample containers, swabs, transport and storage, nucleic acid extraction and PCR protocols were similar for all included patients and all Ct values were reported using the same instrument.
The correlation between VL, severity of illness and viral shedding for respiratory viruses remains a topic of debate. In patients with influenza infection, higher VLs were detected in hospitalized patients compared to ambulatory patients, however, this was generally not associated with worse outcomes [14, 15]. In contrast, there is evidence that among patients infected with the Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome virus (SARS), a higher VL was an independent risk factor for death [16, 17]. This study consisted predominantly of acutely infected patients with a median of 4 days since symptom onset to PCR. We demonstrated that the Ct value of nasopharyngeal swab samples obtained on initial presentation can act as a predictor of patient outcomes during hospitalization. Many studies correlated a higher SARS-CoV-2 viral load with increased severity of illness [3, 5, 6, 10, 18, 19]. A recent systematic review of 18 studies also supported that lower Ct values are associated with worse outcomes [20], and this study adds to the growing body of medical knowledge that a high initial nasopharyngeal viral load is an independent predictor of in-hospital mortality and intubation.
Higher plasma SARS-CoV-2 viral load may be associated with increased risk of death and higher levels of inflammatory markers such as lower lymphocyte count, higher C-reactive protein (CRP), and interleukin-6 levels [4]. We demonstrated that a higher initial nasopharyngeal viral load is associated with elevated ferritin and CRP levels as compared to low viral load on presentation. This raises the question of whether stratifying patients by levels of viremia and severity of inflammation can aid in determining which patients may benefit more from antiviral therapy versus anti-inflammatory therapy [4]. It remains unclear whether nasopharyngeal Ct values correlate closely with levels of plasma viremia as no quantitative SARS-CoV-2 PCRs received U.S Food and Drug Administration emergency use authorization, making their use limited to select centers with test availability. Proving a strong correlation between nasopharyngeal Ct values and levels of viremia may further support the use of the nasopharyngeal Ct as a prognostic marker for hospitalized patients with COVID-19 infection.
Despite comparable pandemic conditions, the 30-day in-hospital mortality in this study was 28.6%, which is significantly higher than the mortality rate reported by a similar cohort study by Magleby et al. from New York (19.2%) which included a total of 678 SARS-CoV-2 infected patients between March 30 –April 30, 2020 [3]. It is worth noting that both studies spanned a period when Michigan and New York city witnessed their first peak in newly diagnosed COVID-19 cases coinciding on April 7 and April 12 respectively [7]. Reasons for higher mortality seen in our cohort may be related to differences in underlying comorbidities, age, and decreased access to healthcare in the Detroit population. Notably, African Americans comprised 78.5% of our patient population, compared to 14% as reported by Magleby et al. [3] however, ethnicity itself was not an independent predictor of mortality in either study. Moreover, recent studies investigated the impact of race, socioeconomic and insurance status on COVID-19 outcomes in Detroit and found no association between these factors and COVID-19 mortality [21, 22]. The authors however reported that advanced age and comorbidities on admission were associated with more adverse outcomes [21, 22]. Remarkably, Tejada C et al. recently reported that all patients in a cohort of 25 African American renal transplant recipients who were hospitalized for COVID-19 had full recovery despite the presence of comorbidities and seemingly worse prognosis [23]. This was attributed to the use of steroids and calcineurin inhibitors which may have had an inhibitory effect on immune cell activation and concomitant cytokine dysregulation [23]. Rather than attributing higher death rates to intrinsic biologic susceptibilities, improving access to healthcare and managing comorbidities may eliminate ethnic differences in COVID-19 mortality [24, 25].
This study has some limitations. First, this is a retrospective cohort study, therefore, patients were not followed-up after discharge to document deaths that may have occurred outside the hospital. Moreover, given the retrospective nature of the study, some data may have been missed or misclassified during extraction from medical records, however, we performed constant queries during data abstraction to guarantee the accuracy of the data. Secondly, only the Cepheid Xpert Xpress SARS-CoV-2 PCR was used, therefore, our results may not be generalizable to all PCR platforms. Lastly, our data was obtained prior to the availability of vaccines and prior to the emergence of new variants.
In this study, we report an independent association between initial VL and risk for intubation and mortality in a cohort composed predominantly of SARS-CoV-2 infected African American patients. Our data suggest that the initial Ct value reported on hospital admission may serve as an important risk stratification tool by allowing physicians to identify patients who are at increased risk of developing adverse outcomes secondary to COVID-19 infection. Importantly, trends in the initial cycle threshold values over time may serve as a useful marker to assess the tempo or the direction of the pandemic in the region and may have an impact on public health measures, infection control and clinical management of infected patients.
Supporting information
S1 Table
Univariate analysis of factors associated with increased risk for intubation and mortality.(DOCX)
Acknowledgments
Data collection team: Siri Sarvepalli, Vichar Trivedi, Adnan Halboni, Joseph Sebastian, Ishita Datta, Shiva Bongu, Zachary Cantor, Jiping Zhou, Jennifer Young, Jana Dbaibou, Bhavana Tetali, Tyler Mumm.
Data Availability
The data underlying this study are available publicly at https://doi.org/10.6084/m9.figshare.15085656.v3.
References
Decision Letter 0
5 May 2021
PONE-D-21-08808
SARS-CoV-2 infection: Initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic
PLOS ONE
Dear Dr. Chandrasekar,
Thank you for submitting your manuscript to PLOS ONE. After careful consideration, we feel that it has merit but does not fully meet PLOS ONE’s publication criteria as it currently stands. Therefore, we invite you to submit a revised version of the manuscript that addresses the points raised during the review process.
Please submit your revised manuscript by Jun 19 2021 11:59PM. If you will need more time than this to complete your revisions, please reply to this message or contact the journal office at gro.solp@enosolp. When you're ready to submit your revision, log on to https://www.editorialmanager.com/pone/ and select the 'Submissions Needing Revision' folder to locate your manuscript file.
Please include the following items when submitting your revised manuscript:
A rebuttal letter that responds to each point raised by the academic editor and reviewer(s). You should upload this letter as a separate file labeled 'Response to Reviewers'.
A marked-up copy of your manuscript that highlights changes made to the original version. You should upload this as a separate file labeled 'Revised Manuscript with Track Changes'.
An unmarked version of your revised paper without tracked changes. You should upload this as a separate file labeled 'Manuscript'.
If you would like to make changes to your financial disclosure, please include your updated statement in your cover letter. Guidelines for resubmitting your figure files are available below the reviewer comments at the end of this letter.
If applicable, we recommend that you deposit your laboratory protocols in protocols.io to enhance the reproducibility of your results. Protocols.io assigns your protocol its own identifier (DOI) so that it can be cited independently in the future. For instructions see: http://journals.plos.org/plosone/s/submission-guidelines#loc-laboratory-protocols. Additionally, PLOS ONE offers an option for publishing peer-reviewed Lab Protocol articles, which describe protocols hosted on protocols.io. Read more information on sharing protocols at https://plos.org/protocols?utm_medium=editorial-email&utm_source=authorletters&utm_campaign=protocols.
We look forward to receiving your revised manuscript.
Kind regards,
Tai-Heng Chen, M.D.
Academic Editor
PLOS ONE
Journal Requirements:
When submitting your revision, we need you to address these additional requirements.
1) Please ensure that your manuscript meets PLOS ONE's style requirements, including those for file naming. The PLOS ONE style templates can be found at
https://journals.plos.org/plosone/s/file?id=wjVg/PLOSOne_formatting_sample_main_body.pdf and
2) PLOS requires an ORCID iD for the corresponding author in Editorial Manager on papers submitted after December 6th, 2016. Please ensure that you have an ORCID iD and that it is validated in Editorial Manager. To do this, go to ‘Update my Information’ (in the upper left-hand corner of the main menu), and click on the Fetch/Validate link next to the ORCID field. This will take you to the ORCID site and allow you to create a new iD or authenticate a pre-existing iD in Editorial Manager. Please see the following video for instructions on linking an ORCID iD to your Editorial Manager account: https://www.youtube.com/watch?v=_xcclfuvtxQ
3) Thank you for including your ethics statement: "The Institutional Review Board reviewed the study protocol and ethical approval for the conduction of this study was granted (IRB No. RR19393). The study was conducted under a waiver of informed consent. Data were analyzed anonymously".
Please amend your current ethics statement to include the full name of the ethics committee/institutional review board(s) that approved your specific study.
Once you have amended this/these statement(s) in the Methods section of the manuscript, please add the same text to the “Ethics Statement” field of the submission form (via “Edit Submission”).
For additional information about PLOS ONE ethical requirements for human subjects research, please refer to http://journals.plos.org/plosone/s/submission-guidelines#loc-human-subjects-research.
4) We noted in your submission details that a portion of your manuscript may have been presented or published elsewhere. [poster presented at the ESCMID Conference]. Please clarify whether this [conference proceeding or publication] was peer-reviewed and formally published. If this work was previously peer-reviewed and published, in the cover letter please provide the reason that this work does not constitute dual publication and should be included in the current manuscript.
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #1: Yes
Reviewer #2: Partly
Reviewer #3: Yes
**********
2. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #1: Yes
Reviewer #2: No
Reviewer #3: I Don't Know
**********
3. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #1: Yes
Reviewer #2: No
Reviewer #3: Yes
**********
4. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #1: Yes
Reviewer #2: Yes
Reviewer #3: Yes
**********
5. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #1: The present study was enrolled 461 SARS-CoV-2 infected patients who were admitted in hospital between April 4- June 5, 2020 to analyze the effect of initial viral load on the intubation and mortality of the patients, and drew the conclusion that low initial viral load was related to less chances of intubation and mortality, and coinciding with the descending trend of the pandemic. The data was analyzed with different but appropriate statistic ways in accord with their types, while the difference among the groups were given strong supports to the conclusion of the study. The article was written in a formal way but easy for understanding. All the tables and graphs are neat and well explained, only the image resolution is needed to get improved, especially Fig 3. Moreover, there is a missing part of sign “A” in Fig 3.
Reviewer #2: Said El Zein et al have studied the correlation of nasopharyngeal SARS-CoV-2 viral load and a series of factors ( signs, comorbidities, outcome...) of 461 hospitalized patients during April- June 2020. These patients were mostly of one ethnicity ( African-Americans) which makes the study more interesting. They found that The viral load ( stratified in 3 categories) is independently correlated with the outcome of the patients.
The subject is interesting, the number of cases studied is high enough, the findings are reasonable. However, I have the following comments and questions.
1- I am not familiar with the RT-PCR kit used in this study, but most of the kits that I have evaluated use internal control (IC) genes too. The Ct value of IC is a good indicator of the quality of the biological samples ( with respect to collecting enough biological material as well as lacking PCR inhibitors). It is more reasonable to consider the difference between the Ct of IC and SARS-CoV-2 gene than considering the latter as an indicator of viral load. Even if the authors prefer to use absolute viral load as their main variable, comparing the IC Cts may still be helpful to exclude any variation or change in sample collection technique during the this study, which may be another reason why the VLs declined over time.
2- using Ct as a correlate to the viral load needs more validation. Basically, as it is well mentioned by the authors, these kits have been developed as qualitative tests. Using just 2 standard points ( in this case 10000 and 100000 VPs) is not enough to use the results as an even semi-quantitative test. I would suggest using at least 4 standard concentrations of the virus to see if the test is linear and efficient in a broader range. It is note worthy that the observed difference between the 2 standard concentration is not 3.3 ( as it is written in the manuscript) but it is 3.6 which might have larger impacts in lower or higher viral loads.
3- It is not clear to me how the standard virus sample has been quantified. Does it come from a virus culture or a clinical sample? Is it quantified based on genetic material copy number or based on infectious particle counting by one of virus titration methods?
4- Following the previous comments on reliability of the quantification method, in my opinion choosing the cutoffs of 26 and 36 are not well justified. We have found several COVID cases with Ct values below 20. On the other hand the intercept of the standard curve of the majority of QPCR kits is slightly higher than 35 ( usually below 40), so the readouts above 35 are not usually reliable, or could be due to non-infectious viral debris.
5- Based on the previous comments and uncertainties regarding the QPCR results, I would recommend statistical re-analyzing the data with parametric test as well. In those analyses the absolute value of Ct ( or delta Ct, in case the Ct of the internal controls are available) could be used. Then the author may come to similar conclusion, or find out choosing different cutoffs for stratification is more suitable, or just report the results of the parametric analysis.
6-Based on results presented in table 1, it seems to me that Age and Viral load are rather highly correlated. In table 2 both have very close ORs on Mortality rate as well ( 5.50 vs 5.42). I wonder what is the correlation of these two variable with each other, and whether they are not covariates. Have the author tested other variables in different age groups ( or again, using age as a parametric variable against other variables)? In the Statistical Analysis section there is a reference to table S1, which I could not find it in the manuscript or its supplementary materials.
7- in table 1, the average level of Troponin is 40, while the average of each VL group is below 29. I wonder if it is correct.
Reviewer #3: The manuscript describes a cohort study on relation among initial viral load in nasopharyngeal covid-19 samples and severity of the disease as well as status of pandemic. The subject is well designed and performed although some points need authors’ attention:
1- An important factor influencing the outcome of the disease and the results of this study is the type of treatment and should be considered in all interpretations. Type of antiviral or anti-inflammatory drugs, interval time between PCR and starting the treatment should be considered in analysis.
2- An explanation of the division of weeks according to pandemic conditions should be given at the first appearance in the text.
3- In comparing with the report of Magleby et al. in addition to the reasons given for the discrepancy with the results of the present study, the authors should also examine and discuss the pandemic conditions in two different studied time periods. Similarly, due to the fact that pandemic peaks are not the same in different countries, this point should be considered in comparison with the data of other countries.
4 -The sentence in line 97 and 98 needs reference.
**********
6. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #1: No
Reviewer #2: No
Reviewer #3: No
[NOTE: If reviewer comments were submitted as an attachment file, they will be attached to this email and accessible via the submission site. Please log into your account, locate the manuscript record, and check for the action link "View Attachments". If this link does not appear, there are no attachment files.]
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com/. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Registration is free. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email PLOS at gro.solp@serugif. Please note that Supporting Information files do not need this step.
Author response to Decision Letter 0
18 May 2021
Dear editor
Tai-Heng Chen, M.D.
PLOS One
Thank you for considering our manuscript for publication in your esteemed journal.
We would also like to thank the reviewers for their valuable and constructive comments.
Kindly find below our point-by-point response to the academic editor and reviewers’ comments:
Academic editor
1. Please ensure that your manuscript meets PLOS ONE's style requirements, including those for file naming.
We reviewed PLOS ONE’s style requirements and made changes accordingly.
2. PLOS requires an ORCID iD for the corresponding author in Editorial Manager on papers submitted after December 6th, 2016.
The ORCID iD for the corresponding author was linked to the submission
3. Thank you for including your ethics statement: "The Institutional Review Board reviewed the study protocol and ethical approval for the conduction of this study was granted (IRB No. RR19393). The study was conducted under a waiver of informed consent. Data were analyzed anonymously".
Please amend your current ethics statement to include the full name of the ethics committee/institutional review board(s) that approved your specific study.
Once you have amended this/these statement(s) in the Methods section of the manuscript, please add the same text to the “Ethics Statement” field of the submission form (via “Edit Submission”).
Thank you for your comment. We added a section to the manuscript titled “Ethics Statement” (Line 130-132) and included the full name of the ethics committee that approved our study. We also amended this statement in the “Ethics Statement” field of the submission system
4) We noted in your submission details that a portion of your manuscript may have been presented or published elsewhere. [poster presented at the ESCMID Conference]. Please clarify whether this [conference proceeding or publication] was peer-reviewed and formally published. If this work was previously peer-reviewed and published, in the cover letter please provide the reason that this work does not constitute dual publication and should be included in the current manuscript.
Thank you for your comment. A portion of the results discussing the trend in Ct value over time was presented as a poster presentation at the virtual ECCVID, 2020 meeting. The results have never been published elsewhere, either as part of a conference proceeding or as a peer-reviewed publication. We attached the poster presentation in the submission system for your kind reference.
Reviewers' comments:
Reviewer # 1:
1. The present study was enrolled 461 SARS-CoV-2 infected patients who were admitted in hospital between April 4- June 5, 2020 to analyze the effect of initial viral load on the intubation and mortality of the patients and drew the conclusion that low initial viral load was related to less chances of intubation and mortality and coinciding with the descending trend of the pandemic. The data was analyzed with different but appropriate statistic ways in accord with their types, while the difference among the groups were given strong supports to the conclusion of the study. The article was written in a formal way but easy for understanding. All the tables and graphs are neat and well explained, only the image resolution is needed to get improved, especially Fig 3. Moreover, there is a missing part of sign “A” in Fig 3.
Thank you for your comments and for your observation. We split figure 3A into figure 3 and figure 4 respectively. Figure resolutions were much better maintained when graphs were included separately.
Reviewer #2:
1. I am not familiar with the RT-PCR kit used in this study, but most of the kits that I have evaluated use internal control (IC) genes too. The Ct value of IC is a good indicator of the quality of the biological samples (with respect to collecting enough biological material as well as lacking PCR inhibitors). It is more reasonable to consider the difference between the Ct of IC and SARS-CoV-2 gene than considering the latter as an indicator of viral load.
2. Even if the authors prefer to use absolute viral load as their main variable, comparing the IC Cts may still be helpful to exclude any variation or change in sample collection technique during this study, which may be another reason why the VLs declined over time.
Dear reviewer,
Thank you for your feedback. We hope that the following response provides an answer to both comments #1 and #2.
The Cepheid assay used in this study does not have a control to check for specimen quality by looking for human genomic material. However, it contains a Sample Processing Control (SPC) in the cartridge utilized by the GeneXpert Xpress System instrument. The SPC is present to control for adequate processing of the sample and to monitor for the presence of potential inhibitor(s) in the RT-PCR reaction. The SPC also ensures that the RT-PCR reaction conditions (temperature and time) are appropriate for the amplification reaction and that the RT-PCR reagents are functional.
The SPC should be positive in a negative sample and can be negative or positive in a positive sample. Unfortunately, comparison between the target Ct value and specimen processing control Ct value is not practical or meaningful, as the Ct value of the SPC is impacted by the quantity of SARS CoV-2 in the patient sample and can be even negative in a positive SARS-CoV-2 test. Kindly see table from package insert ( visible in the word document uploaded with this submission)
3. Using Ct as a correlate to the viral load needs more validation. Basically, as it is well mentioned by the authors, these kits have been developed as qualitative tests. Using just 2 standard points ( in this case 10000 and 100000 VPs) is not enough to use the results as an even semi-quantitative test. I would suggest using at least 4 standard concentrations of the virus to see if the test is linear and efficient in a broader range. It is noteworthy that the observed difference between the 2 standard concentration is not 3.3 (as it is written in the manuscript) but it is 3.6 which might have larger impacts in lower or higher viral loads.
Thank you for your comment.
We used a serial dilution of commercially available, quantified SARS-CoV-2 to better understand the performance of our Cepheid SARS-CoV-2 PCR. Our data showed a linear relationship between the viral concentration and the Ct values over four “1/10” dilutions of standard with an r2 value of 0.9.
In the manuscript, we intended to explain that theoretically, a 3.3 difference in Ct value between two concentrations occurs when the r2 value is 1.0. For r2 values less than 1.0 (eg;0.9 such as in our study), the 3.3 difference between the Ct value of two concentrations that are 10 fold different may be little higher or lower.
We clarified this point in the methods section, lines 102-106 and rearranged the format of our sentence to avoid any confusions. We also added a reference at the end of the sentence.
4. It is not clear to me how the standard virus sample has been quantified. Does it come from a virus culture or a clinical sample? Is it quantified based on genetic material copy number or based on infectious particle counting by one of virus titration methods?
Thank you for your question. We used quantified SARS-CoV-2 material for better understanding of the relationship between Ct values and the virus concentration in the patient sample. The quantified material was obtained from a commercial source. We changed the wording in the manuscript to read as: Using a commercial quantified standard, we performed studies to determine the Ct value at different target concentrations (Line 104)
5. Following the previous comments on reliability of the quantification method, in my opinion choosing the cutoffs of 26 and 36 are not well justified. We have found several COVID cases with Ct values below 20. On the other hand the intercept of the standard curve of the majority of QPCR kits is slightly higher than 35 (usually below 40), so the readouts above 35 are not usually reliable, or could be due to non-infectious viral debris.
We agree with your comment as we also have seen many positive SARS-CoV-2 cases with Ct values below 20. As a CLIA certified, CAP approved laboratory, we must follow the guidelines for reporting test result set by the manufacturer and approved by FDA for the SARSCoV-2 assay that has received FDA emergency use authorization. While there are reports that specimens positive for SARS-CoV-2 with high Ct value may be non-infectious, we still report the test as positive regardless of the Ct value. The lab leaves the interpretation of the test results with high Ct to the clinicians and medical teams. Moreover, we excluded asymptomatic patients who were admitted to the hospital for reasons other than COVID-19 infection and who were found to be COVID-19 positive on screening (for example pre-operatively) . (Fig1).
6. Based on the previous comments and uncertainties regarding the QPCR results, I would recommend statistical re-analyzing the data with parametric test as well. In those analyses the absolute value of Ct ( or delta Ct, in case the Ct of the internal controls are available) could be used. Then the author may come to similar conclusion, or find out choosing different cutoffs for stratification is more suitable, or just report the results of the parametric analysis.
Thank you for your constructive comment. We agree that re-analyzing the data by using the Ct value as a continuous variable is extremely important. We added a paragraph to the methods (lines 123-127 and results (lines 205-209) sections describing our statistical methods and results (as detailed below). We also added an extra table (Table 3) to the main manuscript as we believe that these findings strengthen our results.
We performed a Shapiro-Wilk test of normality on the continuous variables age and Ct values. Both did not assume a normal distribution. We then proceeded to approximate their correlation independently with mortality using a Mann-Whitney U test. A multiple linear regression model was then performed.
Age and Ct value were independently associated with in-hospital mortality (p<0.001). On multiple linear regression, both age (ß= 0.182, p<0.001) and Ct value (ß= -0.251, p<0.001) were significantly associated with mortality, such that Death = 0.431 + 0.006*Age - 0.016*Ct value. The negative association between Ct and mortality reflects the fact that lower Ct values indicates higher viral load.
7. Based on results presented in table 1, it seems to me that Age and Viral load are rather highly correlated. In table 2 both have very close ORs on Mortality rate as well ( 5.50 vs 5.42). I wonder what is the correlation of these two variable with each other, and whether they are not covariates. Have the author tested other variables in different age groups ( or again, using age as a parametric variable against other variables)? In the Statistical Analysis section there is a reference to table S1, which I could not find it in the manuscript or its supplementary materials.
Thank you for your comment. There was an inverse association (p<0.001) between age and Ct value such that Ct value = 39.3 – 0.088*Age. These two variables do seem to be related, however, after controlling for age in our model, a higher viral load was still associated with increased mortality. Given that the main goal of our study was to analyze the association between viral load and outcomes (mortality and intubation) and explore the potential use of Ct value in predicting the course of the pandemic, we elected not to explore the relationship between Ct and age or age and other variables.
The supplementary table was not uploaded separately as part of the supplementary material (by mistake); It was instead included at the bottom of the manuscript below the references section. We re-uploaded the table separately as part of the supplementary material with this revised submission.
8. In table 1, the average level of Troponin is 40, while the average of each VL group is below 29. I wonder if it is correct.
Thank you for your observation and thorough review of the manuscript. The value was indeed incorrect. The corrected troponin value is 28 (11-80) ; this was adjusted in Table 1.
Reviewer #3:
The manuscript describes a cohort study on relation among initial viral load in nasopharyngeal covid-19 samples and severity of the disease as well as status of pandemic. The subject is well designed and performed although some points need authors’ attention:
1. An important factor influencing the outcome of the disease and the result of this study is the type of treatment and should be considered in all interpretations. Type of antiviral or anti-inflammatory drugs, interval time between PCR and starting the treatment should be considered in analysis.
Dear reviewer,
Thank you for your constructive comment. We added a paragraph in the results section to address your comment (Lines 212-221). We did collect data on 3 medications: steroids, tocilizumab and remdesivir. At the time of the study, little evidence was available to support the use of these medications in patients with COVID-19 infection. A minority of patients received tocilizumab or remdesivir. Corticosteroids were prescribed to 52.3% (n=102) of patients with low viral load, 73.2% (n=123) of patients with intermediate viral load and 78.6% (n=77) of those with high viral load. Patients who received steroids had higher mortality rates compared to patients who did not (35.8% vs 15.1%, p < 0.011). Since corticosteroids were generally reserved for more severe cases, the increased prevalence of their use in higher viral load categories reflects the described relationship between viral load and mortality highlighted our study. No conclusions regarding the effect of corticosteroids on mortality could be drawn.
On a related note, given that we only included Ct values that were obtained on initial admission to the hospital and prior to any therapeutic interventions, the Ct values should not be affected by any type of therapy that patients received during their hospitalization.
2. An explanation of the division of weeks according to pandemic conditions should be given at the first appearance in the text.
Thank you for your comment. We added a brief sentence describing the pandemic condition in Michigan as it relates to the weeks of our study. (lines 90-94)
3- In comparing with the report of Magleby et al. in addition to the reasons given for the discrepancy with the results of the present study, the authors should also examine and discuss the pandemic conditions in two different studied time periods. Similarly, due to the fact that pandemic peaks are not the same in different countries, this point should be considered in comparison with the data of other countries.
Thank you for your comment. We added a sentence mentioning that pandemic conditions between our study and Magleby et al. were comparable as both studies spanned a period when Michigan and New York city witnessed their first peak in newly diagnosed COVID-19 cases coinciding on April 7 and April 12 respectively (Lines 271-274)
Regarding the paragraph discussing the trend in Ct values in Italy, the authors of the study did confirm that Ct values were collected during the month of April, which represented the peak of the pandemic in that region of Italy and compared these values to those obtained, in May, a time when the pandemic in Italy became less severe. This is also comparable to our study as we followed Ct values between April 4 (peak of the pandemic) until June 5, a time when the pandemic severity had been weaning down gradually.
4 -The sentence in line 97 and 98 needs reference.
Thank you for your comment; We added 2 references (now line 101)
Sincerely,
Pranatharthi Chandrasekar, MD
Division of Infectious Diseases
Wayne State University School of Medicine, Detroit, Michigan
Phone: 313-745-7105 / Fax: 313-993-0302
Attachment
Submitted filename: Response to reviewers.docx
Decision Letter 1
16 Jun 2021
PONE-D-21-08808R1
SARS-CoV-2 infection: Initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic
PLOS ONE
Dear Dr. Chandrasekar,
Thank you for submitting your manuscript to PLOS ONE. After careful consideration, we feel that it has merit but does not fully meet PLOS ONE’s publication criteria as it currently stands. Therefore, we invite you to submit a revised version of the manuscript that addresses the points raised during the review process.
Please submit your revised manuscript by Jul 31 2021 11:59PM. If you will need more time than this to complete your revisions, please reply to this message or contact the journal office at gro.solp@enosolp. When you're ready to submit your revision, log on to https://www.editorialmanager.com/pone/ and select the 'Submissions Needing Revision' folder to locate your manuscript file.
Please include the following items when submitting your revised manuscript:
A rebuttal letter that responds to each point raised by the academic editor and reviewer(s). You should upload this letter as a separate file labeled 'Response to Reviewers'.
A marked-up copy of your manuscript that highlights changes made to the original version. You should upload this as a separate file labeled 'Revised Manuscript with Track Changes'.
An unmarked version of your revised paper without tracked changes. You should upload this as a separate file labeled 'Manuscript'.
If you would like to make changes to your financial disclosure, please include your updated statement in your cover letter. Guidelines for resubmitting your figure files are available below the reviewer comments at the end of this letter.
If applicable, we recommend that you deposit your laboratory protocols in protocols.io to enhance the reproducibility of your results. Protocols.io assigns your protocol its own identifier (DOI) so that it can be cited independently in the future. For instructions see: http://journals.plos.org/plosone/s/submission-guidelines#loc-laboratory-protocols. Additionally, PLOS ONE offers an option for publishing peer-reviewed Lab Protocol articles, which describe protocols hosted on protocols.io. Read more information on sharing protocols at https://plos.org/protocols?utm_medium=editorial-email&utm_source=authorletters&utm_campaign=protocols.
We look forward to receiving your revised manuscript.
Kind regards,
Tai-Heng Chen, M.D.
Academic Editor
PLOS ONE
Journal Requirements:
Please review your reference list to ensure that it is complete and correct. If you have cited papers that have been retracted, please include the rationale for doing so in the manuscript text, or remove these references and replace them with relevant current references. Any changes to the reference list should be mentioned in the rebuttal letter that accompanies your revised manuscript. If you need to cite a retracted article, indicate the article’s retracted status in the References list and also include a citation and full reference for the retraction notice.
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. If the authors have adequately addressed your comments raised in a previous round of review and you feel that this manuscript is now acceptable for publication, you may indicate that here to bypass the “Comments to the Author” section, enter your conflict of interest statement in the “Confidential to Editor” section, and submit your "Accept" recommendation.
Reviewer #2: All comments have been addressed
Reviewer #3: All comments have been addressed
**********
2. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #2: Partly
Reviewer #3: Yes
**********
3. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #2: I Don't Know
Reviewer #3: I Don't Know
**********
4. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #2: No
Reviewer #3: Yes
**********
5. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #2: Yes
Reviewer #3: Yes
**********
6. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #2: Thank you for addressing all comments. However, I still can not see the evidence of any standard curve to partially validate the quantification of VL by the QPCR. IT is mentioned that 1 to 10 dilution was used to draw such a standard curve and R^2 was 0.9, but no data has been provided. The source or specifications of the commercial virus sample is not clear as well.
I am not sure, but I think the added statistical analysis is not accurate. I appreciate testing for normality of data and comparing age and Ct in outcome groups by Mann-Withney U test, but I do not understand how and on what variables the linear regression model was fitted. the equation for table 3, Death = 0.431 + 0.006*Age - 0.016*Ct value, looks like a logistic regression equation to me, whereas by "Death" the authors mean the probability of death, but it is not mentioned in the method section. Beside, in the same table, while age and Ct value are parametric variables, CKD must be nominal and it is not clear to me by which method this p value has been calculated.
Other answers are satisfactory
Reviewer #3: My comments have been addressed by the authors, but regarding the reviewer #2 comments 1 &2 it is suggested that brief explanations similar to the response to the reviewer about the Cepheid GenExpert instrument system be given in the text.
**********
7. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #2: No
Reviewer #3: No
[NOTE: If reviewer comments were submitted as an attachment file, they will be attached to this email and accessible via the submission site. Please log into your account, locate the manuscript record, and check for the action link "View Attachments". If this link does not appear, there are no attachment files.]
While revising your submission, please upload your figure files to the Preflight Analysis and Conversion Engine (PACE) digital diagnostic tool, https://pacev2.apexcovantage.com/. PACE helps ensure that figures meet PLOS requirements. To use PACE, you must first register as a user. Registration is free. Then, login and navigate to the UPLOAD tab, where you will find detailed instructions on how to use the tool. If you encounter any issues or have any questions when using PACE, please email PLOS at gro.solp@serugif. Please note that Supporting Information files do not need this step.
Author response to Decision Letter 1
17 Jul 2021
Dear editor
Tai-Heng Chen, M.D.
PLOS One
Thank you for considering our manuscript for publication.
We would also like to thank the reviewers for their valuable and constructive comments.
Kindly find below in bold our point-by-point response to the academic editor and reviewers’ comments:
Reviewer #2:
1. Thank you for addressing all comments. However, I still can not see the evidence of any standard curve to partially validate the quantification of VL by the QPCR. IT is mentioned that 1 to 10 dilution was used to draw such a standard curve and R^2 was 0.9, but no data has been provided. The source or specifications of the commercial virus sample is not clear as well.
Thank you for your comment. Please find below our answer to comments #1 and #2 respectively:
Source and specifications of the commercial virus sample:
In order to determine the performance of Cepheid Genexpert SARS-CoV-2 assay we used quantified standard materials that were obtained from Exact Diagnostics (www.exactdiagnostics.com). The product from Exact diagnostics is EDX SARS-CoV-2 Standard that is manufactured with synthetic RNA transcripts containing five gene targets (E, N, ORF1ab, RdRP and S Genes of SARS-CoV-2). The product allows laboratories to validate/verify the entire process of the assay including extraction, amplification, and detection. The EDX SARS-CoV-2 Standard contains E, N, ORF1ab, RdRP and S genes that are each quantitated at 200,000 cp/mL using Bio-Rad Digital Droplet PCR (ddPCR).
The other commercially available source that we used was Zeptometrix NATtrol™ (SARS-CoV-2) External positive Control. It was used as well for the validation/verification studies and in determination of the lower limit of detection of our SARS-CoV-2 assay.
This control is a quantified source for SARS-CoV-2 and has a concentration of 50,000 copies/ml.
We also received SARS-CoV-2 virus from BEI resources repository (www.niaid.nih.gov/research/bei-resources-repository after signing a material transfer agreement. Being able to obtain the SARS-CoV-2 was a big factor in quick validation/verification of our SARS-CoV-2 assay at the very early stage of COVID-19 pandemic which allowed our laboratory to start testing patient samples for COVID-19 in March of 2020.
Standard curve:
We are attaching a picture of the standard curve in the submission system as a supplementary file and the figure can be also seen in the response to reviewers word document attached with this revision.
After repeating the experiment, we have made few changes pertaining to the correlation between the viral loads and corresponding Ct values. The changes made are highlighted below, and these do not affect our results or conclusions. (Lines 110-114)
At 2x102, 2x103 and 2x104 viral particles, the Ct values were 43, 39.4 and 36.7 respectively. We designated high, intermediate, and low VL samples to have a Ct value of ≤ 25, 26-36, and ≥ 37 consecutively, therefore, assuming a linear relationship between the Ct value and target concentration, samples with a Ct value of 26 should have a VL of 2x107 while samples with a Ct value of 36 should have a VL of 2x 104 approximately.
The viral load categories (low, intermediate and high) were chosen based on the Ct values of <25, 26-36 and > 37 respectively. Other comparable categories of viral load are used in the literature. (Rabaan AA, et al. Diagnostics (Basel). 2021;11(6):1091 and Rao SN, et al. Infect Dis Ther 2020;9(3):573-586. 10.1007/s40121-020-00324-3.
Correlating a Ct value to a viral load in our manuscript is used only to provide perspective to the readers. Moreover, the association between Ct value and mortality was also demonstrated on multivariate logistic regression when Ct values were analyzed as continuous variables.
2. I am not sure, but I think the added statistical analysis is not accurate. I appreciate testing for normality of data and comparing age and Ct in outcome groups by Mann-Withney U test, but I do not understand how and on what variables the linear regression model was fitted. the equation for table 3, Death = 0.431 + 0.006*Age - 0.016*Ct value, looks like a logistic regression equation to me, whereas by "Death" the authors mean the probability of death, but it is not mentioned in the method section. Beside, in the same table, while age and Ct value are parametric variables, CKD must be nominal and it is not clear to me by which method this p value has been calculated.
Based on your recommendations, we performed a multivariate logistic regression model using age and Ct value as continuous variables and added a paragraph to the manuscript. (Lines 234-240).
Using multiple logistic regression, both age and Ct value were found to be significantly associated with mortality, such as every 1-year increase in age results in 3.5% increase in odds of death and every 1 unit increase in Ct value decreases the odds of death by 8%. The negative association between Ct and mortality reflects the fact that lower Ct values indicate higher viral load (Table 3 - can be seen in the main body of the manuscript and word document of the response to reviewers attached with this submission)
Reviewer #3:
My comments have been addressed by the authors, but regarding the reviewer #2 comments 1 &2 it is suggested that brief explanations similar to the response to the reviewer about the Cepheid GenExpert instrument system be given in the text.
Thank you for your feedback. We added a paragraph to the methods section briefly explaining how our assay does not contain an internal control but instead, the SPC utilized by the GeneXpert Xpress System instrument serves to confirm adequate processing of the sample and to ensure that the RT-PCR reaction conditions are appropriate (Lines 102- 105).
Sincerely,
Pranatharthi Chandrasekar, MD
Division of Infectious Diseases
Wayne State University School of Medicine, Detroit, Michigan
Phone: 313-745-7105 / Fax: 313-993-0302
Attachment
Submitted filename: Response to reviewers.docx
Decision Letter 2
28 Jul 2021
SARS-CoV-2 infection: Initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic
PONE-D-21-08808R2
Dear Dr. Chandrasekar,
We’re pleased to inform you that your manuscript has been judged scientifically suitable for publication and will be formally accepted for publication once it meets all outstanding technical requirements.
Within one week, you’ll receive an e-mail detailing the required amendments. When these have been addressed, you’ll receive a formal acceptance letter and your manuscript will be scheduled for publication.
An invoice for payment will follow shortly after the formal acceptance. To ensure an efficient process, please log into Editorial Manager at http://www.editorialmanager.com/pone/, click the 'Update My Information' link at the top of the page, and double check that your user information is up-to-date. If you have any billing related questions, please contact our Author Billing department directly at gro.solp@gnillibrohtua.
If your institution or institutions have a press office, please notify them about your upcoming paper to help maximize its impact. If they’ll be preparing press materials, please inform our press team as soon as possible -- no later than 48 hours after receiving the formal acceptance. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information, please contact gro.solp@sserpeno.
Kind regards,
Tai-Heng Chen, M.D.
Academic Editor
PLOS ONE
Reviewers' comments:
Reviewer's Responses to Questions
Comments to the Author
1. If the authors have adequately addressed your comments raised in a previous round of review and you feel that this manuscript is now acceptable for publication, you may indicate that here to bypass the “Comments to the Author” section, enter your conflict of interest statement in the “Confidential to Editor” section, and submit your "Accept" recommendation.
Reviewer #2: All comments have been addressed
Reviewer #3: All comments have been addressed
**********
2. Is the manuscript technically sound, and do the data support the conclusions?
The manuscript must describe a technically sound piece of scientific research with data that supports the conclusions. Experiments must have been conducted rigorously, with appropriate controls, replication, and sample sizes. The conclusions must be drawn appropriately based on the data presented.
Reviewer #2: Yes
Reviewer #3: Yes
**********
3. Has the statistical analysis been performed appropriately and rigorously?
Reviewer #2: Yes
Reviewer #3: Yes
**********
4. Have the authors made all data underlying the findings in their manuscript fully available?
The PLOS Data policy requires authors to make all data underlying the findings described in their manuscript fully available without restriction, with rare exception (please refer to the Data Availability Statement in the manuscript PDF file). The data should be provided as part of the manuscript or its supporting information, or deposited to a public repository. For example, in addition to summary statistics, the data points behind means, medians and variance measures should be available. If there are restrictions on publicly sharing data—e.g. participant privacy or use of data from a third party—those must be specified.
Reviewer #2: Yes
Reviewer #3: Yes
**********
5. Is the manuscript presented in an intelligible fashion and written in standard English?
PLOS ONE does not copyedit accepted manuscripts, so the language in submitted articles must be clear, correct, and unambiguous. Any typographical or grammatical errors should be corrected at revision, so please note any specific errors here.
Reviewer #2: Yes
Reviewer #3: Yes
**********
6. Review Comments to the Author
Please use the space provided to explain your answers to the questions above. You may also include additional comments for the author, including concerns about dual publication, research ethics, or publication ethics. (Please upload your review as an attachment if it exceeds 20,000 characters)
Reviewer #2: Thank you for revising the statistical analysis. I have just one further recommendation, in real time PCR analyses, the standard curve graph (here presented in the supplement) is usually drawn as Log of concentration in the X axis and Ct values in the Y axis. Though the statistical meaning is the same, this is more common because in the resulting regression equation, the slope could be easily converted to the efficiency of the PCR.
Reviewer #3: (No Response)
**********
7. PLOS authors have the option to publish the peer review history of their article (what does this mean?). If published, this will include your full peer review and any attached files.
If you choose “no”, your identity will remain anonymous but your review may still be made public.
Do you want your identity to be public for this peer review? For information about this choice, including consent withdrawal, please see our Privacy Policy.
Reviewer #2: Yes: Kayhan Azadmanesh MD. Ph.D.
Reviewer #3: Yes: Dr T Bamdad
Acceptance letter
8 Sep 2021
PONE-D-21-08808R2
SARS-CoV-2 infection: Initial viral load (iVL) predicts severity of illness/outcome, and declining trend of iVL in hospitalized patients corresponds with slowing of the pandemic
Dear Dr. Chandrasekar:
I'm pleased to inform you that your manuscript has been deemed suitable for publication in PLOS ONE. Congratulations! Your manuscript is now with our production department.
If your institution or institutions have a press office, please let them know about your upcoming paper now to help maximize its impact. If they'll be preparing press materials, please inform our press team within the next 48 hours. Your manuscript will remain under strict press embargo until 2 pm Eastern Time on the date of publication. For more information please contact gro.solp@sserpeno.
If we can help with anything else, please email us at gro.solp@enosolp.
Thank you for submitting your work to PLOS ONE and supporting open access.
Kind regards,
PLOS ONE Editorial Office Staff
on behalf of
Dr. Tai-Heng Chen
Academic Editor
PLOS ONE
Articles from PLOS ONE are provided here courtesy of PLOS
Full text links
Read article at publisher's site: https://doi.org/10.1371/journal.pone.0255981
Read article for free, from open access legal sources, via Unpaywall:
https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0255981&type=printable
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/113606956
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1371/journal.pone.0255981
Article citations
Association of COVID-19 with acute and post-acute risk of multiple different complications and mortality in patients infected with omicron variant stratified by initial disease severity: a cohort study in Hong Kong.
BMC Med, 22(1):461, 14 Oct 2024
Cited by: 0 articles | PMID: 39402606 | PMCID: PMC11476291
SARS-CoV-2 viremia but not respiratory viral load is associated with respiratory complications in patients with severe COVID-19.
BMC Pulm Med, 24(1):366, 29 Jul 2024
Cited by: 0 articles | PMID: 39080682 | PMCID: PMC11288013
A retrospective analysis of the influencing factors of nucleic acid CT value fluctuation in COVID-19 patients infected with Omicron variant virus in Changchun city.
Front Public Health, 12:1377135, 14 Jun 2024
Cited by: 0 articles | PMID: 38947348
Analyzing factors affecting positivity in drive-through COVID-19 testing: a cross-sectional study.
Virol J, 21(1):111, 14 May 2024
Cited by: 0 articles | PMID: 38745200 | PMCID: PMC11094999
Host factors of SARS-CoV-2 in infection, pathogenesis, and long-term effects.
Front Cell Infect Microbiol, 14:1407261, 22 May 2024
Cited by: 0 articles | PMID: 38846354
Review
Go to all (28) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Quantitative detection of SARS-CoV-2 RNA in nasopharyngeal samples from infected patients with mild disease.
J Med Virol, 93(4):2439-2445, 05 Jan 2021
Cited by: 7 articles | PMID: 33368332
Relationship of the cycle threshold values of SARS-CoV-2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19.
Int J Infect Dis, 101:160-166, 28 Sep 2020
Cited by: 39 articles | PMID: 32992013 | PMCID: PMC7521909
SARS-CoV-2 viral load in nasopharyngeal swabs is not an independent predictor of unfavorable outcome.
Sci Rep, 11(1):12931, 21 Jun 2021
Cited by: 19 articles | PMID: 34155307 | PMCID: PMC8217169
Quantifying SARS-CoV-2 viral load: current status and future prospects.
Expert Rev Mol Diagn, 21(10):1017-1023, 09 Aug 2021
Cited by: 11 articles | PMID: 34369836 | PMCID: PMC8425446
Review Free full text in Europe PMC