| PMC full text: | Published online 2021 Sep 23. doi: 10.3389/fpls.2021.668020
|
Figure 3
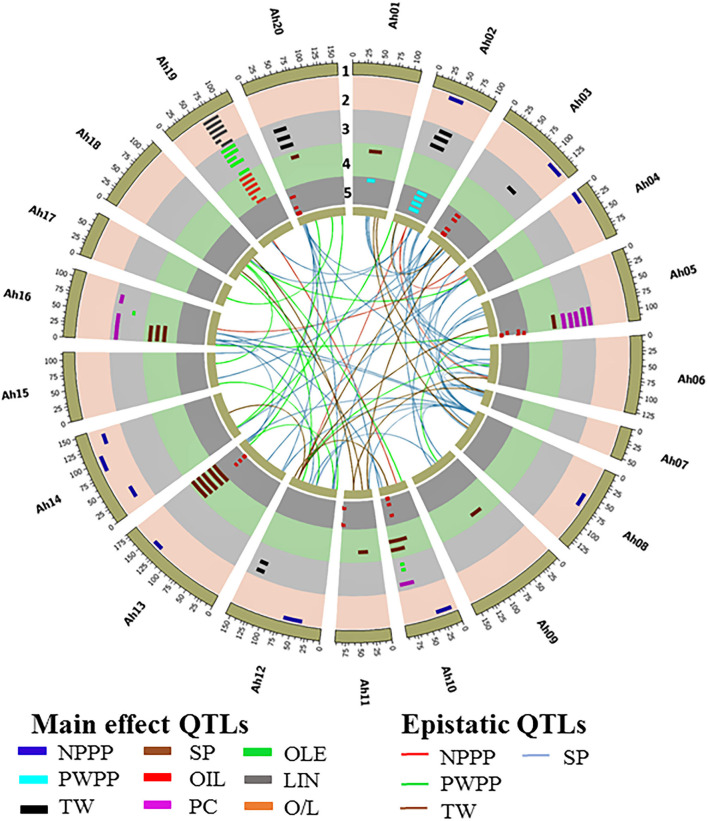
Circos plot illustrating main effect and epistatic (QTL × QTL) QTLs identified for productivity and quality traits in ML population of TMV 2 and TMV 2-NLM of peanut. The tracks from outside to inside indicates (1) 20 chromosomes of tetraploid genome Arachis hypogaea, (2) main effect QTLs for number of pods per plant (NPPP) and linoleic acid (IAN) qNPPP–Ah02-1, qNPPP–Ah04-1, qNPPP–Ah08-1, qNPPP–Ah10-1, qNPPP–Ah12-1, qNPPP–Ah13-1, qNPPP–Ah14-1,qNPPP–Ah14-2, qNPPP–Ah14-3, qLIN–Ah19-1, qLIN–Ah19-2, qLIN–Ah19-3, qLIN–Ah19-4, qLIN–Ah19-5, qLIN–Ah19-6, qLIN–Ah19-7, qLIN–Ah19-8, qLIN–Ah19-9, qLIN–Ah19–10, qLIN–Ah19-11, qLIN–Ah19-12, qLIN–Ah19-13, (3) main effect QTLs for test weight (TW), protein content (PC), and oleic acid content (OLE) qTW–Ah02-1, qTW–Ah02-2, qTW–Ah02-3, qTW–Ah02-4, qTW–Ah03-1, qPC–Ah05-1, qPC–Ah05-2, qPC–Ah05-3, qPC–Ah10-1, qOLE–Ah10-1, qTW–Ah12-1, qTW–Ah12-2, qTW–Ah12-3, qTW–Ah12-4, qPC–Ah16-1, qPC–Ah16-2, qPC–Ah16-3, qOLE–Ah16-1, qOLE–Ah19-1, qOLE–Ah19-2, qOLE–Ah19-3, qOLE–Ah19-4, qOLE–Ah19-5, qOLE–Ah19-6, qOLE–Ah19-7, qOLE–Ah19-8, qOLE–Ah19-9, qOLE–Ah19-10, qOLE–Ah19-11, qOLE–Ah19-12, qTW–Ah20-1, qTW–Ah20-2, qTFV–Ah20-3, (4) main effect QTLs for shelling percentage (SP) and oleic to linoleic ratio (0/L) qSP–Ah01-1, qSP–Ah02-1, qSP–Ah05-1, q5P–Ah09-1, qSP–Ah10-1, qSP–Ah10-2, qSP–Ah11-1, OP–Ah13-1, OP-A/216-1, qO/L–Ah19-1, qO/L–Ah19-2, qO/L–Ah19-3, qO/L–Ah19-4, q0/L–Ah19-5, qO/L–Ah19-6, qO/L–Ah19-7, qO/L–Ah19-8, qO/L–Ah19-9, qO/L–Ah19-10, qO/L–Ah19-11, qO/L–Ah19-12, qO/L–Ah19-13, qSP–Ah20-1, (5) main effect QTLs for oil content (OIL) and pod weight per plant (PWPP). qPWPP–Ah01-1, qPWPP–Ah02-1, qOIL–Ah03-1, qOIL–Ah03-2, qOIL–Ah03-3, qOIL–Ah03-4, qOIL–Ah05-1, qOIL–Ah05-2, qOIL–Ah10-1 qOIL–Ah11-1, qOIL–Ah13-1, qOIL–Ah20-1, qOIL–Ah20-2. Innermost links connecting between the loci indicates the epistatic QTLs for NPPP, PWPP, TW, and SP.