Abstract
Free full text

Quantitation of Hepatitis B Virus Genomic DNA by Real-Time Detection PCR
Abstract
Quantitation of hepatitis B virus (HBV) DNA in serum is a useful method for the monitoring of HBV replication. We attempted to develop a quantitative assay system for HBV DNA that is more sensitive, accurate, and reproducible than existing systems. We detected HBV DNA by real-time detection PCR (RTD-PCR) based on Taq Man chemistry. The efficacy of this assay was evaluated by quantitatively measuring sequential levels of synthetic DNA and DNA in clinical serum samples. The detection limit of this system was as few as 10 DNA copies/reaction. A linear standard curve was obtained between 101 and 108 DNA copies/reaction. The coefficient of variation for both intra- and interexperimental variability indicated remarkable reproducibility. This system detected HBV DNA in 100% of chronic hepatitis B patients tested and never detected HBV DNA in healthy volunteers who were negative for HBV markers. These observations suggest that RTD-PCR is an excellent candidate for a standard HBV quantification method.
Hepatitis B virus (HBV) is a major causative agent of chronic hepatitis and can cause liver cirrhosis and hepatocellular carcinoma (1). The prevalence of HBV infection is epidemiologically associated with that of hepatocellular carcinoma (16). Control of HBV infection is, therefore, an important goal for public health in areas of endemicity.
Antiviral treatment consisting of interferon (IFN) or lamivudine (3TC) is now available for chronic hepatitis B (2, 16). Accurate quantification of HBV DNA is essential for monitoring the efficacy of these antiviral treatments. HBV replication often correlates with hepatitis activity, and effective antiviral treatments are known to induce rapid decreases in serum viral load (11). The DNA polymerase assay has been widely used for monitoring antiviral treatments. In this assay, levels of DNA polymerase are quantified in vitro by the incorporation of radio-labeled deoxynucleoside monophosphates (8), which is difficult to standardize among laboratories (3). Therefore, the assay has been replaced by a simpler and more accurate HBV DNA quantification system that uses a branched-DNA (bDNA) probe (6, 9). However, this method is not sensitive enough to monitor serum virus levels in patients undergoing antiviral treatment.
Currently, HBV DNA is quantified by PCR. Although monitoring by PCR is more sensitive than detection using the bDNA probe, contamination, poor quantitation, and poor reproducibility limit its clinical applicability.
In this report, we show our results of a highly sensitive assay for HBV DNA by real-time detection PCR (RTD-PCR) based on Taq Man chemistry (5). We have evaluated the efficacy of this system using synthetic standard HBV DNA and several clinical samples and have assessed the correlation between the results obtained by this assay and those obtained using the bDNA assay. We have also demonstrated the potential clinical usefulness of this RTD-PCR assay by monitoring HBV DNA levels in a patient currently undergoing antiviral treatment for acute exacerbation of his HBV carrier status.
CASE REPORT
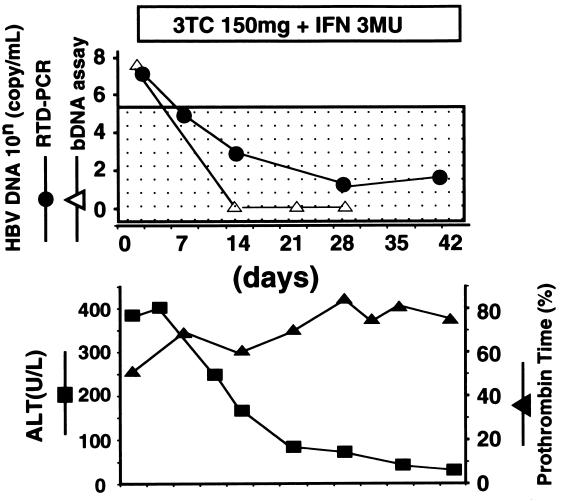
A 40-year-old male was admitted to our hospital due to acute reactivation of persistent HBV infection. He was an asymptomatic HBV carrier with antibodies to HBe antigen. His HBV carrier status was diagnosed based on the positive results for HBV surface antigen (HBsAg) lasting more than 6 months and high-titered antibodies to hepatitis B core antigen. At admission, he was icteric and his prothrombin time (PT) level was 50%. After the start of antiviral treatment, consisting of IFN and 3TC, his PT level increased and his serum alanine aminotransferase (ALT) level decreased. Although the HBV DNA level rapidly decreased below the detection limit of the bDNA assay, the RTD-PCR using primers and probe set 1 (see Materials and Methods) detected and tracked changes in the HBV DNA level during the clinical course. The patient recovered from acute exacerbation with normalization of ALT and PT levels (Fig. (Fig.1).1).
MATERIALS AND METHODS
Preparation of standard HBV DNA.
We subcloned an HBV genome insert (nucleotides [nt] 1 to 2182) from pGEM7 into pBlueScript II SK(+) (Stratagene, La Jolla, Calif.). The recombinant plasmid was purified and subsequently quantified by measuring the optical density at 260 nm.
Oligonucleotide primers and probes.
RTD-PCR was performed with three sets of PCR primers and a probe; two sets were located in the HBV surface gene and the remaining one was located in the X gene. The criteria for primer and probe sets are as follows: first, a highly conserved sequence homology region; then, sufficient sensitivity; and finally, distance within the HBV genome, as with the HBV S and X genes. Set 1 of primers and a probe, derived from the S gene, consisted of a forward primer, HBSF1 (nt 166 to 186), 5′-CACATCAGGATTCCTAGGACC-3′; a reverse primer, HBSR1 (nt 339 to 321), 5′-GGTGAGTGATTGGAGGGTTG-3′; and a Taq Man probe, HBSP1 (nt 242 to 267), 5′-CAGAGTCTAGACTCGTGGTGGACTTC-3′; set 2 consisted of HBSF2 (nt 406 to 426), 5′-CTTCATCCTGCTGCTATGCCT-3′; HBSR2 (nt 646 to 627), 5′-AAAGCCCAGGATGATGGGAT-3′; and HBSP2 (nt 461 to 488), 5′-ATGTTGCCCGTTTGTCCTCTAATTCCA-3′. Set 3 of primers and a probe, derived from the X gene, consisted of a forward primer, HBXF1 (nt 1414 to 1435), 5′-ACGTCCTTTGTTTACGTCCCGT-3′; a reverse primer, HBXR1 (nt 1744 to 1723), 5′-CCCAACTCCTCCCAGTCCTTAA-3′; and a Taq Man probe, HBXP1 (nt 1681 to 1705), 5′-TGTCAACGACCGACCTTGAGGCATA-3′. A reporter dye (6-carboxy-fluorescein) was covalently attached to the 5′ end, and a quencher dye (6-carboxy-tetramethyl-rhodamine) was incorporated into the 3′ end of the probe sequence. A passive reference dye was included in the PCR buffer.
Quantification of HBV DNA by RTD-PCR.
The principle of RTD-PCR is as follows. If the target of interest is present during PCR, the probe specifically anneals between the forward and reverse primers. The 5′-3′ exonuclease activity of Taq polymerase cleaves the probe between the reporter and the quencher. This results in an increase in fluorescence of the reporter that is proportional to the amount of product accumulated. Normalized reporter signal (Rn) is calculated by dividing the amount of reporter signal by the amount of passive reference signal (4). ΔRn represents the amount of normalized reporter signal minus the amount of reporter signal before PCR. ΔRn increases during PCR as the target is amplified, until the reaction approaches a plateau (Fig. (Fig.2)2) (4). Following amplification, real-time data acquisition and analysis are performed. In this present study, the relative fluorescent emission threshold, determined from the baseline of the first 15 cycles, was set 10 standard deviations above the mean baseline emission calculated from cycles to 1 to 15. Once the threshold was chosen, the point at which the amplification plot crossed the threshold was defined as the threshold cycle (Ct). The Ct represents the threshold of sequence detection and is also dependent on the starting quantity of HBV DNA.
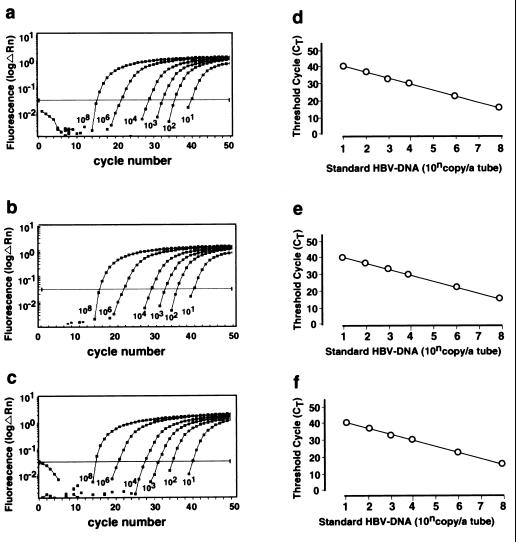
Amplification plots and standard curves of synthetic HBV DNA based on three sets of primers and probe. Set 1 (a and d) and set 2 (b and e) were located in the S region, and set 3 (c and f) was located in the X region. Serial 10-fold dilutions of standard HBV DNA from 101 to 108 copies/reaction tube were prepared. (a to c) ΔRn was plotted against each cycle number. ΔRn increases during PCR as the amplicon copy number increases until the reaction reaches a plateau. The number of copies of each standard HBV DNA are shown. (d to f) Ct was plotted against each copy number. Ct represents the PCR cycle at which reporter signal can first be detected.
Total DNA was extracted from 100 μl of serum with the SMI test EX-RandD (Sumitomo Metal Industries, Tokyo, Japan) used according to the manufacturer’s instructions. Purified DNA was resuspended in 20 μl of distilled water. A 10-μl aliquot of DNA solution (50-μl serum equivalent) was used for RTD-PCR, which was performed with a PCR core reagent kit with an ABI 7700 sequence detector system (Perkin Elmer, Foster City, Calif.). Amplification reaction mixtures (50 μl) contained 10 μl of DNA solution, 5 μl of 10× Taq Man buffer A, 200 μM dATP, 200 μM dCTP, and 200 μM dGTP, 500 μM dUTP, 3.5 mM MgCl2, 200 nM forward primer, 200 nM reverse primer, 300 nM Taq Man probe, 1.25 U of Ampli Taq Gold DNA polymerase, and 0.5 U of Amp Erase uracil N-glycosylase (UNG). Thermal cycling conditions were as follows: initial activation of UNG at 50°C for 2 min followed by activation of Taq Gold and inactivation of UNG at 95°C for 10 min. Subsequently, 53 cycles of amplification were performed at 95°C for 20 s and 60°C for 1 min. Quantification of HBV DNA by RTD-PCR was carried out as a blind test.
bDNA hybridization assay and serology.
HBV DNA in serum was quantitated by the signal amplification method with enzymatically labeled bDNA (Quantiplex HBV DNA; Chiron Corp., Emeryville, Calif.) according to the manufacturer’s instructions. The quantification limit of Quantiplex HBV DNA contains approximately 7 × 105 DNA copies/ml (6).
Specimens.
We examined 46 serum samples collected from patients with chronic hepatitis B and 23 serum samples collected from healthy volunteers. All chronic hepatitis B patients had a positive result for HBsAg and high-titered antibodies to HBV core antigen by enzyme-linked immunosorbent assay (ELISA) (Abbott Laboratories, North Chicago, Ill.). All healthy volunteers had normal serum ALT levels and were negative for HBsAg by ELISA as well as for antibodies to HBV by ELISA (International Reagents Corporation, Kobe, Japan). The presence of HBV DNA in these samples was then assessed by RTD-PCR and bDNA signal amplification assays.
Serial clinical samples were obtained from a 40-year-old male patient with acute exacerbation of chronic HBV infection (Case Report). The patient was treated with 3 × 106 U of beta IFN (Feron; Torey Medical, Tokyo, Japan) daily for 2 weeks and three times a week for 6 weeks in combination with daily 3TC (150 mg).
RESULTS
We first examined RTD-PCR’s sensitivity to and its ability to quantitate synthetic HBV DNA. RTD-PCR detected synthetic HBV DNA in the range of 101 to 108 copies/reaction with each primer and probe set (Fig. (Fig.2).2). A linear relationship was obtained between the Ct and the number of copies of standard HBV DNA (r > 0.99) (Fig. (Fig.22).
Intraexperimental variability was studied by using six samples collected from chronic hepatitis B patients and two samples from healthy volunteers. These eight samples were also assayed in quadruplicate in one experiment. The results of quantitation, along with standard deviations and coefficients of variation (CVs), are shown in Table Table1.1.
TABLE 1
Intraexperimental variability in this studya
studya
| Sample no. | HBV DNA (copies/ml) in expt:
| Mean | SD | CV (%) | |||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||
| 1 | 3.40 × × 102 102 | 2.20 × × 102 102 | 2.70 × × 102 102 | 4.70 × × 102 102 | 3.25 × × 102 102 | 0.141 | 5.67 |
| 2 | 2.30 × × 103 103 | 3.80 × × 103 103 | 3.70 × × 103 103 | 3.50 × × 103 103 | 3.30 × × 103 103 | 0.112 | 3.19 |
| 3 | 2.60 × × 106 106 | 2.50 × × 106 106 | 2.30 × × 106 106 | 2.10 × × 106 106 | 2.38 × × 106 106 | 0.041 | 0.65 |
| 4 | 4.0 × × 107 107 | 3.60 × × 107 107 | 2.40 × × 107 107 | 3.30 × × 107 107 | 3.30 × × 107 107 | 0.096 | 1.27 |
| 5 | 1.40 × × 109 109 | 1.50 × × 109 109 | 1.50 × × 109 109 | 2.00 × × 109 109 | 1.60 × × 109 109 | 0.069 | 0.75 |
| 6 | 6.40 × × 108 108 | 6.80 × × 108 108 | 7.20 × × 108 108 | 5.20 × × 108 108 | 6.40 × × 108 108 | 0.062 | 0.70 |
| 7 |  NDb NDb | ND | ND | ND | |||
| 8 | ND | ND | ND | ND | |||
Interexperimental variability was studied in four independent experiments using the eight sera mentioned above. Results of these experiments are shown in Table Table2.2.
TABLE 2
Interexperimental variability in this studya
studya
| Sample no. | HBV DNA (copies/ml) on day:
| Mean | SD | CV (%) | |||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||
| 1 | 4.70 × × 102 102 | 2.30 × × 102 102 | 2.50 × × 102 102 | 2.80 × × 102 102 | 3.08 × × 102 102 | 0.139 | 5.64 |
| 2 | 1.30 × × 103 103 | 6.50 × × 102 102 | 2.80 × × 103 103 | 8.30 × × 102 102 | 1.40 × × 103 103 | 0.279 | 9.07 |
| 3 | 2.60 × × 106 106 | 2.40 × × 106 106 | 2.10 × × 106 106 | 4.40 × × 106 106 | 2.88 × × 106 106 | 0.141 | 2.19 |
| 4 | 4.00 × × 107 107 | 3.20 × × 107 107 | 5.60 × × 107 107 | 4.00 × × 107 107 | 4.20 × × 107 107 | 0.100 | 1.32 |
| 5 | 1.50 × × 109 109 | 1.50 × × 109 109 | 1.90 × × 109 109 | 6.40 × × 109 109 | 2.83 × × 109 109 | 0.302 | 3.23 |
| 6 | 6.80 × × 108 108 | 2.40 × × 109 109 | 7.20 × × 108 108 | 5.20 × × 108 108 | 1.08 × × 109 109 | 0.296 | 3.30 |
| 7 |  NDb NDb | ND | ND | ND | |||
| 8 | ND | ND | ND | ND | |||
The presence or quantity of HBV DNA in the sera of the 25 of 46 patients and 23 healthy volunteers was determined by RTD-PCR using set 1 and set 3 primers and probes and by bDNA assay. The quantity of HBV DNA in the sera of the remaining 21 patients was determined by RTD-PCR using the set 2 primers and probe and by bDNA assay. RTD-PCR using the primers and probes located in the S or X regions detected the HBV genome in all samples. On the other hand, 9 of the 46 samples contained HBV DNA at levels below the quantitation limit of the bDNA assay (Fig. (Fig.3a3a to c). A significant correlation was found between results obtained by RTD-PCR using the primers and probe sets located in the S (set 1) and X (set 3) regions (r = 0.974) (Fig. (Fig.3d).3d). A significant correlation was also found between the results of the bDNA assay and those of RTD-PCR with primer and probe sets located in the S (set 1 or 2) or X (set 3) region, with correlation coefficients of 0.961 (Fig. (Fig.3a),3a), 0.972 (Fig. (Fig.3b),3b), and 0.926 (Fig. (Fig.3c),3c), respectively. HBV DNA was not detected in any serum samples derived from healthy volunteers by the RTD-PCR assay with any of three primer-probe sets.
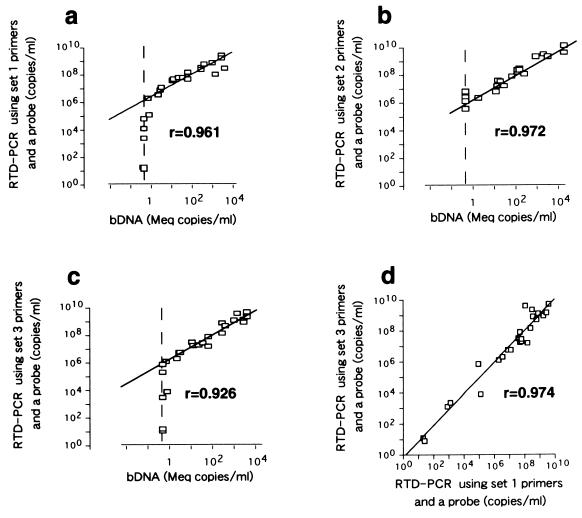
The correlation between serum HBV DNA levels determined by RTD-PCR and those determined by bDNA assay was studied with samples collected from 46 patients with chronic hepatitis B. (a and c) Correlation between serum HBV DNA levels in 25 of 46 patients determined by bDNA assay and RTD-PCR using set 1 and set 3 primers and probes. (b) Correlation between serum HBV DNA levels determined by bDNA assay and RTD-PCR using set 2 primers and probe. (d) Correlation between serum HBV DNA levels determined by RTD-PCR using primers and probes located in the S region (set 1) and the X region (set 3). Correlation coefficients are indicated in each panel. Broken lines indicate the detection limit of the bDNA assay.
DISCUSSION
We have developed a highly sensitive and quantitative RTD-PCR assay for the detection of HBV DNA. Furthermore, the assay is less laborious than competitive PCR. The entire process can be completed within 5 h, including extraction of DNA from the patient’s serum. The RTD-PCR assay is performed by a single step, requiring a single tube, a single enzyme, and a single set of primers with a target-specific fluorogenic probe. Post-PCR data analysis of the RTD-PCR can be performed by using a computer connected to an ABI 7700 sequence detector without gel electrophoresis and probe hybridization (12, 13).
The RTD-PCR showed a close correlation between Ct and a starting quantity of HBV DNA ranging from 101 to 108 copies/reaction. The sensitivity of the RTD-PCR is similar to that of nested PCR. The high sensitivity and good linearity over a wide dynamic range are explained as follows. As shown in a previous study of PCR kinetics, a logarithmic plot of copy numbers in a series of standard HBV DNA samples versus Ct yields a straight line, resulting in good linearity over a wide dynamic range for the RTD-PCR assay (7). In addition, the sensitive fluorescence detection system allows the Ct to be observed while the PCR amplification is still in the exponential phase. Furthermore, none of the components in the reaction mixture is limiting during exponential-phase amplification. Therefore, determining the Ct is more reliable than determining the presence or amount of PCR product (4, 5).
RTD-PCR detected HBV DNA in 8 of the 46 patients with chronic hepatitis B who were negative for HBV DNA by the bDNA assay and did not detect HBV DNA in any of the 23 healthy volunteers. RTD-PCR is estimated to be 104 to 105 times more sensitive than the bDNA assay (Fig. (Fig.3a3a to c). In this limited study, RTD-PCR showed a good sensitivity and specificity.
Ten years has passed since PCR was first applied to the detection of HBV DNA (10, 15). Since then, PCR has been regarded as a sensitive and useful assay for the detection of HBV DNA. However, a PCR method suitable for standardization has not been established until now. A program for quality assurance in the detection of HBV DNA revealed considerable variability in sensitivity and specificity among the 39 participating laboratories around the world (14). The study showed that the main problem in the detection of HBV DNA is false positivity. To avoid contamination of serum samples and previously amplified PCR products, stringent precautions must be routinely implemented. Reaction mixtures for PCR are made in a DNA- and RNA-free working space, and the other steps of RTD-PCR are performed in an amplicon-free working space. The chance of contamination is reduced via monitoring and calculation of fluorescent signals in a single sample tube with a closed optical cap. The use of UNG can also reduce the possibility of contamination.
Changes in serum virus levels frequently induce fluctuation of serum ALT levels. Thus, accurate quantitation of the serum virus level is useful in monitoring the clinical course of patients with active hepatitis B. A reliable quantitative assay should be able to detect small changes in viral level. In accordance with this, the inter- and intraexperimental variabilities of our RTD-PCR were extremely small compared with those of previously reported quantitative methods (12, 13). The CV of quantitation was less than 9.07% throughout the quantitative dynamic range. Therefore, a greater-than-twofold change in HBV DNA levels, as quantified by RTD-PCR, is regarded as significant. In the HBV carrier examined in the present study, RTD-PCR was able to detect the precise change in HBV DNA level and revealed an association between HBV DNA and ALT levels which the bDNA assay failed to confirm (Fig. (Fig.11).
Our study shows that the RTD-PCR assay for HBV DNA is superior to other methods for quantitation of HBV DNA in sensitivity, specificity, simplicity, and reproducibility. The usefulness of this RTD-PCR assay should be further investigated with a large number of samples to ascertain whether it can be accepted as a standard HBV assay. This assay is a candidate for a standard HBV detection system using PCR. This assay may be especially useful in cases of spontaneous reactivation of HBV carriers and acute exacerbation following immunosuppressive treatment, in predicting chronicity following acute infection, and in monitoring the therapeutic effect of antiviral treatments.
ACKNOWLEDGMENTS
We are grateful to Tomoko Takeuchi and Asao Katsume for their helpful comments and suggestions.
This work was supported in part by a Grant-in-Aid for Specially Promoted Research on Viral Diseases from the Tokyo Metropolitan Government, a grant from the Ministry of Education, Science, and Culture of Japan, and a grant from the Ministry of Health and Welfare of Japan.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.37.9.2899-2903.1999
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/37/9/2899.full.pdf
Free to read at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/abstract/37/9/2899
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/reprint/37/9/2899.pdf
Free after 4 months at intl-jcm.asm.org
http://intl-jcm.asm.org/cgi/content/full/37/9/2899
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jcm.37.9.2899-2903.1999
Article citations
Multiscale modeling of HBV infection integrating intra- and intercellular viral propagation to analyze extracellular viral markers.
PLoS Comput Biol, 20(3):e1011238, 11 Mar 2024
Cited by: 2 articles | PMID: 38466770 | PMCID: PMC10957078
Modeling suggests that virion production cycles within individual cells is key to understanding acute hepatitis B virus infection kinetics.
PLoS Comput Biol, 19(8):e1011309, 03 Aug 2023
Cited by: 3 articles | PMID: 37535676 | PMCID: PMC10426918
MNAzymes and gold nanoparticles as isothermal signal amplification strategy for visual detection of miRNA.
Mikrochim Acta, 190(8):292, 17 Jul 2023
Cited by: 3 articles | PMID: 37458796 | PMCID: PMC10352400
Residual risk of mother-to-child transmission of HBV despite timely Hepatitis B vaccination: a major challenge to eliminate hepatitis B infection in Cambodia.
BMC Infect Dis, 23(1):261, 26 Apr 2023
Cited by: 3 articles | PMID: 37101167 | PMCID: PMC10131410
Viral Diagnosis of Hepatitis B and Delta: What We Know and What Is Still Required? Specific Focus on Low- and Middle-Income Countries.
Microorganisms, 10(11):2096, 22 Oct 2022
Cited by: 1 article | PMID: 36363693 | PMCID: PMC9694472
Review Free full text in Europe PMC
Go to all (181) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Quantitative detection of hepatitis B virus DNA in serum by a new rapid real-time fluorescence PCR assay.
J Viral Hepat, 8(6):465-471, 01 Nov 2001
Cited by: 51 articles | PMID: 11703579
Development and assessment of a novel real-time PCR assay for quantitation of HBV DNA.
J Virol Methods, 103(2):201-212, 01 May 2002
Cited by: 45 articles | PMID: 12008014
High-throughput quantitative analysis of hepatitis B virus DNA in serum using the TaqMan fluorogenic detection system.
Hepatology, 32(3):626-629, 01 Sep 2000
Cited by: 100 articles | PMID: 10960459