Abstract
Free full text

A Subset of Tumor-Derived Mutant Forms of p53 Down-Regulate p63 and p73 through a Direct Interaction with the p53 Core Domain
Abstract
The p53 protein is related by sequence homology and function to the products of two other genes, p63 and p73, that each encode several isoforms. We and others have discovered previously that certain tumor-derived mutants of p53 can associate and inhibit transcriptional activation by the α and β isoforms of p73. In this study we have extended these observations to show that in transfected cells a number of mutant p53 proteins could bind and down-regulate several isoforms not only of p73 (p73α, -β, -γ, and -δ) but also of p63 (p63α and -γ; ΔNp63α and -γ). Moreover, a correlation existed between the efficiency of p53 binding and the inhibition of p63 or p73 function. We also found that wild-type p63 and p73 interact efficiently with each other when coexpressed in mammalian cells. The interaction between p53 mutants and p63 or p73 was confirmed in a physiological setting by examining tumor cell lines that endogenously express these proteins. We also demonstrated that purified p53 and p73 proteins interact directly and that the p53 core domain, but not the tetramerization domain, mediates this interaction. Using a monoclonal antibody (PAb240) that recognizes an epitope within the core domain of a subset of p53 mutants, we found a correlation between the ability of p53 proteins to be immunoprecipitated by this antibody and their ability to interact with p73 or p63 in vitro and in transfected cells. Based on these results and those of others, we propose that interactions between the members of the p53 family are likely to be widespread and may account in some cases for the ability of tumor-derived p53 mutants to promote tumorigenesis.
The p53 family consists of three distinct genes (p53, p63, and p73) that share significant homology and whose products can function as sequence-specific transcriptional activators (29, 31, 47, 51, 52, 61). Proteins encoded by these three genes also share the same modular organization which consists of an amino-terminal transactivation domain, a central sequence-specific DNA binding domain, and a C-terminal tetramerization domain. Both p63 and p73 genes, however, encode multiple isoforms that vary in their N and C termini. In some cases the use of a cryptic promoter generates potentially transcriptionally inert isoforms that lack the transactivation domain located in the N terminus (ΔN-) of p73 and p63 proteins (ΔNp73α, ΔNp63α, ΔNp63β, and ΔNp63γ). Both p63 and p73 genes also generate several forms with various C-terminal extensions (p73α, -β, -γ, -δ, and - , and p63α, -β, and -γ) that are produced by alternative splicing (7, 13).
, and p63α, -β, and -γ) that are produced by alternative splicing (7, 13).
At a functional level, it has been shown that ectopic expression of p73 and p63 can transactivate endogenous targets of p53, such as the cell cycle inhibitor p21 (29, 31, 49), as well as p21-promoter-containing reporters (29, 49, 61). We have found that p73 proteins can activate other p53 target promoters (14) such as the proapoptotic genes Bax (41), IGF-BP3 (4), and cyclin G (45). However, in a more physiological context Zhu et al. have found significant differences in the abilities of induced p53 and p73 proteins to activate several targets (69). One of the cellular functions of p53 is to induce apoptosis in response to genotoxic stress, such as damaged DNA (reviewed in references 20, 32, and 35). Similarly, it has been found that overexpression of both p73 and p63 can inhibit cell growth by inducing apoptosis (29, 47, 61, 69).
Despite the studies mentioned above, it is still not fully understood whether and when p63 or p73 causes cells to arrest growth or to undergo apoptosis. In contrast to the more ubiquitous expression of p53, p63 and p73 have restricted tissue expression patterns (47, 51, 61), which suggests that p63 and p73 may have a role in the development of specific tissues. Results obtained from transgenic knockout mice support this assumption. Transgenic p73−/− mice harbor developmental problems in their nervous and immune systems (63) and p63−/− mice present severe defects in skin and limb development (62). The role of p63 in limb formation is conserved, since mutations in human p63 have been associated with hand and foot developmental malformations (8, 25). The homology between p53 and its relatives suggests also that p63 and p73 might have a role in cellular stress response. Recently, it has been shown that p73 is activated upon DNA-damaging treatments, such as cisplatin or γ-radiation, through a c-abl-dependent pathway (1, 19, 59, 66). In contast to the better-studied p53, however, it is still unclear what the involvement of p63 or p73 is in tumorigenesis. One of the initial studies reported that the p73 gene is located in a region of chromosome 1p36.1 that is frequently lost during neuroblastoma formation. Multiple studies have since assessed the status of p63 and p73 genes in different tumors in terms of mutation or loss of heterozygosity, in some cases reaching contradictory conclusions. Several studies have described a frequent loss of heterozygosity in neuroblastoma (15, 23, 26, 33), gastric cancer (23, 65), ovarian cancer (42), and lung cancer (43). However, only three missense point p73 mutations (P405R, P425L, and R269Q) have been found among almost 1,000 tumors screened. Similarly, only a few mutations have been found in p63. In fact, multiple studies now show that in neuroblastoma (33), colorectal cancer (56), breast cancer (67), bladder cancer (64), and hepatocellular carcinoma (57), there is an overexpression of what is likely to be wild-type p73. While there may be an apparent inconsistency in the results described above, the fact that the mouse p73 gene generates ΔN isoforms that lack the transactivation domain and potentially exert a dominant negative effect on p53 may explain how overexpression could affect p53-mediated tumor suppression (63). Indeed, a p73 variant that lacks the transactivation domain has been identified in neuroblastoma (7). More recently, overexpression of the ΔNp63 isoforms has also been observed in bladder carcinomas (48), nasopharyngeal carcinomas (11), and squamous-cell carcinomas of the head and neck (44, 60).
The percent identity between the tetramerization domains of p53, p63, and p73 initially suggested the possibility that these proteins may form heterotetramers, and Kaghad et al. (31) reported that p73β but not p73α can interact modestly with p53 in a yeast two-hybrid assay. We previously showed that two p53 tumor-derived mutants, R175H and R248W, were able to interact with p73α. More recently, Marin et al. (37) reported interactions between mutant forms of p53 and p73α and -β that were at least partially dependent on the presence of a polymorphism (arginine [R] versus proline [P]) on p53 at amino acid 72, in which R72 favors binding to p73. These various studies did not address the question of what part of these proteins is involved in their heterotypic associations. Davison et al., using purified oligomerization domains of p53, p63, and p73, failed to find any interaction between this region of p53 with its homologues and only weak binding between p63 and p73 oligomerization domains (12). A more recent study has described the ability of two p53 mutants to interact with different p73 isoforms and has shown that various deleted forms of p53 can interact with p73, leading to the suggestion that the p53 core domain is involved (55).
Given the potential importance of interactions between members of the p53 family, we have extended our initial observations to determine which isoforms of p63 and p73 interact with p53 and, using purified proteins, to gain insight into the characteristics of p53 that govern these interactions.
MATERIALS AND METHODS
Cell culture.
H1299, HI293, and HT29 cells were obtained from the American Type Culture Collection. HACAT cells were a gift from P. Hall. Cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) in the presence of a mixture of 5% CO2 and 95% air at 37°C. SF9 insect cells were grown in TC100 medium (GIBCO BRL) at 30°C. A H1299 cell line expressing tetracycline-regulated mutant R175H (9) was cultured in the presence of puromycin (2 μg/ml), neomycin (500 μg/ml), and tetracycline (5 μg/ml).
Expression vectors.
pC53-SN3, pC53-175, pC53-220, pC53-249, pC53-277, and pC53-283 express full-length p53, p53R175H, p53Y220C, p53R249S, p53C277Y, and p53R283H cDNA, respectively, from the CMV promoter (16). PC53-HA expresses full-length HA-p53 cDNA from the CMV promoter in pCDNA3. Expression of human HA-p73α, HA-p73α R192H, HA-p73β, HA-p73γ, HA-p73δ, Myc-p63α, Myc-p63γ, Myc-ΔNp63α, and Myc-ΔNp63γ cDNA from the CMV promoter was as described previously (13, 61). PBacp73α and pBacp73β were obtained by subcloning a HindIII/XhoI fragment of the CMVp73 expression constructs into the FastBac vector (GIBCO BRL, Gaithersburg, Md.).
Expression and purification of the p53 and p73 proteins.
Infection, expression, and purification of p53 proteins from insect cells were done as previously described (28). Baculoviruses that expressed mutant p53 143A, 175H, 248W, or R283; the p53 core domain (amino acids 96 to 312); or p53 lacking the N terminus (Δ96), C terminus (Δ30), or oligomerization domain (ΔDD; deletion from amino acids 334 to 356) were used to infect and purify p53 proteins (27, 28). The bacterially expressed and purified core domain of p53 R248W was a kind gift from N. Pavletich. Expression and purification of p53 R248W from bacteria were performed as described previously (53). Preparation and amplification of the p73 baculoviruses using the FastBac system were done as indicated by the manufacturer (GIBCO-BRL). Purification of p73 proteins was performed as described previously for p53. SF9 cells freshly seeded (1 h) at 90% confluence in 20-cm plates (between 20 and 40 plates) were infected for 1 h with 500 μl of virus diluted in 2 ml of medium for each plate. After 48 h, cells were removed from the plates in their medium and washed two times in phosphate-buffered saline. After the second wash, cells were lysed for 30 min in buffer A (50 mM Tris-HCl [pH 8], 0.5% NP-40, 150 mM NaCl, 1 mM dithiothreitol [DTT], 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% aprotinin). The soluble fraction was isolated by centrifugation at 4°C for 30 min at 20,000 rpm in a Sorvall RC-5B refrigerated centrifuge and added to protein A-Sepharose beads (1 to 2 ml) cross-linked to hemagglutinin (HA) monoclonal antibody (MAb) 12CA5 which were rocked overnight at 4°C. Beads were then washed two times in buffer B (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.5% NP-40, 100 mM NaCl, 1 mM DTT, 10% glycerol, 0.5 mM PMSF) and two times in buffer C (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl, 1 mM DTT, 10% glycerol, 0.5 mM PMSF) and then poured into a 5-ml syringe. Beads were washed with three column volumes of buffer D (20 mM Tris-HCl, 1 mM EDTA, 10% glycerol, 250 mM NaCl, 1 mM DTT), and p73 proteins were eluted with an HA peptide (10 μg/ml final concentration) in an elution buffer containing 20 mM Tris-HCl, 1 mM EDTA, 10% glycerol, 500 mM NaCl, and 1 mM DTT. Fractions containing p73 were pooled according to their similarities in concentration and purity.
Reporter vectors.
p21min-luc contains a duplex oligonucleotide encoding the p53-responsive cis-acting element from p21, cloned upstream of the minimal c-fos promoter (positions −53 to +42) in pGL3-OFLUC (kindly provided by N. Clarke). The synthesized oligonucleotides (Operon Technologies, Inc.) encoding the p21 cis-acting p53-responsive element were as follows: 5′ GATCCTCGAGGAACATGTCCCAACATGTTGCTCGAG 3′ and 5′ GATCCTCGAGCAACATGTTGGGACATGTTCCTCGAG 3′. The resulting plasmid was sequenced for the orientation and insert number of the oligoduplex.
Transfection and luciferase assays.
H1299 cells (American Type Culture Collection) were maintained in DMEM supplemented with 10% FBS in 5% CO2 at 37°C. Cells were transfected by a lipopolyamine-based (Transfectam) protocol, as described previously (19). Briefly, cells were grown in a solution of DMEM and 10% FBS and transfected with various amounts of DNA. The precipitate was left on the cells for 6 h, after which time fresh DMEM and 10% FBS were added for the periods indicated. For luciferase assays, cells were seeded in 12-well, 3.8-cm2 plates and transfected with one of the expression vectors (200 ng of each) and two different reporter constructs (250 ng of each), a CMV-expressed luciferase cDNA from renilla and a p53-responsive luciferase cDNA from the firefly. Luciferase activity was measured in each well 24 h later by a Dual-Luciferase Reporter Gene assay (Promega).
Preparation of whole-cell extracts and immunoprecipitation analysis.
H1299 cells in 6-cm plates were transfected with the indicated plasmids (8 μg) and harvested 48 h later. SF9 cells were infected with 50 μl of virus in six-well plates. Cells were lysed in 300 μl of lysis buffer (20 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5% NP-40, 150 mM NaCl, 1 mM DTT, 10% glycerol, and protease inhibitor) and the extracts were centrifuged at 13,000 rpm in a Heraeus Biofuge centrifuge at 4°C for 12 min to remove cell debris. Protein concentrations were determined using the colorimetric assay (Bio-Rad Laboratories, Richmond, Calif.). p53 proteins (400 to 750 μg of whole-cell extract) were incubated with 100 μg of p53 MAbs (MAb240, MAb1801, and DO11) and p73 proteins with anti-HA MAb 16B12 (BAbCo; 1 mg/ml; final dilution, 1/150) followed by rocking at 4°C for 1 h. After incubation, 25 μl of protein G-Sepharose beads (Pharmacia; 50% slurry) was added, and the samples were rocked at 4°C for 1 h and then washed four times with 1 ml of wash buffer (20 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM DTT, 10% glycerol). Excess liquid was aspirated, and 35 μl of 2× sample buffer (54) was added. Samples were then heated to 95°C for 10 min and centrifuged for 3 min at 13,000 rpm in a Heraeus Biofuge centrifuge at 4°C followed by electrophoresis through a 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel. Protein gels were transferred to nitrocellulose membranes (Schleicher & Schuell, Inc.). For p53 detection, a mixture of p53 MAb-containing supernatants (MAb421, MAb1801, and MAb240) was used, with each supernatant at a 1/4 dilution; for HA-p73 and HA-p53 detection (Fig. (Fig.1C),1C), the 16B12 MAb (BAbCo; 1 mg/ml) was used at a dilution of 1/1,000. Myc-p63 proteins were detected with a myc epitope MAb (Zymed Laboratories; 1/1,000 dilution). HSC70 was immunoprecipitated and detected using an MAb (W27 [Santa Cruz]; immunoprecipitation, 1/100; Western, 1/1,000). Proteins were visualized with an enhanced chemiluminescence detection system (Amersham).

p53 mutants interact with p73 and p63 isoforms in mammalian cells. Wild-type p53 (p53 wt) or mutant p53 R175H (p53 175) were coexpressed in H1299 cells along with p73 isoforms (α, β, γ, or δ) (panel A) or p63 isoforms p63α and ΔNp63α (panel B), or p63γ and ΔNp63γ (panel C). After 48 h, cells were lysed and p53 proteins were immunoprecipitated with a p53 MAb (PAb1801), as described in Materials and Methods. The immune complexes were then run on a 10% SDS-PAGE gel, transferred to nitrocellulose, and subjected to Western blotting. HA-tagged p73, Myc-tagged p63, and p53 proteins, respectively, were detected with an HA epitope MAb (HA11, BAbCo; 1/1,000 dilution), a Myc MAb (Zymed Laboratories; 1/1,000 dilution) and a mixture of p53 MAbs (MAb1801, MAb421, and DO11; hybridoma supernatant, 1/5 dilution). A secondary anti-mouse antibody recognizing only the λ light chain of the immunoglobulin (R8-140, PharMingen; 1/1,000 dilution) was used to visualize p53 proteins. Ten percent of the extract (±75 μg) used in the immunoprecipitations was loaded in the right-hand panels, as indicated in the figure. IP, immunoprecipitated.
In vitro immunoprecipitations.
Interactions with purified proteins were assessed in a buffer containing 2 μg of bovine serum albumin (BSA)/μl, 20 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5% NP-40, 250 mM NaCl, 1 mM DTT, 10% glycerol, and protease inhibitor. Antibodies and protein G-Sepharose were preincubated in the same buffer for 30 min before use, and proteins were added to the bead-antibody mixture and incubated at 4°C for 1 h. The samples were washed four times with 1 ml of wash buffer (20 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM DTT, 10% glycerol), the excess liquid was aspirated, and 30 μl of 2× sample buffer (54) was added. Samples were heated to 95°C for 5 min, centrifuged for 3 min at 13,000 rpm, and electrophoresed through a 10% SDS-polyacrylamide gel. An anti-p53 MAb recognizing the core domain was used to immunoprecipitate the core domains of p53 (DO11, Zymed Laboratories; 1/150 dilution).
RESULTS
A p53 mutant interacts with multiple p73 and p63 isoforms.
To extend previous studies by this group and others, we first examined the ability of wild-type p53 and a common hot-spot p53 mutant, R175H, to associate with a number of p73 isoforms after transfection into H1299 cells, specifically p73α, p73β, p73γ, and p73δ. As shown in Fig. Fig.1A,1A, these four p73 isoforms could be coimmunoprecipitated with mutant p53 but not with wild-type p53 when similar levels of both forms of p53 were expressed. To further expand on this observation, we then examined whether either wild-type or mutant p53 could associate with several p63 isoforms (p63α and -γ or ΔNp63α and -γ) in H1299 cells. Similarly to what had been observed with p73, p53 (R175H) coimmunoprecipitated with all p63 isoforms (Fig. (Fig.1B1B and C). Wild-type p53 did not coprecipitate detectably with p63 (Fig. (Fig.1B1B and C) or precipitated rather weakly with p63γ (data not shown). In contrast, p53 (R175H) bound more efficiently to p63α and -γ isoforms than to the corresponding ΔNp63 isoforms. Comparing the quantities of proteins obtained after immunoprecipitation to the corresponding protein in 10% of total cell extract (Fig. (Fig.1B1B and C, right lanes), it appeared that p53 R175H binds similarly to p73α and to p63α and -γ (between 10 to 30% of estimated total immunoreactive p63 protein), although its affinity for the ΔNp63 isoforms was significantly reduced (less than 2% of the total immunoreactive p63 protein).
P73 and p63 interact in H1299 cells.
Whereas wild-type p53 protein interacts poorly if at all with wild-type p73 (14) (Fig. (Fig.1A)1A) or p63 proteins (Fig. (Fig.1B1B and C), we were interested in determining whether p63 and p73 proteins can interact in mammalian cells. After transfection of p73 and p63 expression vectors into H1299 cells, we were able to detect an interaction between wild-type p73 and wild-type p63α or ΔNp63α isoforms (Fig. (Fig.2,2, left lanes). P63 interacted more efficiently with p73α mutated at codon 292 (R292H). When comparing the amount of protein present in 10% of total cell extract (Fig. (Fig.2,2, right lanes), we estimate that 5 to 20% of p63 proteins are found in a complex with wild-type p73, and between 10 and 40% are found in a complex with p73 R292H. Under our conditions, coexpression of these proteins did not affect their protein levels (Fig. (Fig.2,2, right lanes).
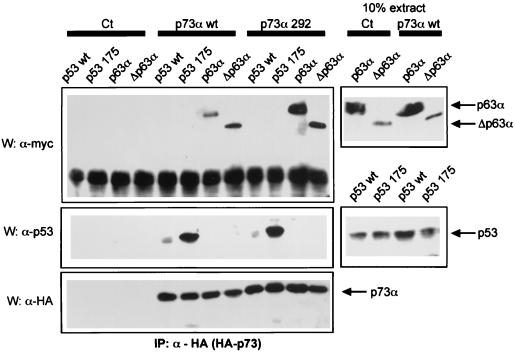
p63 and p73 interact in mammalian cells. Wild-type p73α (p73α wt) or mutant p73α R292H (p73α 292) were coexpressed in H1299 cells along with p53 proteins (wild-type or mutant R175H) or p63 isoforms (p63α and ΔNp63α). After 48 h, cells were lysed and p73α proteins were immunoprecipitated with an HA epitope MAb (HA11, Babco; 1/150 dilution). The immune complexes were then resolved on a 10% SDS–PAGE gel, transferred to nitrocellulose, and subjected to Western blotting. HA-tagged p73 and myc-tagged p63 and p53 proteins were then detected as described in Fig. Fig.1.1. Right panels show 10% of the extract (~75 μg) used for the immunoprecipitation. IP, immunoprecipitated.
Based on these results, we propose that interactions occur between p73 and p63 but that wild-type p53 does not associate with p63 or p73 proteins. Since mutation of some residues in the core domain of p53 facilitates interaction with either p63 or p73, this suggests that characteristics of mutant p53 distinct from the oligomerization domain are involved in its interactions with p63 or p73 proteins.
Interactions between endogenously expressed mutant p53, p63, and p73 proteins.
To assess the biological relevance of the association between mutant forms of p53 and p53 family members, we examined whether the complexes could be seen in cells that express endogenous p53, p63, and p73 proteins. We used two cell lines expressing p73 and either wild-type p53 (293 cells) or mutant forms of p53 (mutated at position 273 in HT29 cells; mutated at position 179 in HACAT cells). HACAT cells also express p63 proteins (22). We also used H1299 cells that express endogenous p73 proteins and tetracycline-regulated mutant p53 (R175H) (9). Proteins were immunoprecipitated using either a p73 MAb (MAb2; PharMingen), a p53 MAb (PAb1801), or as a control, a MAb directed against simian virus 40 T antigen (PAb419). In H1299 p53-175 cells, HT29 cells, and HACAT cells, p53 mutants and p73 were coimmunoprecipitated (Fig. (Fig.3A3A and B, left lanes). In contrast, no significant coimmunoprecipitation between wild-type p53 and p73 in 293 cells could be demonstrated. We were also able to detect p63 proteins in p53 immunoprecipitates from extracts of HACAT cells (Fig. (Fig.3B,3B, right lanes). Thus, consistent with results from transfected cells, in two cell lines endogenously expressed mutant p53 protein bound to p73 or p63, while in a third cell line wild-type p53 protein interacted extremely poorly with p73.
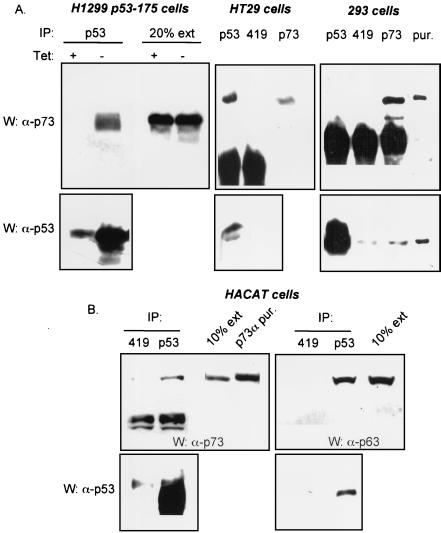
Interaction between endogenously expressed mutant p53, p63 and p73. (A) Extracts (1.5 mg) of H1299 cells expressing a tetracycline-regulated p53 R175H (left panel), HT29 cells (middle panel), or 293 cells (right panel), were immunoprecipitated with a p53 MAb (1801) or a p73 MAb (ER15, Oncogene; 1/100 dilution). The immune complexes were then resolved on a 10% SDS–PAGE gel, transferred to nitrocellulose, and subjected to Western blotting. As a control, a MAb recognizing the simian virus 40 large T antigen (419) was used. p73 and p53 proteins were then detected with a p73 polyclonal antibody and a mixture of p53 MAbs (PAb1801, PAb421, and PAbDO11, hybridoma supernatants 1/5), respectively. The secondary anti-mouse antibody used to detect p53 proteins recognizes only the immunoglobulin λ light chain. The left panel shows 20% of the H1299 p53-175 cell extract used for the immunoprecipitation. In the right panel, purified p73α protein (50 ng) and purified p53 protein (50 ng) were loaded to indicate the position of the proteins (pur). All three cell lines expressed various detectable levels of p73 proteins. HT29 cells express endogenous p53 mutant (273H) and 293 cells express wild-type p53. (B) p53 proteins present in HACAT cells were immunoprecipitated as described in panel A. p63 was detected with a p63 polyclonal antibody (gift of P. Hall). p73 and p53 proteins were detected as described in panel A. 10% of the HACAT cell extract used for the immunoprecipitation was loaded to indicate the relative amount of protein found in complex with p53. In the left panel, 50 ng of purified p73α protein (pur) was loaded as a marker for the position of the proteins. HACAT cells express endogenous p53 mutant (179Y). IP, immunoprecipitated.
The ability of p53 mutants to interact with p73 or p63 correlates with their ability to inhibit p73 or p63 transcriptional activity.
We previously showed that two tumor-derived mutant forms of p53 that bind to p73, specifically R175H and R248W, can inhibit the ability of p73 to transactivate reporter genes containing p53 response elements. We wished to assess whether binding to p73 by mutant p53 correlates with down-regulation of p73. To approach this problem, p73α or p73β was cotransfected into H1299 cells along with a number of p53 tumor-derived mutants (Y220C, R249S, C277Y, and R283H, as well as R175H) to determine their relative abilities to bind to and down-regulate p73. P53 was immunoprecipitated using a p53 MAb (PAb 1801) (Fig. (Fig.4A,4A, left lower lanes), and the presence of HA-tagged p73 in the complex was detected by Western blotting (Fig. (Fig.4A,4A, left upper lanes). P73α interacted with three of these p53 tumor-derived mutants, specifically Y220C, R249S, and R283H, although marked variations in efficiency were observed. Y220C interacted the most efficiently and to an extent similar to that of R175H, while R249S and R283H displayed a much weaker affinity for p73α. Comparable interactions occurred between the same p53 mutants and p73β. p53 mutated at position 277 (C277Y) did not coimmunoprecipitate with p73α or p73β. Note that expression of p53 mutants did not vary significantly, and coexpression of p73 with p53 mutants did not affect their expression levels (Fig. (Fig.4,4, right lanes). Comparing the quantities of proteins obtained after immunoprecipitation to the corresponding protein in 5% of total cell extract (Fig. (Fig.4A,4A, right lanes), it appeared that p53 R175H and Y220C bind similarly to p73α (between 5 and 30% of estimated total immunoreactive p73 protein, depending on the experiment).
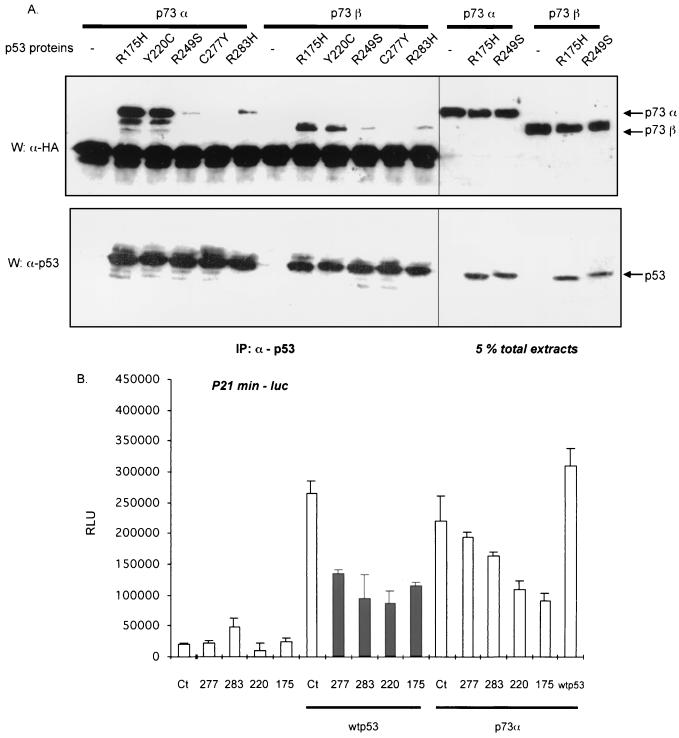
Ability of p53 mutants to interact with p73 correlates with their ability to inhibit p73 transcriptional activity. (A) p73α or p73β was coexpressed in H1299 cells along with various p53 mutants (R175H, Y220C, R249S, C277Y, or R283H). Transfection, immunoprecipitation, and detection of p53 and p73 proteins were performed as described in Fig. Fig.1.1. The lanes on the right show 5% of the extract (~40 μg) used in the immunoprecipitations. (B) Indicated combination of p53 (wild-type, wt; C277Y, R283H, 283; Y220C, 220; R175H, 175) and p73 (p73α) expressing vectors were cotransfected into H1299 cells along with a p53-target reporter gene (p21min-luc). Cells in 12-well plates were transfected with 200 ng of each expression vector and reporter genes. After 12 h, fresh medium was added, and 24 h later cells were harvested. Diagrams represent means in relative luminescence units (RLU) and bars are standard deviations. Each diagram shows a representative experiment out of three, each done in triplicate. IP, immunoprecipitated.
To assess whether coimmunoprecipitation correlates with the ability of p53 mutants to repress transcriptional activation by p73, we cotransfected a p53 target reporter gene containing the p53 binding site from the p21 gene upstream of the minimal c-fos promoter (p21min-luc), along with the p53 and p73 expression vectors. As expected, the four p53 mutants by themselves had little or no effect on the reporter gene activity (Fig. (Fig.4B)4B) but all efficiently inhibited wild-type p53 activity. However, the ability of these mutants to down-regulate p73-dependent transcription varied. Two p53 mutants, R175H and Y220C, inhibited p73 activity by more than 50%; the p53 mutant R283H reduced p53 transactivation by ~20%; and C277Y had no significant effect. Thus, there is a correlation between the ability of p53 tumor-derived mutants to inhibit p73 functions and their capacity to interact with p73. While the correlation was not quantitatively proportional to the extent of binding, it may well be that the amount of p73 found in p53 immunoprecipitates was an underrepresentation of the amount of the two proteins in complex, because the conditions of immunoprecipitation and washing of immunoprecipitates may well have led to a loss of associated proteins.
As shown above, the p53 mutant (R175H) is also able to interact with p63 proteins, and so we tested the same set of mutant p53 proteins for their ability to bind to and affect transactivation by p63. Using an approach similar to that described above, we found that p63γ interacted well with p53 R175H, p53 Y220C, and p53 R248W (Fig. (Fig.5A).5A). Similar results were obtained with p63α (see Fig. Fig.5A).5A). P53 mutants R249S and R283H displayed a lesser affinity for p63, and no interaction with either wild-type p53 or mutant p53 C277Y was detected. Again, under these experimental conditions similar levels of p53 or p63 proteins were expressed (Fig. (Fig.5A,5A, right and bottom lanes). We estimate that the level of interaction between p63 proteins and p53 mutants is about 10%.
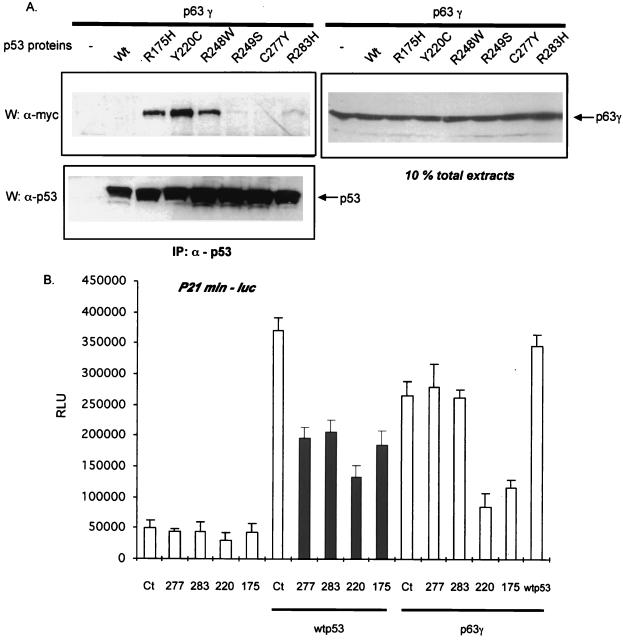
Ability of p53 mutants to interact with p63 correlates with their ability to inhibit p63 transcriptional activity. (A) p63γ was coexpressed in H1299 cells along with various p53 mutants (R175H, Y220C, R249S, C277Y, or R283H). Transfection, immunoprecipitation, and detection of p53 and p63 proteins were performed as described in Fig. Fig.1.1. The right lanes show 10% of the extract (~75 μg) used for the immunoprecipitations. (B) Indicated combination of p53 (wild-type, wt; C277Y, 277; R283H, 283; Y220C, 220; R175H, 175) and p63 (p63γ) expressing vectors were cotransfected into H1299 cells along with a p53-target reporter gene (p21min-luc). Transfection and luciferase assays were performed as in Fig. Fig.4.4. Diagrams represent means in RLU, and bars are standard deviations. Each diagram shows a representative experiment out of three, each done in triplicate. IP, immunoprecipitated.
To determine whether p53 mutants can also block p63 function, as before, we coexpressed p53 mutants and p63γ in H1299 cells and tested the transcriptional activity of p63γ on a p53 target reporter gene (p21minluc). As was the case with p73, we observed that those p53 mutants that bind p63 efficiently reduced the activity of p63 on this reporter gene (Fig. (Fig.5B).5B). Thus, not only is there a correlation between the ability of a class of tumor-derived mutants to bind p63 and to inhibit p63 function, but the same mutants repress (or do not repress) both p63 and p73.
Interaction between p53 and p73 in insect cells does not require the p53 oligomerization domain.
To gain further insight into possible cross talk between the p53 family members, we studied interactions between purified proteins. We first checked whether the binding between p73 and p53 mutants could occur in insect cells. SF9 cells were coinfected with baculoviruses expressing p53 variants and p73α, and then p53 proteins were immunoprecipitated with a p53 MAb (DO11) followed by assessment of the amount of p73 in immunoprecipitates by Western blotting using an anti-HA antibody (Fig. (Fig.6).6). Confirming the data obtained in mammalian cells, we observed that p73 and p53 (R175H) were able to interact in insect cells, while virtually no interaction between wild-type p53 and p73 was observed. p53 variants lacking the oligomerization domain (ΔDDp53), the 96 N-terminal amino acids (Δ96), or the C-terminal 30 amino acids (Δ30) also tested in this way showed that the interaction between p53 and p73 did not require the oligomerization domain or either the N or C terminus of p53. These results were surprising, however, because all of these p53 proteins contain a wild-type central core domain. This finding suggested that deletion of several regions of p53 could alter the conformation of this protein in a manner that favors its interaction with p73 protein. This altered conformation may have features in common with tumor-derived mutant forms of p53 that are recognized by p63 and p73.
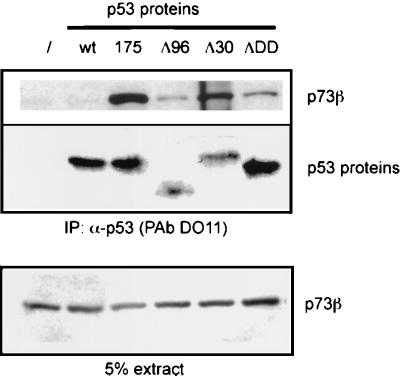
Interaction between p53 and p73 in insect cells does not require the oligomerization domain. SF9 insect cells were coinfected with baculoviruses expressing p73β along with wild-type p53 or p53 variants (R175H; Δ96, deletion of the first 96 amino acids; Δ30, deletion of the last amino acids; ΔDD, deletion of the oligomerization domain). After 48 h, cells were lysed, p53 proteins were immunoprecipitated, and proteins were detected as described in Fig. Fig.1.1. In the bottom panel, 5% of the extract (~20 μg) used for the immunoprecipitation was loaded.
Purified p73 and p53 proteins interact directly in vitro.
Since our previous experiments studying the association between p53 and its relatives used either mammalian or insect cell extracts, they did not reveal whether the interactions detected were direct or not. We therefore immunopurified p73 and p53 proteins and found that p73α and p73β were able to bind directly and efficiently with the p53 mutants tested (Fig. (Fig.7A).7A). It was unanticipated that purified wild-type p53 would be able to interact as strongly with p73 as mutant proteins in this assay. To understand the discrepancy between the results with wild-type p53 in cell extracts or other purification, we considered the possibility that purified wild-type p53 preparations may contain partially unfolded or conformationally altered protein such that they now have features in common with mutant p53.

Purified p73 and p53 proteins interact directly in vitro. (A) In vitro coimmunoprecipitation of purified p53 proteins (143, 175, 248, and 273) and p73β. Proteins (~25 ng each) were mixed in 100 μl of a binding buffer containing BSA (2 μg/μl) and p53 proteins were immunoprecipitated with a p53 MAb (1801) as described in Materials and Methods. Proteins were detected as described in Fig. Fig.1.1. On the right side of the panel, 10% of the input used for the immunoprecipitation was loaded (p73β input). (B) Comparative immunoprecipitation of purified p53 proteins (wild type and R175H) by three different antibodies (PAb240, -421, or -1801). Beads alone were used as an additional control. Detection of p53 was performed as described in Fig. Fig.1.1. In the right lane, 20% of the input used for the immunoprecipitations is shown. (C) Comparative immunoprecipitation of H1299-expressed p53 (wt or R175H) by PAb1801 or PAb240. p53 expression vectors were transfected in H1299. After 48 h, cells were lysed and p53 proteins were immunoprecipitated with PAb240, PAb421 or the beads alone. Proteins were then detected as described in Fig. Fig.1.1. In the right panel, 20% of the extract used in the immunoprecipitations is shown. (D) Purified p53 wild-type proteins were first immunodepleted with PAb240 (Pre-cl.), then incubated with purified p73β, and finally immunoprecipitated with PAb1801. The immune complexes were separated into two aliquots, resolved by SDS-PAGE, transferred to nitrocellulose, and subjected to Western blotting, as described in Fig. Fig.1.1. Half of the blot was used to detect p73 proteins and half was used to detect immunoprecipitated p53 proteins. p53 proteins levels were compensated prior to immunodepletion in order to keep a equivalent amount of p53 protein in each condition. As an additional control, beads alone were use to preclear the purified p53 proteins. IP, immunoprecipitated.
To study this possibility, we employed an MAb, PAb240, that recognizes an epitope within the core DNA binding domain and has been used extensively to assess mutant p53 conformation (2, 3, 18). Wild-type p53 or R175H p53 proteins were compared for their ability to be immunoprecipitated with PAb240 when purified from insect cells or when present in extracts of H1299 cells that had been transfected with p53 expression vectors (Fig. (Fig.7B7B and C). It is interesting that that a fraction of the purified wild-type p53 protein could be immunoprecipitated with PAb240 (Fig (Fig7B),7B), and in fact the amount of PAb240-reactive wild-type p53 was comparable in relative proportions to the amount of purified p53 (R175H) that could be recognized by this antibody. In contrast, in transfected H1299 cells, while a significant quantity of R175H p53 was immunoprecipitated by PAb240, virtually no wild-type p53 protein was recognized by the same antibody (Fig. (Fig.7C).7C). These results, which showed that a portion of our purified preparations of wild-type p53 is in a mutant conformation, suggested that this PAb240-reactive fraction may be the same as that which interacted with p73 in vitro. To support this hypothesis, we immunodepleted the PAb240-reactive portion of the wild-type p53 preparation and then assessed whether the remainder would be able to interact with p73. Indeed, even after compensating for p53 protein levels, we observed a marked reduction in the ability of PAb240-depleted wild-type p53 to interact with p73 (Fig. (Fig.7D).7D). These results strongly indicate that the portion of the wild-type p53 fraction that interacts with p73 is in a mutant conformation.
The core domain of p53 mediates its interaction with p73 proteins.
The data described above suggest that the core domain in p53 is responsible for interaction with p73 proteins. To confirm this hypothesis, we performed in vitro immunoprecipitations using purified p73 proteins and various truncations of purified p53 proteins (Fig. (Fig.8A).8A). As observed in insect cells, the oligomerization domain and the last 30 C-terminal amino acids are not necessary for the interaction (Fig. (Fig.8B).8B). It was most relevant that the p53 core domain was sufficient to allow the interaction with p73 proteins. By itself, the C terminus containing the oligomerization domain was not able to interact with p73 proteins. We extended these results by using a core domain mutated at position 248 (R248W) purified from bacteria (Fig. (Fig.8D).8D). p53 proteins were immunoprecipitated with the DO11 p53 MAb, and p73 proteins were detected by Western blotting. As observed with baculovirus-expressed and purified proteins, bacterially expressed full-length or core domain p53 proteins interact directly with p73. We extended these results by showing that expression of the mutated p53 (R175H) core domain interacts with p73β when coexpressed in H1299 cells (data not shown). However, the very low expression level of this construct prevented us from demonstrating that the core domain is sufficient to inhibit p73 transactivation function. Bullock et al. showed that the isolated core domain is more susceptible to unfolding than is intact p53, which may account for its instability in vivo (5).
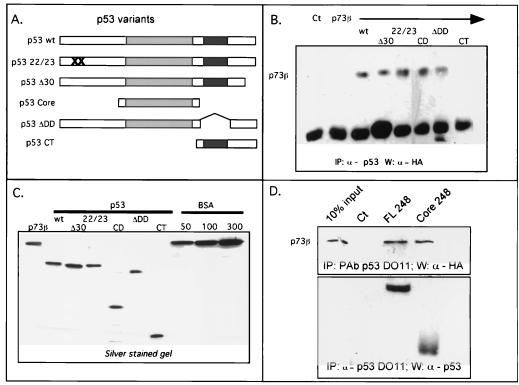
Core domain of p53 mediates the interaction with p73. (A) Schematic representation of p53 variants purified from baculovirus-infected cells: p53 wt is the full-length p53 protein; p53 22/23 harbors a double mutation, L22Q and W23S (marked by two crosses), in the MDM2 recognition region; p53 Δ30 lacks the 30 last C-terminal amino acids; p53 Core has the amino acids from 96 to 312, lacking both N and C termini; p53 ΔDD lacks the oligomerization domain (amino acids 334 to 356); p53 CT has the amino acids from 312 to 396 and contains the oligomerization domain and the extreme C terminus. The shaded and the black boxes indicate the p53 core and oligomerization domains, respectively. (B) In vitro coimmunoprecipitation between p73 and p53 variants. p53 proteins were immunoprecipitated with a mixture of p53 MAbs that recognize the core domain (PAb1620, PAb240), the N terminus (PAb1801) or the C terminus (PAb421). Detection of p53 and p73 proteins by Western blot were performed as described in Fig. Fig.7A.7A. Approximately 50 ng of purified p73β and 50 ng of purified p53 variants were used. (C) Silver-stained gel showing purified p73β and p53 variants used in panel B. BSA protein levels with concentrations in nanograms are shown on the right, as indicated. (D) In vitro coimmunoprecipitation of p73β with full-length p53 R248W or a mutated p53 core domain (mutation R248W). Immunoprecipitation and detection of p73 and p53 proteins were performed as described in panel B. IP, immunoprecipitated.
p53 mutants strongly interacting with p73 or p63 are reactive with PAb240.
Our data showed that the core domain of p53 is necessary and sufficient for its interaction with p73. To further understand characteristics of mutant p53 that affect its interaction with p53, we returned to our observation that PAb240-reactive purified p53 binds to p73. In fact it has been reported that several tumor-derived mutations are located at residues that make direct contact with the DNA, while others affect the overall structure of the core domain (10). It has also been postulated that the PAb240 antibody recognizes the conformational but not the contact class of mutants (10, 18, 34, 58). Based on this supposition, we tested whether there is a correlation between the ability of p53 mutants to interact with p73 or p63 and their ability to be recognized by PAb240.
H1299 cells were transfected with p73α or p63α and various p53 mutants. Half of the extract was used for immunoprecipitation with PAb240 and half with PAb1801. In each case immune complexes were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose, and the presence of p73 or p63 in the complex was assessed by Western blotting. Confirming again the experiments shown in Fig. Fig.44 and and5,5, immunoprecipitation with PAb1801 showed that p73 associated most efficiently with R175H, Y220C, and R248W (Fig. (Fig.9A9A and B, upper lanes). P73 was found in PAb240 immunoprecipitates and most prominently with these same three p53 mutants, although a small amount of R249S was also coprecipitated. It is important that Western blot analysis of p53 proteins revealed that, although differing in extent of interaction (the mutant R248W is less recognized than R175H and Y220C), these three mutants are the ones that are detectably immunoprecipitated by PAb240 (Fig. (Fig.9A9A and B, bottom left lanes). As shown in Fig. Fig.99 (bottom right lane) protein levels of these mutants do not account for the differences. Taken together, our results indicate a correlation between the conformation of p53 that is recognized by PAb240 and the ability of p53 to bind to p73.
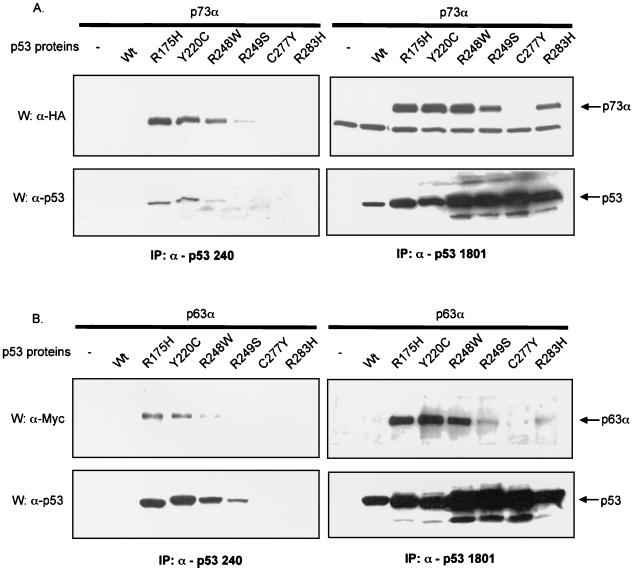
Correlation between p53 mutants strongly interacting with p73 or p63 and PAb240 reactivity. Comparative coimmunoprecipitation of various p53 mutants with p53 homologues by PAb1081 or PAb240. p53 and p73α (panel A) or p63α (panel B) expressing vectors were transfected into H1299 cells. After 48 h, cells were lysed and half of the extracts were used to immunoprecipitate p53 with PAb1801 and the other half to immunoprecipitate with PAb240. Detection of p53, p63, and p73 by Western blot analysis was performed as described in Fig. Fig.1.1. IP, immunoprecipitated.
Analysis of HSP protein binding to p53 and p73 proteins.
It has been documented that a subset of mutant forms of p53 can interact with members of the heat shock protein family, including HSC70 (24, 49). In fact, Gannon et al. reported that there is a correlation between the abilities of mutant forms of p53 to interact with PAb240 and with HSC70 (18). It was therefore of interest to determine whether those mutants in our study that bind p73 and PAb240 also interact with HSC70 under our conditions (Fig. (Fig.10).10).

HSP70 proteins interact with p53 mutants and p73 proteins. p53 or p73 variants (either wild type or mutants, as indicated) were expressed in H1299 cells. Cells were lysed after 48 h, and extracts were either probed for the indicated protein after Western blotting (10%) or immunoprecipitated with anti-HSP70 MAb (IP: HSP70) followed by Western blotting with antibodies for HSP70, p53, or p73α. IP, immunoprecipitated.
p53 mutants were expressed as before in H1299 cells, and after lysis, endogenous HSC70 was immunoprecipitated. Immune complexes were resolved by SDS-PAGE and transferred to nitrocellulose, and the presence of p53 in the complexes was assessed by Western blotting. Expression levels of p53 variants were checked (Fig. (Fig.10A,10A, bottom). As described in the literature, wild-type p53 did not interact with HSC70 (Fig. (Fig.10A).10A). However, three p53 mutants (R175R, Y220C, and R249S) were able to bind strongly to HSC70. Three other mutants either bound weakly to HSC70 (R248W) or showed no binding (C277Y and R283H). Similar results were obtained when the reverse experiment was done, i.e., immunoprecipitating p53 and probing for HSC70.
To further characterize the relationship between heat shock protein binding and p73, we decided to examine whether p73 itself could be coprecipitated with HSC70. Wild-type or mutant forms of p73 were introduced into H1299 cells, and transfected cell extracts were immunoprecipitated with anti-HSC70 antibody. In contrast to the situation with p53, it was surprising that both forms of p73 were found in the HSP70 precipitates, and, in fact, there did not seem to be relatively greater amounts of mutant p73 protein in the immune complexes. While this supports the possibility that mutant forms of p53 may interact with p73 through HSC70 interactions, as discussed below, our results indicated that this is not likely to be the sole factor that regulates interactions between p53 and p73.
Table Table11 summarizes our data comparing the binding of p53 mutants to p73, PAb240, and HSC70. Two mutants that bound strongly to p73 (R175H and Y220C) also bound well to both PAb240 and HSC70, while the only mutant that we tested that could not bind detectably to p73 was not immunoprecipitated by either PAb240 or anti-HSC70 antibody. Nevertheless, results with the other mutants showed that there was not a strong correlation between these three interactions. Most striking was the fact that mutant R249S, which bound only weakly to p73, was efficiently coprecipitated with HSC70. Furthermore, mutant R283H that was not efficiently coprecipitated with p73 did not interact detectably with HSC70, while mutant R248W bound well to p73 but only weakly to HSC70 and PAb240. Taken together, our data indicated that PAb240 binding better reflects the interactions between mutant p53 and p73 than does HSC70 binding, although it is acknowledged that quantitative differences existed in the extents of PAb240 binding. Thus, there must be other determinants that affect the interactions between mutant p53 and p73.
TABLE 1
Interactions of p53 mutants with p73, PAb240, and HSP70
| p53 mutanta | Strength of binding interactionb with
| ||
|---|---|---|---|
| p73 | PAb240 | HSP70 | |
| Wild type (P) | − | − | − |
| R175H (R) | +++ | +++ | +++ |
| Y220C (P) | +++ | +++ | +++ |
| R248W (P) | +++ | + | + |
| R249S (P) | + | − | +++ |
| C277Y (P) | − | − | − |
| R283H (P) | + | − | − |
DISCUSSION
The discovery in 1997 of two p53-related genes, each of which encoded multiple isoforms, added a new level of complexity to the p53 field (30, 36, 46, 59). For one thing, this presented the possibility that interactions exist between members of this complex family and that cross-regulation may occur. Indeed, we and others found that certain p53 tumor-derived mutants can associate with some p73 isoforms and inhibit their functions (14, 37, 55). Here we have extended our analysis of the interactions between p53 family members. We found that (i) a specific set of p53 mutants has the ability to interact not only with p73 variants but also with several isoforms of p63, (ii) there is a correlation between the efficiency of p53 binding and down-regulation of p63 or p73 functions, (iii) the p53 core domain alone is sufficient for direct interactions with p73, and (iv) to some extent there is a correlation between the ability of p53 mutants to interact with p63 and p73 and their recognition by the p53 conformation sensitive MAb PAb240.
Multiple interactions and cross talk between members of the p53 family.
We observed that there are interactions not only between mutant forms of p53 and p63 or p73 but also between wild-type p63 and p73 proteins. Thus, possible interactions between different p53 family members have been expanded as well as the potential network that involves these transcription factors. What is the physiological relevance of these heterologous interactions? It will be difficult to assess the impact of interactions between transcriptionally competent p53 family members without information about the specific targets of p63 or p73 and an appropriate functional assay. However, such analysis is possible with the heterologous interactions involving p53 mutants and p63 or p73. While many of our experiments relied on transient transfection assays, our group and others have also found that endogenously expressed p53 family members can coassociate, as evidenced by coimmunoprecipitation from cell extracts. In fact, with HACAT cells we discovered that both p63 and p73 are present in p53 immunoprecipitates. Thus, there is a physiological basis for the transfection experiments that were performed. Also relevant is the fact that there is a strong correlation between the extent of interaction of mutant p53 with p63 or p73 family members and down-regulation of their activity. Therefore, the mechanism by which p53 mutants affect p63 and p73 is most likely explained by a direct interaction between them, rather than by indirect “squelching” or competition by p53 proteins. Since there are p53 mutants that neither bind nor repress p63 and p73, this suggests that only a subset of p53 mutations in tumors may actively regulate the transcriptional program of other family members. Nonetheless, it is striking that each mutant p53 variant interacts (or does not interact) with all p63 and p73 isoforms that were tested.
The interaction with and regulation of p63 and p73 by mutant p53 might contribute to tumorigenesis. The tissue and stage-specific expression pattern of p63 and p73 isoforms would imply, however, that only certain tumors would express both p53 mutants and other p53 family members. Nevertheless, it is becoming clear that many types of tumors overexpress wild-type p73 or p63 protein even when compared to the related normal tissue. The ability of tumors to tolerate high levels of p63 or p73 may be related in some cases to the ability of mutant p53 to keep them in check. Marin et al. (37) have proposed a role for the interaction of several p53 mutants with p73 in the establishment of squamous cell cancers. In their study the ability of p53 to interact with p73 and possibly favor cancer formation depends on both missense mutation in the core domain and the presence of a specific polymorphism at position 72 (R versus P). In our experiments we were not able to fully corroborate this observation because, of the three best interacting mutants, two are variants of P72 (Y220C and R248W). Differences between the data of Marin et al. and ours may be related to the different cell types used or to conditions of expression and immunoprecipitation. Whatever the basis for these discrepancies, there are likely to be multiple determinants which dictate whether or not p53 family members interact. Table Table22 summarizes data from studies by this group and others on a number of p53 mutants and their relative interactions with p53 homologues. Compiling such information and correlating it with clinical studies might eventually help to identify the p53 characteristics that are critical for these interactions.
TABLE 2
Interactions between wild-type or mutant p53 and its homologues
| Protein | Type of binding | Homologue(s) demonstrating indicated strengtha of binding to p53
| ||
|---|---|---|---|---|
| − | + | +++ | ||
| p73 | In vitro | 248(P), 273(P) | WT(P), 143A(P), 175P(R) | |
| Ectopic | 248W(P/R),b 273H,c 281G,bWT(P), 277Y(P) | 143A(P),b 173L(P),b 175P(P),b 179Q(P/R),b249S(P), 283R(P) | WT(R),b 143A(R), 173L(R) 175P(R),b 281G,d175H(R), 220C(P), 248W(P) | |
| Endogenous | 151S,b 179Y(R),b 206F,b 194F,d 175H,d175H, 179Y, 273H | |||
| p63 | In vitro | |||
| Ectopic | WT(P), 277Y(P) | 249S(P), 283R(P) | 143A(P), 175H(R), 220C(P), 248R(P) | |
| Endogenous | 179Y | |||
Considerable insight into the natural functions of p63 and p73 has come from the study of mice that are genetically engineered to be null for these genes. With respect to p63−/− mice, since these animals die so soon after birth, it is not possible to assess whether loss of p63 would predispose mice for cancer (38, 62). However, the p73−/− mice are relatively long lived and are not particularly cancer prone (63). It is possible that the roles of p63 and p73 in mice and humans differ. In murine cancers both of the p53 alleles are usually deleted or not expressed, while in humans there is frequent expression (often high) of at least one p53 gene containing a missense mutation. In addition, it should be noted that removal of the whole p73 gene leads to the deletion of both the full-length and ΔN isoforms. These two sets of proteins may have opposite effects regarding p53. The ΔN isoforms may down-regulate wild-type p53 (61), while mutant forms of p53 may correspondingly repress activation by full-length p63 and p73. In fact, a number of publications show that the ΔNp63 isoforms are overexpressed in several tumors, suggesting that these proteins exert a positive effect on tumor development. Thus, deletion of the whole gene may not impact tumorigenesis because two opposing types of regulation have been deleted. Selective deletion of the full-length isoform may eventually help us to understand the role of p53 homologues in tumorigenicity and how the interaction with p53 mutants is relevant.
How do mutant forms of p53 interact with other family members?
We sought to gain more insight into the mode by which p53 recognizes these proteins. A recent study using mammalian extracts expressing several deleted forms of p53 or p73 concludes that the core domain of p53 and the core-tetramerization domain of p73, respectively, are required for their interaction (12). Here we used purified p53 and p73 proteins and now demonstrate that the p53 core domain alone binds directly to p73β isoforms. Our data also indicate that one characteristic of p53 required for its binding to p73 and p63 is recognition by the MAb PAb240. The epitope recognized by this antibody, amino acids 212 to 217 (58), lies within the hydrophobic interior of the β sandwich component of the core domain (10). It was proposed by Cho et al. that recognition by PAb240 reflects an unfolded state of the p53 core domain in which its epitope becomes unmasked (10). Tumor-derived mutations in the core can, in some cases, cause conformational changes that may be propagated a long distance, leading to exposure of this epitope. Bullock et al. analyzed the integrity of several mutant core domains in terms of unfolding at physiological temperatures and solvent exposure (6). Using these and other criteria, Bullock et al. concluded that mutants R175H and Y220C are globally denatured. In fact, our results show that these two mutants bind well to p73 in vivo and are efficiently immunoprecipitated by PAb240 and anti-HSC70 antibodies, thus suggesting that extensive unfolding is a key determinant. However, HSC70 binding, which should be a strong predictor of unfolding, does not appear to be a necessary component of the ability of mutant p53 to be associated with p63 and p73; Bullock et al. (6) showed that another mutant, R249S, is not globally unfolded, yet R249S binds as efficiently to HSC70 as do R175H and Y220C, while interacting only weakly with p63 or p73. Furthermore, R248W, which interacts well with p63 and p73, does not interact with HSC70. Thus, there is not a strong correlation between unfolding as evidenced by HSC70 binding and the ability of mutant p53 to down-regulate p73. At this point our data show a stronger correlation between the PAb240 reactivity of mutants and their ability to interact with p53 relatives, although it is acknowledged that a still wider range of mutants needs to be examined. Deletion of the p53 N or C terminus or its tetramerization domain allows p53 to bind p73 even though its core domain is of the wild type, suggesting that comparable conformational changes can also be propagated from outside the core. However, it is also possible that the regions outside the core domain cause steric hindrance, preventing access to the central portion of p53. Indeed, a recent report showed similarities between p53 proteins mutated in the core domain and wild-type p53 deleted of its proline-rich region in the N terminus (50). It is interesting that wild-type p53 can be recognized by PAb240 in a cell cycle-dependent manner (39, 68), when it forms heterooligomers with mutant p53 (40), or when it is bound to DNA (21). We cannot therefore exclude the possibility that, under some conditions, interaction between wild-type p53 and p53 homologues could occur in vivo. Whatever the basis for the interaction, the DNA binding activity of p73 is impaired when p53 mutants are cotranslated in vitro (37) or coexpressed in insect cells (M. Lokshin, C. Gaiddon, and C. Prives, unpublished data).
There is still much to learn about the features of p63 and p73 that are involved in their interactions with mutant p53. In this regard, the fact that p53 bound more poorly to ΔN forms of p63 than to full-length p63 suggested that although the core domain can bind directly, the N terminus of at least p63 plays an auxiliary role. The N terminus may either facilitate core interactions of p63 (or p73) with p53 or may possess a binding surface for p53. Alternatively, it may serve to bring p53 family members into a complex through common N-terminal interactions with other cellular proteins such as Mdm2 or with components of the transcriptional machinery.
Our current working hypothesis is that mutant p53 down-regulation of p63 and p73 may help to propagate tumor development in some situations. Although this has not been definitively proven, since our data suggest that interactions between p53 family members are widespread, it is highly likely that cross-regulation can occur in some cases. By gaining insight into the mechanism by which mutant p53 interacts with its relatives, it is hoped that it might be possible eventually to develop reagents that disrupt this interaction and permit p63 or p73 proteins to function, like wild-type p53, to counteract tumor growth by causing growth arrest or cell death.
ACKNOWLEDGMENTS
We are grateful to Ella Freulich for excellent technical assistance and to C. Di Como and K. McKinney for providing purified p73 antibody, support, and helpful discussion. We also thank W. Kaelin for the ER15 p73 monoclonal antibody, G. Melino and V. De Laurenzy for the p73 expression vectors, F. McKeon for the p63 expression vectors, N. Pavletich for the purified p53 R248W core domain, and P. Hall for the p63 polyclonal antibody.
This work was supported by NIH grant CA77742. C.G. was supported by a fellowship from the Human Frontiers Foundation (LT0776/1997M).
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.21.5.1874-1887.2001
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc86759?pdf=render
HAL Open Archive
https://www.hal.inserm.fr/inserm-02360323
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.21.5.1874-1887.2001
Article citations
From regulation to deregulation of p53 in hematologic malignancies: implications for diagnosis, prognosis and therapy.
Biomark Res, 12(1):137, 14 Nov 2024
Cited by: 0 articles | PMID: 39538363
Review
TP53: the unluckiest of genes?
Cell Death Differ, 23 Oct 2024
Cited by: 0 articles | PMID: 39443700
Review
Predictive value of TP53 RNAscope®in situ hybridization and p53 immunohistochemistry for TP53 mutational status in canine diffuse large B-cell lymphoma.
Vet Q, 44(1):1-9, 16 Sep 2024
Cited by: 0 articles | PMID: 39282821 | PMCID: PMC11407423
Activating transcription factor 3 mediates apoptosis and cell cycle arrest in TP53-mutated anaplastic thyroid cancer cells.
Thyroid Res, 17(1):12, 01 Aug 2024
Cited by: 0 articles | PMID: 39085957 | PMCID: PMC11292864
Anticancer therapeutic strategies for targeting mutant p53-Y220C.
J Biomed Res, 38(3):222-232, 01 May 2024
Cited by: 0 articles | PMID: 38738269 | PMCID: PMC11144932
Go to all (310) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53.
J Biol Chem, 274(26):18709-18714, 01 Jun 1999
Cited by: 103 articles | PMID: 10373484
p63 and p73 transactivate differentiation gene promoters in human keratinocytes.
Biochem Biophys Res Commun, 273(1):342-346, 01 Jun 2000
Cited by: 87 articles | PMID: 10873608
Evolution of functions within the p53/p63/p73 family.
Ann N Y Acad Sci, 926:90-100, 01 Jan 2000
Cited by: 72 articles | PMID: 11193045
Review
Evidence for a distinct inhibitory factor in the regulation of p53 functional activity.
J Biol Chem, 276(30):27999-28005, 29 May 2001
Cited by: 8 articles | PMID: 11382762
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: CA77742
Grant ID: R01 CA077742





