Abstract
Background
A rapid increase in coronavirus disease 2019 (Covid-19) cases due to the omicron (B.1.1.529) variant of severe acute respiratory syndrome coronavirus 2 in highly vaccinated populations has aroused concerns about the effectiveness of current vaccines.Methods
We used a test-negative case-control design to estimate vaccine effectiveness against symptomatic disease caused by the omicron and delta (B.1.617.2) variants in England. Vaccine effectiveness was calculated after primary immunization with two doses of BNT162b2 (Pfizer-BioNTech), ChAdOx1 nCoV-19 (AstraZeneca), or mRNA-1273 (Moderna) vaccine and after a booster dose of BNT162b2, ChAdOx1 nCoV-19, or mRNA-1273.Results
Between November 27, 2021, and January 12, 2022, a total of 886,774 eligible persons infected with the omicron variant, 204,154 eligible persons infected with the delta variant, and 1,572,621 eligible test-negative controls were identified. At all time points investigated and for all combinations of primary course and booster vaccines, vaccine effectiveness against symptomatic disease was higher for the delta variant than for the omicron variant. No effect against the omicron variant was noted from 20 weeks after two ChAdOx1 nCoV-19 doses, whereas vaccine effectiveness after two BNT162b2 doses was 65.5% (95% confidence interval [CI], 63.9 to 67.0) at 2 to 4 weeks, dropping to 8.8% (95% CI, 7.0 to 10.5) at 25 or more weeks. Among ChAdOx1 nCoV-19 primary course recipients, vaccine effectiveness increased to 62.4% (95% CI, 61.8 to 63.0) at 2 to 4 weeks after a BNT162b2 booster before decreasing to 39.6% (95% CI, 38.0 to 41.1) at 10 or more weeks. Among BNT162b2 primary course recipients, vaccine effectiveness increased to 67.2% (95% CI, 66.5 to 67.8) at 2 to 4 weeks after a BNT162b2 booster before declining to 45.7% (95% CI, 44.7 to 46.7) at 10 or more weeks. Vaccine effectiveness after a ChAdOx1 nCoV-19 primary course increased to 70.1% (95% CI, 69.5 to 70.7) at 2 to 4 weeks after an mRNA-1273 booster and decreased to 60.9% (95% CI, 59.7 to 62.1) at 5 to 9 weeks. After a BNT162b2 primary course, the mRNA-1273 booster increased vaccine effectiveness to 73.9% (95% CI, 73.1 to 74.6) at 2 to 4 weeks; vaccine effectiveness fell to 64.4% (95% CI, 62.6 to 66.1) at 5 to 9 weeks.Conclusions
Primary immunization with two doses of ChAdOx1 nCoV-19 or BNT162b2 vaccine provided limited protection against symptomatic disease caused by the omicron variant. A BNT162b2 or mRNA-1273 booster after either the ChAdOx1 nCoV-19 or BNT162b2 primary course substantially increased protection, but that protection waned over time. (Funded by the U.K. Health Security Agency.).Free full text

Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant
Abstract
Background
A rapid increase in coronavirus disease 2019 (Covid-19) cases due to the omicron (B.1.1.529) variant of severe acute respiratory syndrome coronavirus 2 in highly vaccinated populations has aroused concerns about the effectiveness of current vaccines.
Methods
We used a test-negative case–control design to estimate vaccine effectiveness against symptomatic disease caused by the omicron and delta (B.1.617.2) variants in England. Vaccine effectiveness was calculated after primary immunization with two doses of BNT162b2 (Pfizer–BioNTech), ChAdOx1 nCoV-19 (AstraZeneca), or mRNA-1273 (Moderna) vaccine and after a booster dose of BNT162b2, ChAdOx1 nCoV-19, or mRNA-1273.
Results
Between November 27, 2021, and January 12, 2022, a total of 886,774 eligible persons infected with the omicron variant, 204,154 eligible persons infected with the delta variant, and 1,572,621 eligible test-negative controls were identified. At all time points investigated and for all combinations of primary course and booster vaccines, vaccine effectiveness against symptomatic disease was higher for the delta variant than for the omicron variant. No effect against the omicron variant was noted from 20 weeks after two ChAdOx1 nCoV-19 doses, whereas vaccine effectiveness after two BNT162b2 doses was 65.5% (95% confidence interval [CI], 63.9 to 67.0) at 2 to 4 weeks, dropping to 8.8% (95% CI, 7.0 to 10.5) at 25 or more weeks. Among ChAdOx1 nCoV-19 primary course recipients, vaccine effectiveness increased to 62.4% (95% CI, 61.8 to 63.0) at 2 to 4 weeks after a BNT162b2 booster before decreasing to 39.6% (95% CI, 38.0 to 41.1) at 10 or more weeks. Among BNT162b2 primary course recipients, vaccine effectiveness increased to 67.2% (95% CI, 66.5 to 67.8) at 2 to 4 weeks after a BNT162b2 booster before declining to 45.7% (95% CI, 44.7 to 46.7) at 10 or more weeks. Vaccine effectiveness after a ChAdOx1 nCoV-19 primary course increased to 70.1% (95% CI, 69.5 to 70.7) at 2 to 4 weeks after an mRNA-1273 booster and decreased to 60.9% (95% CI, 59.7 to 62.1) at 5 to 9 weeks. After a BNT162b2 primary course, the mRNA-1273 booster increased vaccine effectiveness to 73.9% (95% CI, 73.1 to 74.6) at 2 to 4 weeks; vaccine effectiveness fell to 64.4% (95% CI, 62.6 to 66.1) at 5 to 9 weeks.
Conclusions
Primary immunization with two doses of ChAdOx1 nCoV-19 or BNT162b2 vaccine provided limited protection against symptomatic disease caused by the omicron variant. A BNT162b2 or mRNA-1273 booster after either the ChAdOx1 nCoV-19 or BNT162b2 primary course substantially increased protection, but that protection waned over time. (Funded by the U.K. Health Security Agency.)
On November 26, 2021, the World Health Organization Technical Advisory Group on SARS-CoV-2 Virus Evolution named the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.529 variant, first detected in Botswana and South Africa, as the omicron variant of concern.1 This classification was based on a rapid increase in confirmed cases of SARS-CoV-2 infection in South Africa, coinciding with an increase in detections of the omicron variant, identification of a number of worrisome mutations, and early evidence of an increased risk of reinfection among recently infected persons.
A large number of mutations have been identified in the omicron variant, including multiple mutations in the receptor-binding domain of the spike protein that have been associated with increased transmissibility and immune evasion after natural infection and vaccination.2 Emerging laboratory data indicate a substantially reduced neutralizing antibody response to the omicron variant as compared with the original strain of SARS-CoV-2 or the delta (B.1.617.2) variant in vaccinated persons, although booster doses improved neutralizing activity.3-5 Neutralizing antibodies correlate with protection against reinfection and vaccine effectiveness against infection; therefore, reduced vaccine effectiveness against the omicron variant is anticipated on the basis of these early laboratory findings.6-8
Coronavirus disease 2019 (Covid-19) vaccines are highly effective against symptomatic disease and, more so, against severe disease and fatal outcomes caused by the original strain of SARS-CoV-2 as well as the alpha (B.1.1.7) variant that predominated in early 2021.9-15 Modest reductions in vaccine effectiveness against infection and mild disease have been observed with the beta (B.1.351) and delta variants, although effectiveness against severe disease has remained high for at least 6 months after primary immunization with two Covid-19 vaccine doses.16-19 Waning of protection has been observed with time since vaccination, especially with the delta variant, which is able to at least partially evade natural and vaccine-induced immunity.20 However, third (booster) doses provide a rapid and substantial increase in protection against both mild and severe disease.19,21-25
In the United Kingdom, cases of infection with the omicron variant were first identified in mid-November 2021 through whole-genome sequencing of polymerase-chain-reaction (PCR)–positive swab samples. Initially, cases occurred primarily in travelers and their close contacts, but community transmission was apparent beginning in late November.26 The U.K. Covid-19 vaccination program has been in place since December 2020 with primary courses of two doses of BNT162b2 (Comirnaty, Pfizer–BioNTech), ChAdOx1 nCoV-19 (Vaxzevria, AstraZeneca), or mRNA-1273 (Spikevax, Moderna) vaccine. Two-dose vaccine uptake is more than 60% in all cohorts of persons who are 20 years of age or older and more than 80% in all cohorts of persons who are 50 years of age or older, with vaccinations now being offered to children 12 years of age or older.27 Booster vaccination with either BNT162b2 vaccine or a half dose (50 μg) of mRNA-1273 vaccine was introduced in September 2021 to adults 50 years of age or older and those in at-risk groups and was expanded in November 2021 to all adults. Initially, boosters were offered 6 months after completion of the primary course. With the emergence of the omicron variant in late November 2021, this interval was reduced to 3 months.28
In this study, we estimated vaccine effectiveness against symptomatic disease caused by the delta and omicron variants after two doses (primary immunization) of BNT162b2, mRNA-1273, or ChAdOx1 nCoV-19 vaccine and after homologous or heterologous booster doses with the same three vaccines.
Methods
Study Design
We used a test-negative case–control design to estimate vaccine effectiveness against symptomatic Covid-19 caused by the omicron variant as compared with the delta variant in persons 18 years of age or older.17 The odds of vaccination in persons with symptomatic, PCR-positive cases of SARS-CoV-2 infection were compared with those in symptomatic persons who tested negative for SARS-CoV-2 in England.
Data Sources
Covid-19 Testing Data
PCR testing for SARS-CoV-2 in England is undertaken by hospital and public health laboratories (Pillar 1) as well as by community testing (Pillar 2). Pillar 2 testing is available to anyone with symptoms consistent with Covid-19 (high temperature, new continuous cough, or loss or change in sense of smell or taste), anyone who is a contact of a person with a confirmed case, care home staff and residents, and persons with a positive rapid lateral-flow antigen test. Lateral-flow tests are freely available to all members of the population for regular home testing. Data on all positive PCR and lateral-flow tests, and on negative Pillar 2 PCR tests from persons with a date of onset of Covid-19 symptoms after November 25, 2020, were extracted up to January 12, 2022 (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Persons who reported symptoms and were tested in Pillar 2 between November 27, 2021, and January 12, 2022, were included in the analysis.
Any negative tests taken within 7 days after a previous negative test, and any negative tests for which the symptom-onset date was within the 10 days after a previous symptom-onset date for a negative test, were dropped because these probably represented the same episode. Negative tests taken within 21 days before a subsequent positive test were also excluded because chances were high that these were false negatives. Positive and negative tests within 90 days after a previous positive test were also excluded; however, when participants had later positive tests within 14 days after a positive test, preference was given to PCR tests and tests from symptomatic persons. For persons who had more than one negative test, one test was selected at random in the study period. Data were restricted to persons who had reported symptoms and gave a symptom-onset date within the 10 days before testing to account for reduced PCR sensitivity beyond this period in an infection event. Only positive tests with sequencing or genotyping information or information on spike gene (S) target–negative status (indicative of probable omicron infection) were included in the final analysis. A small number of positive tests were excluded when sequencing showed neither the delta nor the omicron variant. Finally, only samples obtained on November 27, 2021, or after were retained for analysis because this corresponded to the period when S target–negative status was predictive of the omicron variant.
Vaccination Data
The National Immunization Management System (NIMS) contains demographic information on all persons residing in England who are registered with a general practice physician in that country and is used to record all Covid-19 vaccinations.29 The NIMS was accessed on January 18, 2022, for dates of vaccination and vaccine manufacturer, sex, date of birth, race or ethnic group, and residential address. Addresses were used to determine the index of multiple deprivation (a national indication of level of deprivation that is based on small geographic areas of residence, assessed in quintiles) and were also linked to Care Quality Commission–registered care homes with the use of the unique property-reference number. Data on geographic region (NHS region), clinical risk-group status, status of being in a clinically extremely vulnerable group, and health and social care worker status were also extracted from the NIMS. Clinical risk groups included a range of chronic conditions as described in the Green Book,30 whereas the clinically extremely vulnerable group included persons who were considered to be at the highest risk for severe Covid-19, including those with immunosuppressed conditions and those with severe respiratory disease.31 Booster doses were identified as a third dose given at least 175 days after a second dose and administered after September 13, 2021. Persons with four or more doses of vaccine, a heterologous primary schedule, or fewer than 19 days between their first dose and second dose were excluded.
Identification of Variants and Assignment to Cases
Sequencing of PCR-positive samples was undertaken through a network of laboratories, including the Wellcome Sanger Institute. Whole-genome sequences were assigned to U.K. Health Security Agency definitions of variants on the basis of mutations.32,33 S target status on PCR testing is an alternative approach for identifying each variant because the omicron variant has been associated with S target–negative results on PCR testing with the TaqPath assay, whereas the delta variant almost always has an S target–positive result.26 Approximately 40% of Pillar 2 community testing in England is carried out by laboratories using the TaqPath assay (Thermo Fisher Scientific). Cases were defined as being due to the delta or omicron variant on the basis of whole-genome sequencing, genotyping, or S target status, with sequencing taking priority, followed by genotyping. When subsequent positive tests within 14 days included sequencing or genotyping information or information on S target–negative status, this information was used to classify the variant. A priori, we considered that S target–negative status would be used to define the omicron variant when the variant accounted for at least 80% of S target–negative cases. Beginning on January 10, 2022, delta cases were identified by sequencing and genotyping only because the positive predictive value of S target–negative status to identify the delta variant had decreased and could no longer be used.
Testing data were linked to the NIMS on January 18, 2021, through combinations of the unique individual NHS number, date of birth, surname, first name, and postal code with the use of deterministic linkage. A total of 91.8% of eligible tests could be linked to the NIMS.
Statistical Analysis
Logistic regression was used, with the PCR test result as the dependent variable and case participants being those testing positive (stratified in separate analyses as being infected with either the omicron or delta variant) and controls being those testing negative. Vaccination status was included as an independent variable, and effectiveness was defined as 1 minus the odds of vaccination in case participants, divided by the odds of vaccination in controls.
Vaccine effectiveness was adjusted in logistic-regression models for age (18 to 89 years in 5-year bands, then everyone ≥90 years), sex, index of multiple deprivation (quintile), race or ethnic group, history of foreign travel, geographic region, period (day of test), health and social care worker status, clinical risk-group status, status of being in a clinically extremely vulnerable group, and previously testing positive. These factors were all considered potential confounders and so were included in all models.
Analyses were stratified according to primary immunization course (ChAdOx1 nCoV-19, BNT162b2, or mRNA-1273 vaccine). Any heterologous primary schedules were excluded.
Vaccine effectiveness was assessed for each primary course in intervals of 2 to 4, 5 to 9, 10 to 14, 15 to 19, 20 to 24, and 25 or more weeks after the second dose. Vaccine effectiveness was assessed at 2 to 4, 5 to 9, and 10 or more weeks after a BNT162b2 or mRNA-1273 booster after a ChAdOx1 nCoV-19 or BNT162b2 primary course. In addition, the ChAdOx1 nCoV-19 booster was assessed after a ChAdOx1 nCoV-19 primary course in these postvaccination intervals. In persons with an mRNA-1273 primary course, vaccine effectiveness was assessed after BNT162b2 or mRNA-1273 booster vaccines after 1 week and after 2 to 4 weeks.
Results
Descriptive Characteristics
Within sequenced cases from Pillar 2 testing for which S target testing was performed, the percentage of S target–negative tests that were sequenced as the omicron variant was 50% (6 of 12) on November 25, 2021; 65% (13 of 20) on November 26; 90% (18 of 20) on November 27; 91% (10 of 11) on November 28; and 89% (17 of 19) on November 29. We therefore used cases tested on or after November 27, when the positive predictive value was more than 80%.
A total of 886,774 persons with symptomatic disease who were infected with the omicron variant were identified during the study period by sequencing or genotyping or by S target–negative status, and their tests were linked to the NIMS for vaccination status. During the same period, 204,154 eligible persons infected with the delta variant and 1,572,621 eligible test-negative controls were identified (Fig. S2). The characteristics of the persons tested are shown in Table 1, and vaccination status according to test result is shown in Table 2. The distribution according to sex, geographic region, and race or ethnic group was similar to that in the full national Covid-19 case data, although there was a higher proportion of young adults (Table S1). Infection with the omicron variant 14 or more days after a booster occurred a median of 39 days (range, 14 to 118) after the booster.
Table 1
| Characteristic | Overall (N=2,663,549) | Test-Negative Status (N=1,572,621) | Delta Variant (N=204,154) | Omicron Variant (N=886,774) |
|---|---|---|---|---|
| Percent of all participants | 100 | 59.0 | 7.7 | 33.3 |
| Sex — no. (%) | ||||
| Female | 1,555,857 (58.4) | 955,508 (60.8) | 111,080 (54.4) | 489,269 (55.2) |
| Male | 1,102,797 (41.4) | 614,068 (39.0) | 92,764 (45.4) | 395,965 (44.7) |
| Missing data | 4,895 (0.2) | 3,045 (0.2) | 310 (0.2) | 1,540 (0.2) |
| Age — no. (%) | ||||
| 18 or 19 yr | 86,559 (3.2) | 46,445 (3.0) | 4,055 (2.0) | 36,059 (4.1) |
| 20–24 yr | 279,465 (10.5) | 151,700 (9.6) | 14,071 (6.9) | 113,694 (12.8) |
| 25–29 yr | 336,504 (12.6) | 192,568 (12.2) | 20,227 (9.9) | 123,709 (14.0) |
| 30–34 yr | 355,731 (13.4) | 212,628 (13.5) | 26,484 (13.0) | 116,619 (13.2) |
| 35–39 yr | 327,751 (12.3) | 197,453 (12.6) | 30,097 (14.7) | 100,201 (11.3) |
| 40–44 yr | 283,074 (10.6) | 164,907 (10.5) | 31,936 (15.6) | 86,231 (9.7) |
| 45–49 yr | 244,583 (9.2) | 140,024 (8.9) | 26,862 (13.2) | 77,697 (8.8) |
| 50–54 yr | 231,704 (8.7) | 136,899 (8.7) | 20,326 (10.0) | 74,479 (8.4) |
| 55–59 yr | 193,831 (7.3) | 117,764 (7.5) | 14,943 (7.3) | 61,124 (6.9) |
| 60–64 yr | 131,656 (4.9) | 84,101 (5.3) | 8,187 (4.0) | 39,368 (4.4) |
| 65–69 yr | 80,267 (3.0) | 53,952 (3.4) | 3,251 (1.6) | 23,064 (2.6) |
| 70–74 yr | 55,598 (2.1) | 36,893 (2.3) | 1,846 (0.9) | 16,859 (1.9) |
| 75–79 yr | 30,615 (1.1) | 19,748 (1.3) | 976 (0.5) | 9,891 (1.1) |
| 80–84 yr | 15,333 (0.6) | 10,058 (0.6) | 488 (0.2) | 4,787 (0.5) |
| 85–89 yr | 7,452 (0.3) | 5,069 (0.3) | 279 (0.1) | 2,104 (0.2) |
| ≥90 yr | 3,426 (0.1) | 2,412 (0.2) | 126 (0.1) | 888 (0.1) |
| Race or ethnic group — no. (%)† | ||||
| African | 39,865 (1.5) | 19,439 (1.2) | 1,851 (0.9) | 18,575 (2.1) |
| Caribbean | 23,070 (0.9) | 10,052 (0.6) | 1,527 (0.7) | 11,491 (1.3) |
| Another Black background | 3,955 (0.1) | 1,938 (0.1) | 241 (0.1) | 1,776 (0.2) |
| Arab | 8,743 (0.3) | 5,271 (0.3) | 660 (0.3) | 2,812 (0.3) |
| Bangladeshi | 18,372 (0.7) | 10,836 (0.7) | 1,231 (0.6) | 6,305 (0.7) |
| Chinese | 17,803 (0.7) | 10,967 (0.7) | 1,142 (0.6) | 5,694 (0.6) |
| Indian | 88,486 (3.3) | 57,385 (3.6) | 4,574 (2.2) | 26,527 (3.0) |
| Pakistani | 46,291 (1.7) | 27,417 (1.7) | 3,004 (1.5) | 15,870 (1.8) |
| Another Asian background | 34,867 (1.3) | 19,979 (1.3) | 2,082 (1.0) | 12,806 (1.4) |
| Mixed or multiple ethnic groups | 52,395 (2.0) | 29,291 (1.9) | 3,358 (1.6) | 19,746 (2.2) |
| White | 2,215,756 (83.2) | 1,312,372 (83.5) | 176,390 (86.4) | 726,994 (82.0) |
| Another ethnic background | 18,302 (0.7) | 10,715 (0.7) | 1,180 (0.6) | 6,407 (0.7) |
| Prefer not to say | 95,644 (3.6) | 56,959 (3.6) | 6,914 (3.4) | 31,771 (3.6) |
| NHS region — no. (%) | ||||
| East of England | 287,326 (10.8) | 191,259 (12.2) | 22,695 (11.1) | 73,372 (8.3) |
| London | 411,785 (15.5) | 237,614 (15.1) | 24,571 (12.0) | 149,600 (16.9) |
| Midlands | 455,325 (17.1) | 280,909 (17.9) | 35,860 (17.6) | 138,556 (15.6) |
| North East | 414,437 (15.6) | 232,576 (14.8) | 32,467 (15.9) | 149,394 (16.8) |
| North West | 444,930 (16.7) | 207,429 (13.2) | 33,404 (16.4) | 204,097 (23.0) |
| South East | 406,968 (15.3) | 257,588 (16.4) | 33,720 (16.5) | 115,660 (13.0) |
| South West | 242,772 (9.1) | 165,240 (10.5) | 21,437 (10.5) | 56,095 (6.3) |
| Missing | 6 (<0.1) | 6 (<0.1) | 0 | 0 |
| Index of multiple deprivation — no. (%)‡ | ||||
| 1 | 482,884 (18.1) | 261,116 (16.6) | 37,433 (18.3) | 184,335 (20.8) |
| 2 | 529,594 (19.9) | 305,748 (19.4) | 39,456 (19.3) | 184,390 (20.8) |
| 3 | 540,303 (20.3) | 325,218 (20.7) | 41,308 (20.2) | 173,777 (19.6) |
| 4 | 551,055 (20.7) | 333,456 (21.2) | 42,602 (20.9) | 174,997 (19.7) |
| 5 | 550,254 (20.7) | 341,620 (21.7) | 42,756 (20.9) | 165,878 (18.7) |
| Missing data | 9,459 (0.4) | 5,463 (0.3) | 599 (0.3) | 3,397 (0.4) |
| Previously tested positive — no. (%) | ||||
| No | 2,301,572 (86.4) | 1,312,548 (83.5) | 200,400 (98.2) | 788,624 (88.9) |
| Yes | 361,977 (13.6) | 260,073 (16.5) | 3,754 (1.8) | 98,150 (11.1) |
| Vaccine priority group — no. (%) | ||||
| Health and social care worker | 164,113 (6.2) | 113,183 (7.2) | 5,657 (2.8) | 45,273 (5.1) |
| Clinically extremely vulnerable | 142,002 (5.3) | 93,236 (5.9) | 7,443 (3.6) | 41,323 (4.7) |
| At risk | 482,075 (18.1) | 307,240 (19.5) | 32,490 (15.9) | 142,345 (16.1) |
| Severely immunosuppressed | 22,391 (0.8) | 13,985 (0.9) | 1,191 (0.6) | 7,215 (0.8) |
Table 2
| Vaccination Status, Dose, and Interval after Vaccination | Overall (N=2,663,549) | Test-Negative Status (N=1,572,621) | Delta Variant (N=204,154) | Omicron Variant (N=886,774) |
|---|---|---|---|---|
| number (percent) | ||||
| Unvaccinated | 244,716 (9.2) | 107,238 (6.8) | 36,369 (17.8) | 101,109 (11.4) |
| ChAdOx1 nCoV-19 | ||||
| Dose 1 | ||||
| 0–3 wk | 64 (<0.1) | 33 (<0.1) | 8 (<0.1) | 23 (<0.1) |
| ≥4 wk | 16,150 (0.6) | 8,470 (0.5) | 2,133 (1.0) | 5,547 (0.6) |
| Dose 2 | ||||
| 0 or 1 wk | 406 (<0.1) | 224 (<0.1) | 34 (<0.1) | 148 (<0.1) |
| 2–4 wk | 740 (<0.1) | 476 (<0.1) | 28 (<0.1) | 236 (<0.1) |
| 5–9 wk | 1,484 (0.1) | 894 (0.1) | 85 (<0.1) | 505 (0.1) |
| 10–14 wk | 2,700 (0.1) | 1,659 (0.1) | 274 (0.1) | 767 (0.1) |
| 15–19 wk | 17,775 (0.7) | 10,788 (0.7) | 4,013 (2.0) | 2,974 (0.3) |
| 20–24 wk | 120,979 (4.5) | 68,757 (4.4) | 29,100 (14.3) | 23,122 (2.6) |
| ≥25 wk | 292,794 (11.0) | 147,721 (9.4) | 46,103 (22.6) | 98,970 (11.2) |
| Booster dose | ||||
| Any: 0–6 days | 102,033 (3.8) | 54,613 (3.5) | 13,404 (6.6) | 34,016 (3.8) |
| BNT162b2 | ||||
| 1 wk | 60,212 (2.3) | 40,203 (2.6) | 2,088 (1.0) | 17,921 (2.0) |
| 2–4 wk | 165,248 (6.2) | 114,050 (7.3) | 2,029 (1.0) | 49,169 (5.5) |
| 5–9 wk | 157,008 (5.9) | 98,853 (6.3) | 2,291 (1.1) | 55,864 (6.3) |
| ≥10 wk | 34,081 (1.3) | 18,168 (1.2) | 343 (0.2) | 15,570 (1.8) |
| mRNA-1273 | ||||
| 1 wk | 31,958 (1.2) | 22,722 (1.4) | 845 (0.4) | 8,391 (0.9) |
| 2–4 wk | 76,935 (2.9) | 53,353 (3.4) | 417 (0.2) | 23,165 (2.6) |
| 5–9 wk | 22,745 (0.9) | 13,917 (0.9) | 92 (<0.1) | 8,736 (1.0) |
| ≥10 wk | 80 (<0.1) | 38 (<0.1) | 1 (<0.1) | 41 (<0.1) |
| ChAdOx1 nCoV-19 | ||||
| 1 wk | 158 (<0.1) | 107 (<0.1) | 10 (<0.1) | 41 (<0.1) |
| 2–4 wk | 444 (<0.1) | 294 (<0.1) | 19 (<0.1) | 131 (<0.1) |
| 5–9 wk | 440 (<0.1) | 264 (<0.1) | 12 (<0.1) | 164 (<0.1) |
| ≥10 wk | 84 (<0.1) | 48 (<0.1) | 0 | 36 (<0.1) |
| BNT162b2 | ||||
| Dose 1 | ||||
| 0–3 wk | 12,530 (0.5) | 7,038 (0.4) | 944 (0.5) | 4,548 (0.5) |
| ≥4 wk | 54,183 (2.0) | 29,759 (1.9) | 5,381 (2.6) | 19,043 (2.1) |
| Dose 2 | ||||
| 0 or 1 wk | 9,001 (0.3) | 5,900 (0.4) | 466 (0.2) | 2,635 (0.3) |
| 2–4 wk | 13,125 (0.5) | 9,516 (0.6) | 240 (0.1) | 3,369 (0.4) |
| 5–9 wk | 29,912 (1.1) | 20,163 (1.3) | 981 (0.5) | 8,768 (1.0) |
| 10–14 wk | 91,754 (3.4) | 61,014 (3.9) | 5,562 (2.7) | 25,178 (2.8) |
| 15–19 wk | 243,470 (9.1) | 144,172 (9.2) | 17,077 (8.4) | 82,221 (9.3) |
| 20–24 wk | 138,085 (5.2) | 72,018 (4.6) | 10,348 (5.1) | 55,719 (6.3) |
| ≥25 wk | 94,139 (3.5) | 51,625 (3.3) | 8,531 (4.2) | 33,983 (3.8) |
| Booster dose | ||||
| Any: 0–6 days | 80,592 (3.0) | 44,166 (2.8) | 3,212 (1.6) | 33,214 (3.7) |
| BNT162b2 | ||||
| 1 wk | 44,705 (1.7) | 29,459 (1.9) | 631 (0.3) | 14,615 (1.6) |
| 2–4 wk | 87,980 (3.3) | 64,874 (4.1) | 1,220 (0.6) | 21,886 (2.5) |
| 5–9 wk | 156,929 (5.9) | 110,306 (7.0) | 3,769 (1.8) | 42,854 (4.8) |
| ≥10 wk | 104,325 (3.9) | 61,534 (3.9) | 1,222 (0.6) | 41,569 (4.7) |
| mRNA-1273 | ||||
| 1 wk | 18,221 (0.7) | 12,718 (0.8) | 195 (0.1) | 5,308 (0.6) |
| 2–4 wk | 27,480 (1.0) | 20,045 (1.3) | 147 (0.1) | 7,288 (0.8) |
| 5–9 wk | 8,158 (0.3) | 5,311 (0.3) | 40 (<0.1) | 2,807 (0.3) |
| ≥10 wk | 87 (<0.1) | 53 (<0.1) | 1 (<0.1) | 33 (<0.1) |
| mRNA-1273 | ||||
| Dose 1 | ||||
| 0–3 wk | 2,544 (0.1) | 1,429 (0.1) | 134 (0.1) | 981 (0.1) |
| ≥4 wk | 5,656 (0.2) | 3,122 (0.2) | 448 (0.2) | 2,086 (0.2) |
| Dose 2 | ||||
| 0 or 1 wk | 682 (<0.1) | 467 (<0.1) | 35 (<0.1) | 180 (<0.1) |
| 2–4 wk | 1,104 (<0.1) | 855 (0.1) | 13 (<0.1) | 236 (<0.1) |
| 5–9 wk | 3,150 (0.1) | 2,286 (0.1) | 73 (<0.1) | 791 (0.1) |
| 10–14 wk | 14,324 (0.5) | 9,822 (0.6) | 674 (0.3) | 3,828 (0.4) |
| 15–19 wk | 32,913 (1.2) | 20,017 (1.3) | 1,627 (0.8) | 11,269 (1.3) |
| 20–24 wk | 16,291 (0.6) | 8,961 (0.6) | 1,224 (0.6) | 6,106 (0.7) |
| ≥25 wk | 1,221 (<0.1) | 474 (<0.1) | 16 (<0.1) | 731 (0.1) |
| Booster dose | ||||
| Any: 0–6 days | 8,787 (0.3) | 4,745 (0.3) | 203 (0.1) | 3,839 (0.4) |
| BNT162b2 | ||||
| 1 wk | 3,439 (0.1) | 2,031 (0.1) | 15 (<0.1) | 1,393 (0.2) |
| 2–4 wk | 3,410 (0.1) | 2,063 (0.1) | 8 (<0.1) | 1,339 (0.2) |
| 5–9 wk | 36 (<0.1) | 20 (<0.1) | 0 | 16 (<0.1) |
| ≥10 wk | 3 (<0.1) | 0 | 0 | 3 (<0.1) |
| mRNA-1273 | ||||
| 1 wk | 3,001 (0.1) | 1,851 (0.1) | 14 (<0.1) | 1,136 (0.1) |
| 2–4 wk | 3,067 (0.1) | 1,913 (0.1) | 5 (<0.1) | 1,149 (0.1) |
| 5–9 wk | 11 (<0.1) | 4 (<0.1) | 0 | 7 (<0.1) |
Vaccine Effectiveness, Delta Variant
Vaccine effectiveness against symptomatic disease in persons who received a primary course of the ChAdOx1 nCoV-19, BNT162b2, or mRNA-1273 vaccine according to period after primary immunization with second doses and after a third dose with BNT162b2 or mRNA-1273 is shown in Figure 1 and Table 3 and S2. With the delta variant, vaccine effectiveness after two doses of ChAdOx1 nCoV-19 primary course started at 82.8% (95% confidence interval [CI], 74.5 to 88.4) after 2 to 4 weeks but waned to 43.5% (95% CI, 42.4 to 44.5) after 25 or more weeks. With the messenger RNA (mRNA) vaccines, effectiveness after two doses was higher in all periods, but there was also evidence of waning. After a booster with an mRNA vaccine, vaccine effectiveness reached more than 95% with any of the primary courses and remained high up to the longest follow-up points (maximum, ≥10 weeks).
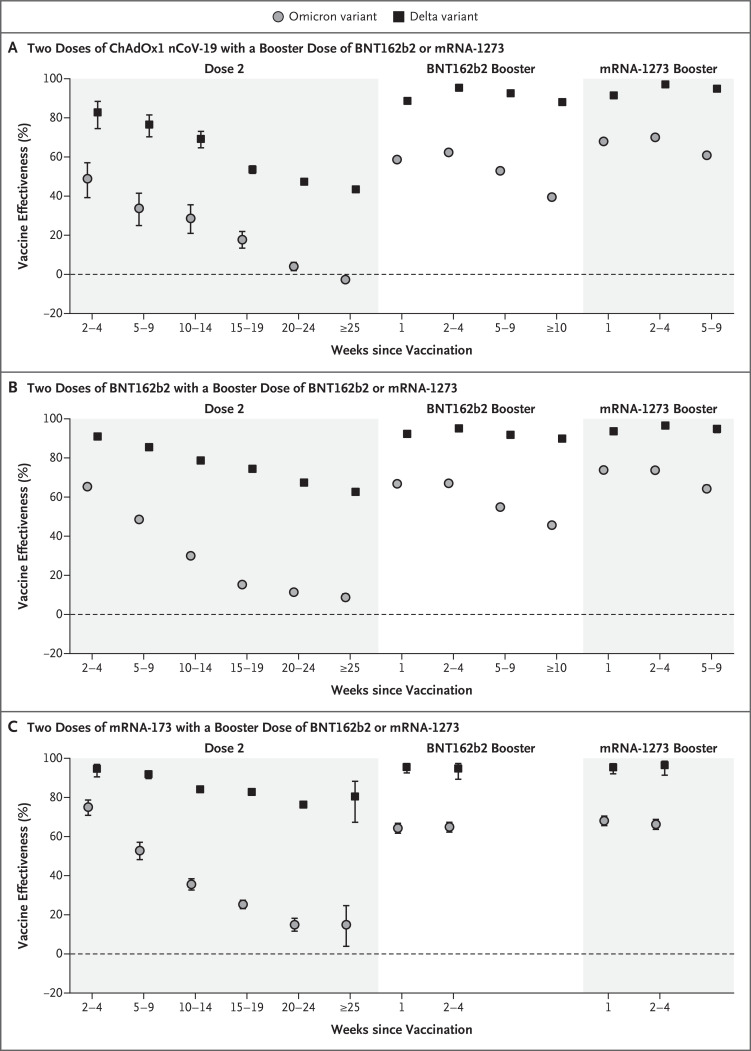
B.1.617.2 is the delta variant, and B.1.1.529 the omicron variant, of the severe acute respiratory syndrome coronavirus 2. 𝙸 bars indicate 95% confidence intervals.
Table 3
| Vaccination Status, Dose, and Interval after Vaccination | Test-Negative Status | Delta Variant | Omicron Variant | ||
|---|---|---|---|---|---|
| Controls | Case Participants | Vaccine Effectiveness (95% CI)† | Case Participants | Vaccine Effectiveness (95% CI)† | |
| no. | no. | % | no. | % | |
| Unvaccinated | 107,238 | 36,369 | Reference | 101,109 | Reference |
| ChAdOx1 nCoV-19 | |||||
| Dose 1 | |||||
| 0–3 wk | 33 | 8 | 23 | ||
| ≥4 wk | 8,470 | 2,133 | 42.9 (39.8 to 45.9) | 5,547 | 17.7 (14.3 to 21.0) |
| Dose 2 | |||||
| 2–4 wk | 476 | 28 | 82.8 (74.5 to 88.4) | 236 | 48.9 (39.2 to 57.1) |
| 5–9 wk | 894 | 85 | 76.5 (70.3 to 81.5) | 505 | 33.7 (25.0 to 41.5) |
| 10–14 wk | 1,659 | 274 | 69.2 (64.7 to 73.1) | 767 | 28.6 (20.9 to 35.6) |
| 15–19 wk | 10,788 | 4,013 | 53.6 (51.6 to 55.5) | 2,974 | 17.8 (13.4 to 21.9) |
| 20–24 wk | 68,757 | 29,100 | 47.4 (46.2 to 48.5) | 23,122 | 4.0 (1.9 to 6.1) |
| ≥25 wk | 147,721 | 46,103 | 43.5 (42.4 to 44.5) | 98,970 | −2.7 (−4.2 to −1.2) |
| Booster dose | |||||
| BNT162b2 | |||||
| 1 wk | 40,203 | 2,088 | 88.7 (88.1 to 89.2) | 17,921 | 58.8 (57.8 to 59.7) |
| 2–4 wk | 114,050 | 2,029 | 95.4 (95.1 to 95.6) | 49,169 | 62.4 (61.8 to 63.0) |
| 5–9 wk | 98,853 | 2,291 | 92.6 (92.2 to 92.9) | 55,864 | 52.9 (52.1 to 53.7) |
| ≥10 wk | 18,168 | 343 | 88.1 (86.7 to 89.3) | 15,570 | 39.6 (38.0 to 41.1) |
| mRNA-1273 | |||||
| 1 wk | 22,722 | 845 | 91.5 (90.9 to 92.1) | 8,391 | 68.0 (67.0 to 68.9) |
| 2–4 wk | 53,353 | 417 | 97.0 (96.7 to 97.3) | 23,165 | 70.1 (69.5 to 70.7) |
| 5–9 wk | 13,917 | 92 | 94.9 (93.8 to 95.9) | 8,736 | 60.9 (59.7 to 62.1) |
| ≥10 wk | 38 | 1 | 41 | ||
| ChAdOx1 nCoV-19 | |||||
| 1 wk | 107 | 10 | 77.1 (55.1 to 88.3) | 41 | 57.7 (37.6 to 71.3) |
| 2–4 wk | 294 | 19 | 82.3 (71.3 to 89.0) | 131 | 55.6 (44.4 to 64.6) |
| 5–9 wk | 264 | 12 | 83.3 (69.7 to 90.8) | 164 | 46.7 (34.3 to 56.7) |
| ≥10 wk | 48 | 0 | 36 | ||
| BNT162b2 | |||||
| Dose 1 | |||||
| 0–3 wk | 7,038 | 5381 | 45.2 (43.3 to 47.1) | 4,548 | 42.8 (40.3 to 45.1) |
| ≥4 wk | 29,759 | 466 | 72.3 (69.4 to 74.9) | 19,043 | 31.5 (29.9 to 33.1) |
| Dose 2 | |||||
| 2–4 wk | 9,516 | 240 | 90.9 (89.6 to 92.0) | 3,369 | 65.5 (63.9 to 67.0) |
| 5–9 wk | 20,163 | 981 | 85.5 (84.5 to 86.5) | 8,768 | 48.7 (47.1 to 50.2) |
| 10–14 wk | 61,014 | 5,562 | 78.7 (78.0 to 79.4) | 25,178 | 30.1 (28.7 to 31.5) |
| 15–19 wk | 144,172 | 17,077 | 74.4 (73.8 to 74.9) | 82,221 | 15.4 (14.2 to 16.6) |
| 20–24 wk | 72,018 | 10,348 | 67.4 (66.5 to 68.2) | 55,719 | 11.5 (10.1 to 12.9) |
| ≥25 wk | 51,625 | 8,531 | 62.7 (61.6 to 63.7) | 33,983 | 8.8 (7.0 to 10.5) |
| Booster dose | |||||
| BNT162b2 | |||||
| 1 wk | 29,459 | 631 | 92.3 (91.6 to 92.9) | 14,615 | 66.9 (66.1 to 67.6) |
| 2–4 wk | 64,874 | 1,220 | 95.1 (94.8 to 95.4) | 21,886 | 67.2 (66.5 to 67.8) |
| 5–9 wk | 110,306 | 3,769 | 91.8 (91.4 to 92.1) | 42,854 | 55.0 (54.2 to 55.8) |
| ≥10 wk | 61,534 | 1,222 | 89.9 (89.2 to 90.5) | 41,569 | 45.7 (44.7 to 46.7) |
| mRNA-1273 | |||||
| 1 wk | 12,718 | 195 | 93.7 (92.7 to 94.6) | 5,308 | 74.0 (73.1 to 74.9) |
| 2–4 wk | 20,045 | 147 | 96.6 (96.0 to 97.1) | 7,288 | 73.9 (73.1 to 74.6) |
| 5–9 wk | 5,311 | 40 | 94.9 (93.0 to 96.2) | 2,807 | 64.4 (62.6 to 66.1) |
| ≥10 wk | 53 | 1 | 33 | ||
| mRNA-1273 | |||||
| Dose 1 | |||||
| 0–3 wk | 1,429 | 134 | 60.1 (51.8 to 66.9) | 981 | 47.9 (43.1 to 52.3) |
| ≥4 wk | 3,122 | 448 | 57.4 (52.6 to 61.8) | 2,086 | 31.9 (27.3 to 36.1) |
| Dose 2 | |||||
| 2–4 wk | 855 | 13 | 94.5 (90.5 to 96.9) | 236 | 75.1 (70.8 to 78.7) |
| 5–9 wk | 2,286 | 73 | 91.8 (89.6 to 93.6) | 791 | 52.8 (48.2 to 57.1) |
| 10–14 wk | 9,822 | 674 | 84.1 (82.7 to 85.3) | 3,828 | 35.6 (32.7 to 38.4) |
| 15–19 wk | 20,017 | 1,627 | 82.8 (81.8 to 83.7) | 11,269 | 25.3 (23.2 to 27.4) |
| 20–24 wk | 8,961 | 1,224 | 76.2 (74.7 to 77.7) | 6,106 | 15.0 (11.6 to 18.2) |
| ≥25 wk | 474 | 16 | 80.4 (67.3 to 88.2) | 731 | 14.9 (3.9 to 24.7) |
| Booster dose | |||||
| BNT162b2 | |||||
| 1 wk | 2,031 | 15 | 95.5 (92.5 to 97.3) | 1,393 | 64.3 (61.7 to 66.8) |
| 2–4 wk | 2,063 | 8 | 94.7 (89.3 to 97.3) | 1,339 | 64.9 (62.3 to 67.3) |
| 5–9 wk | 20 | 0 | 16 | ||
| ≥10 wk | 0 | 0 | 3 | ||
| mRNA-1273 | |||||
| 1 wk | 1,851 | 14 | 95.3 (92.1 to 97.2) | 1,136 | 68.1 (65.6 to 70.5) |
| 2–4 wk | 1,913 | 5 | 96.4 (91.4 to 98.5) | 1,149 | 66.3 (63.7 to 68.8) |
| 5–9 wk | 4 | 0 | 7 | ||
Vaccine Effectiveness, Omicron Variant
Vaccine effectiveness was lower for the omicron variant than for the delta variant at all intervals after vaccination and for all combinations of primary courses and booster doses investigated. Among those who had received two ChAdOx1 nCoV-19 doses, almost no protective effect of vaccination against symptomatic disease caused by the omicron variant was noted from 20 to 24 weeks after the second dose. Among those who had received two BNT162b2 doses, vaccine effectiveness was 65.5% (95% CI, 63.9 to 67.0) 2 to 4 weeks after the second dose, dropping to 15.4% (95% CI, 14.2 to 16.6) after 15 to 19 weeks and dropping further to 8.8% (95% CI, 7.0 to 10.5) after 25 or more weeks. The vaccine effectiveness of two doses of mRNA-1273 vaccine had a similar reduction over time from 75.1% (95% CI, 70.8 to 78.7) after 2 to 4 weeks to 14.9% (95% CI, 3.9 to 24.7) after 25 or more weeks.
Among persons who received ChAdOx1 nCoV-19 as the primary course, from 2 to 4 weeks after a BNT162b2 booster dose, vaccine effectiveness increased to 62.4% (95% CI, 61.8 to 63.0) before waning to 39.6% (95% CI, 38.0 to 41.1) after 10 or more weeks. The mRNA-1273 booster vaccine increased effectiveness to 70.1% (95% CI, 69.5 to 70.7) after 2 to 4 weeks. This waned to 60.9% (95% CI, 59.7 to 62.1) after 5 to 9 weeks. Vaccine effectiveness was lowest among those who received a ChAdOx1 nCoV-19 primary course with a ChAdOx1 nCoV-19 booster vaccine. Waning efficacy was most notable against the omicron variant, for which the vaccine effectiveness was 46.7% (95% CI, 34.3 to 56.7) at 5 to 9 weeks.
Among persons who received BNT162b2 as the primary course, from 2 to 4 weeks after a BNT162b2 booster dose, vaccine effectiveness increased to 67.2% (95% CI, 66.5 to 67.8) before declining to 45.7% (95% CI, 44.7 to 46.7) after 10 or more weeks. The mRNA-1273 booster increased vaccine effectiveness to 73.9% (95% CI, 73.1 to 74.6) after 2 to 4 weeks before decreasing to 64.4% (95% CI, 62.6 to 66.1) after 5 to 9 weeks. After an mRNA-1273 primary course, vaccine effectiveness increased to 64.9% (95% CI, 62.3 to 67.3) at 2 to 4 weeks after a BNT162b2 booster and 66.3% (95% CI, 63.7 to 68.8) at 2 to 4 weeks after an mRNA-1273 booster.
Discussion
Our findings indicate that vaccine effectiveness against symptomatic disease caused by the omicron variant is substantially lower than with the delta variant. After two doses, vaccine effectiveness waned rapidly, with very limited vaccine effects seen from 20 weeks after the second dose of any vaccine. Booster doses resulted in a substantial increase in protection against mild infection; however, waning of protection against symptomatic disease was also seen after booster doses. We are unable to determine protection against severe forms of disease using the test-negative case–control method here owing to the small number of omicron cases resulting in hospitalization so far in our data set and the natural lag between infection and more severe outcomes. Previous experience with the delta variant in the United Kingdom suggested that protection against hospitalization after two doses of vaccine was well maintained.19
These findings are consistent with neutralization data for the omicron variant. South African, German, and U.K. studies indicate a reduction in neutralizing activity by a factor of 20 to 40 in serum specimens obtained from recipients of two doses of BNT162b2 as compared with neutralization against early pandemic viruses and by a factor of at least 10 as compared with neutralization against the delta variant.3-5,34 In serum specimens obtained from recipients of two doses of ChAdOx1 nCoV-19, a greater reduction in neutralizing activity was observed, with a high proportion of postvaccination serum samples having neutralizing activity below the limit of quantification in the assay.34 Higher neutralizing activity was observed after a booster dose.3,4,34
Although a correlation between neutralizing antibody levels and vaccine effectiveness against symptomatic SARS-CoV-2 infection has been observed at a population level,6 a similar correlation with effectiveness against severe disease is much less certain. With previous variants, vaccine effectiveness against severe disease, including hospitalization and death, has been higher and has been retained for a longer period than effectiveness against mild disease.16,19 Cellular immune responses are likely to play a relatively more important role in protection as antibodies wane with time since infection or vaccination; antibody waning may increase the risk of SARS-CoV-2 infection even as cellular immunity probably limits progression to severe disease.35 Cellular immunity is also likely to play an important role in protection against SARS-CoV-2 variants.35 Estimating effectiveness against severe disease caused by the omicron variant will follow a lag; however, on the basis of experience with other variants and early estimates of hospitalization rates, vaccine effectiveness against severe disease is likely to be substantially higher than the estimates against symptomatic disease.19,36
The populations that have received different vaccines as a primary course are different. For example, ChAdOx1 nCoV-19 was the main vaccine used early in the program in care homes and among persons in clinical risk groups. Furthermore, mRNA vaccines were the main vaccines used in persons younger than 40 years of age after the reported association between ChAdOx1 nCoV-19 and vaccine-induced thrombotic thrombocytopenia.37 Although adjustments were made for age and clinical risk factors, these age differences may explain some of the differences in the findings for the primary course — for example, the high vaccine effectiveness against omicron 2 to 9 weeks after the second dose of BNT162b2 is likely to be primarily among recently vaccinated young adults and teenagers. Differences are also noted in populations that have received a booster dose as compared with those who have received only two doses, with the former skewed toward older populations with more coexisting conditions. Persons who have not yet received a booster could have missed the boost for reasons that may be associated with exposure risk; for example, booster vaccination may have been delayed owing to an outbreak in a closed setting. The analysis with ChAdOx1 nCoV-19 boosters is particularly likely to be subject to bias because this vaccine was not recommended as a booster in the United Kingdom; therefore, some misclassification may have occurred, and persons who received ChAdOx1 nCoV-19 are likely to have done so because of contraindications to other vaccines.
The large scale of testing and sequencing in the United Kingdom, as well as the use of a national vaccination register, has enabled rapid evaluation of vaccine effectiveness against symptomatic infection with the omicron variant. Nevertheless, the study has some limitations, and findings should be interpreted with caution. During this early period of circulation of the new variant, a large proportion of cases occurred among travelers. Persons who reported foreign travel in the preceding 2 weeks were excluded from this analysis; however, not all travelers may have been excluded, and contacts of travelers will not have been identified. This group is likely to have different exposure than the wider population and may also have different levels of vaccine coverage. Therefore, residual confounding may be present. Owing to the relatively small number of omicron cases in the United Kingdom, our estimates are subject to considerable uncertainty, and we are unable to break down estimates according to population characteristics that may affect vaccine effectiveness (e.g., age and clinical risk group).19
In this analysis, our comparator group is unvaccinated persons, who make up a very small proportion of persons in several age cohorts. These persons are likely to differ from the general population according to characteristics that could confound our estimates of vaccine effectiveness. In this analysis that covers all ages, this type of confounding may be less of an issue than in analyses restricted to elderly populations. Furthermore, recent analyses using different control groups have shown good concordance between analyses using an unvaccinated control group and analyses estimating relative vaccine effectiveness among persons who have received a booster dose as compared with those who have received two doses. Booster doses have only recently been rolled out in England; therefore, we are able to estimate vaccine effectiveness for only a short period after booster vaccination, and we do not have information on the duration of protection after a booster dose. Some misclassification may also have occurred owing to both imperfect sensitivity and specificity of PCR testing, as well as the use of S target–negative status to identify omicron cases.
Our findings indicate that two doses of vaccination with BNT162b2 or ChAdOx1 nCoV-19 are insufficient to give adequate levels of protection against infection with the omicron variant and mild disease. Boosting with BNT162b2 or mRNA-1273 provided a substantial increase in protection against mild disease, although waning occurred over time. Boosters will probably offer even greater levels of protection against severe and fatal disease. Our findings support maximizing coverage with third doses of vaccine in highly vaccinated populations such as in the United Kingdom. Further follow-up will be needed to assess protection against severe disease and the duration of protection after booster vaccination.
Acknowledgments
We thank the UKHSA Covid-19 Data Science Team and the UKHSA Outbreak Surveillance Team and the staff of NHS England, NHS Digital, and NHS Test and Trace for their roles in developing and managing the Covid-19 testing, variant identification, and vaccination systems and data sets as well as the reporting NHS vaccinators and the staff of NHS laboratories, UKHSA laboratories, and lighthouse laboratories; the staff of the Wellcome Sanger Institute and other laboratories involved in whole-genome sequencing of Covid-19 samples; and the members of the Joint Committee on Vaccination and Immunisation and the U.K. Variant Technical Group for advice and feedback in developing this study.
Notes
Surveillance of coronavirus disease 2019 (Covid-19) testing and vaccination is undertaken under Regulation 3 of the Health Service (Control of Patient Information) Regulations 2002 to collect confidential patient information (www.legislation.gov.uk/uksi/2002/1438/regulation/3/made) under Sections 3(i) (a) to (c), 3(i)(d) (i) and (ii), and 3. The study protocol was subject to an internal review by the Public Health England Research Ethics and Governance Group and was found to be fully compliant with all regulatory requirements. Given that no regulatory issues were identified and that ethics review is not a requirement for this type of work, it was decided that a full ethics review would not be necessary.
This article was published on March 2, 2022, at NEJM.org.
Footnotes
Supported by the U.K. Health Security Agency (UKHSA). Dr. Simons is supported by a doctoral training grant from the Biotechnology and Biological Sciences Research Council (grant number BB/M009513/1).
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
Full text links
Read article at publisher's site: https://doi.org/10.1056/nejmoa2119451
Read article for free, from open access legal sources, via Unpaywall:
https://www.nejm.org/doi/pdf/10.1056/NEJMoa2119451?articleTools=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/123949384
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1056/nejmoa2119451
Article citations
Feasibility of snapshot testing using wearable sensors to detect cardiorespiratory illness (COVID infection in India).
NPJ Digit Med, 7(1):289, 19 Oct 2024
Cited by: 0 articles | PMID: 39427067 | PMCID: PMC11490565
Advancements in the Development of Anti-SARS-CoV-2 Therapeutics.
Int J Mol Sci, 25(19):10820, 09 Oct 2024
Cited by: 0 articles | PMID: 39409149 | PMCID: PMC11477007
Review Free full text in Europe PMC
A Comparison of the Outcomes of COVID-19 Vaccinated and Nonvaccinated Patients Admitted to an Intensive Care Unit in a Low-Middle-Income Country.
Crit Care Res Pract, 2024:9571132, 04 Oct 2024
Cited by: 0 articles | PMID: 39397887 | PMCID: PMC11469933
A prediction of mutations in infectious viruses using artificial intelligence.
Genomics Inform, 22(1):15, 08 Oct 2024
Cited by: 0 articles | PMID: 39380083 | PMCID: PMC11463117
The Influence of Initial Immunosuppression on the Kinetics of Humoral Response after SARS-CoV-2 Vaccination in Patients Undergoing Kidney Transplantation.
Vaccines (Basel), 12(10):1135, 03 Oct 2024
Cited by: 0 articles | PMID: 39460302 | PMCID: PMC11510881
Go to all (1,304) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Effect of mRNA Vaccine Boosters against SARS-CoV-2 Omicron Infection in Qatar.
N Engl J Med, 386(19):1804-1816, 09 Mar 2022
Cited by: 233 articles | PMID: 35263534 | PMCID: PMC8929389
Comparative effectiveness of different primary vaccination courses on mRNA-based booster vaccines against SARs-COV-2 infections: a time-varying cohort analysis using trial emulation in the Virus Watch community cohort.
Int J Epidemiol, 52(2):342-354, 01 Apr 2023
Cited by: 3 articles | PMID: 36655537 | PMCID: PMC10114109
Immunogenicity and reactogenicity against the SARS-CoV-2 variants following heterologous primary series involving CoronaVac, ChAdox1 nCov-19 and BNT162b2 plus BNT162b2 booster vaccination: An open-label randomized study in healthy Thai adults.
Hum Vaccin Immunother, 18(6):2091865, 11 Jul 2022
Cited by: 17 articles | PMID: 35816053 | PMCID: PMC9746495
The Omicron variant wave: Where are we now and what are the prospects?
J Chin Med Assoc, 86(2):135-137, 13 Dec 2022
Cited by: 1 article | PMID: 36524941
Review
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (1)
London Interdisciplinary Doctoral Programme
Prof Jonathan Ashmore, University College London
Grant ID: BB/M009513/1
Medical Research Council (2)
MRC Centre for Global Infectious Disease Analysis
Professor Neil Ferguson, Imperial College London
Grant ID: MR/R015600/1
Investigation of proven vaccine breakthrough by SARS-CoV-2 variants in established UK healthcare worker cohorts: SIREN consortium & PITCH Plus Pathway
Professor Susan Hopkins, Public Health England
Grant ID: MR/W02067X/1
National Institute for Health Research (NIHR) (1)
Grant ID: NIHR201395





