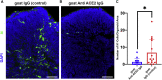
| PMC full text: | Published online 2022 Mar 24. doi: 10.1016/j.stemcr.2022.02.015
|
Figure 4
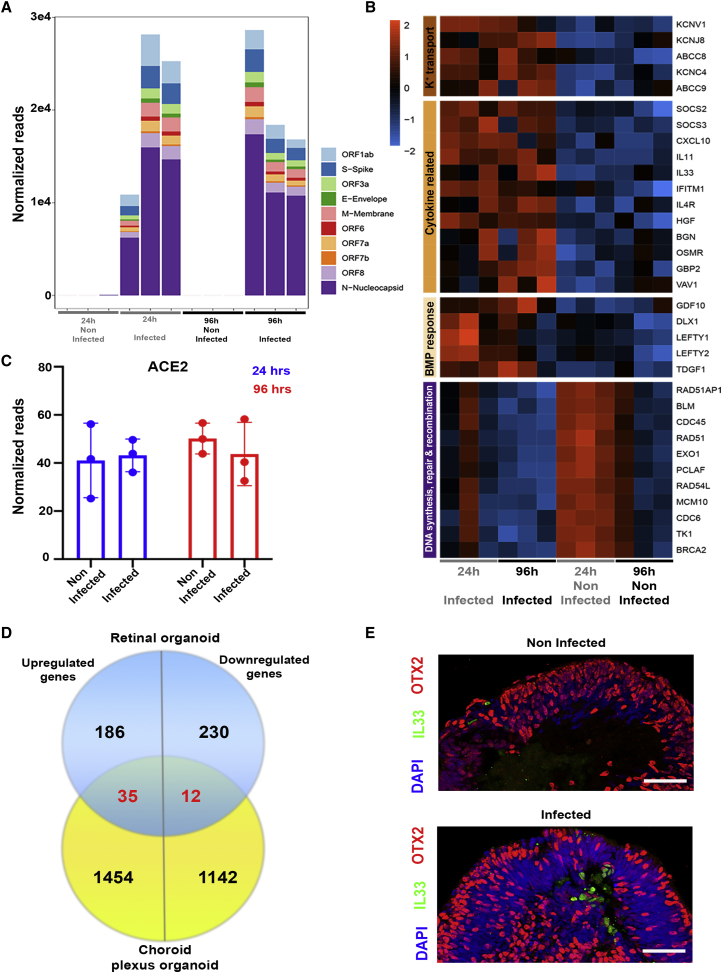
A transcriptomic analysis of SARS-CoV-2-infected retinal organoids
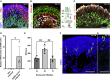
(A) A transcriptomic analysis was conducted using infected retinal organoids, incubated for 24 or 96 h following infection, and their respective controls. N = 3 per group. Each repeat included 5 separately treated organoids. SARS-CoV-2 transcripts were highly expressed in the infected samples.
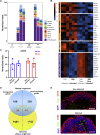
(B) Genes that were significantly differentially expressed (DE) with a fold change of at least 2 (or −2) among all of the samples together were used for Gene Ontology (GO) analysis. A heatmap of genes related to the top enriched categories identified using GO is presented.
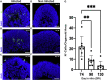
(C) The number of normalized reads of the ACE2 gene in noninfected and infected organoids was extracted from the analysis. A Student’s t test did not identify significant differences between the groups (p = 0.83 [24 h] and 0.48 [96 h]). Error bars represent standard deviation.
(D)The DE genes identified in the retinal organoids 24 h post-infection were compared to those identified in SARS-CoV-2-infected choroid plexus after 24 h. The comparison is presented as a Venn diagram.
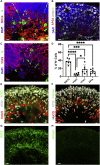
(E) Control and retinal organoids infected on day 74 of differentiation and incubated for 24 h before fixation were stained against IL-33 (green) and OTX2 (red). Scale bars: 50 μm.