Abstract
Background
The mechanisms underlying the known link between overweight/obesity and childhood asthma are unclear. We aimed to identify differentially expressed genes and pathways associated with obesity-related asthma through a transcriptomic analysis of nasal airway epithelium.Methods
We compared the whole transcriptome in nasal airway epithelium of youth with overweight or obesity and asthma with that of youth of normal weight and asthma, using RNA sequencing data from a cohort of 235 Puerto Ricans aged 9-20 years (EVA-PR) and an independent cohort of 66 children aged 6-16 years in Pittsburgh (VDKA). Differential expression analysis adjusting for age, sex, sequencing plate number, and sample sorting protocol, and the first five principal components were performed independently in each cohort. Results from the two cohorts were combined in a transcriptome-wide meta-analysis. Gene enrichment and network analyses were performed on top genes.Results
In the meta-analysis, 29 genes were associated with obesity-related asthma at an FDR-adjusted p <.05, including pro-inflammatory genes known to be differentially expressed in adipose tissue of obese subjects (e.g., CXCL11, CXCL10, and CXCL9) and several novel genes. Functional enrichment analyses showed that pathways for interferon signaling, and innate and adaptive immune responses were down-regulated in overweight/obese youth with asthma, while pathways related to ciliary structure or function were up-regulated. Upstream regulatory analysis predicted significant inhibition of the IRF7 pathway. Network analyses identified "hub" genes like GBP5 and SOCS1.Conclusion
Our transcriptome-wide analysis of nasal airway epithelium identified biologically plausible genes and pathways for obesity-related asthma in youth.Free full text

Differential Gene Expression in Nasal Airway Epithelium from Overweight or Obese Youth with Asthma
Abstract
Background:
The mechanisms underlying the known link between overweight/obesity and childhood asthma are unclear. We aimed to identify differentially expressed genes and pathways associated with obesity-related asthma through a transcriptomic analysis of nasal airway epithelium.
Methods:
We compared the whole transcriptome in nasal airway epithelium of youth with overweight or obesity and asthma with that of youth of normal weight and asthma, using RNA sequencing data from a cohort of 235 Puerto Ricans aged 9 to 20 years (EVA-PR) and an independent cohort of 66 children aged 6 to 16 years in Pittsburgh (VDKA). Differential expression analysis adjusting for age, sex, sequencing plate number, sample sorting protocol, and the first five principal components was performed independently in each cohort. Results from the two cohorts were combined in a transcriptome-wide meta-analysis. Gene enrichment and network analyses were performed on top genes.
Results:
In the meta-analysis, 29 genes were associated with obesity-related asthma at an FDR-adjusted P <0.05, including pro-inflammatory genes known to be differentially expressed in adipose tissue of obese subjects (e.g., CXCL11, CXCL10, and CXCL9) and several novel genes. Functional enrichment analyses showed that pathways for interferon signaling, and innate and adaptive immune responses were down-regulated in overweight/obese youth with asthma, while pathways related to ciliary structure or function were up-regulated. Upstream regulatory analysis predicted significant inhibition of the IRF7 pathway. Network analyses identified “hub” genes like GBP5 and SOCS1.
Conclusion:
Our transcriptome-wide analysis of nasal airway epithelium identified biologically plausible genes and pathways for obesity-related asthma in youth.
INTRODUCTION
Asthma and obesity are public health problems in the United States, particularly in minority groups such as Puerto Ricans and African Americans1,2. Overweight and obesity are associated with childhood asthma3–8, and obesity-related asthma is increasingly recognized as a distinct asthma phenotype9–15 with worse asthma severity16–20 and reduced response to bronchodilators and inhaled corticosteroids21,22.
The mechanisms underpinning obesity-related asthma remain unclear23. Various “omics” approaches have been used to investigate overweight/obesity and asthma, including genome-wide association sudies24, analysis of gene expression or DNA methylation in peripheral blood mononuclear cells (PBMCs)25,26, and similar studies in CD4+ T cells27,28. To date, most studies have focused on blood biomarkers due to easy access, while only a few have examined airway epithelium, a tissue highly relevant to asthma.
Gene expression in the nasal epithelium is highly correlated with that in bronchial epithelium29, and thus the nasal airway epithelium can serve as a surrogate marker for transcriptomics in the lower airways. We recently showed that transcriptomic profiles in nasal epithelium accurately differentiate youth with atopic asthma from non-atopic controls30. Based on our prior findings, we hypothesized that transcriptomic profiles in nasal epithelium would differentiate youth with overweight/obesity and asthma (“OOA”) from those with normal weight and asthma (“NWA”). To test this hypothesis, we conducted a transcriptome-wide association study (TWAS) in nasal airway epithelium from youth in Puerto Rico and Pittsburgh (PA), which are at high risk for both overweight or obesity and asthma.
METHODS
Please also see the Online Supplement.
The Epigenetic Variation and Childhood Asthma in Puerto Ricans Study (EVA-PR)
Details on subject recruitment and study procedures for EVA-PR, a case-control study of asthma in Puerto Rico, have been previously described31. In brief, youth ages 9 to 20 years were recruited in San Juan and Caguas (Puerto Rico) from February 2014 to May 2017 using a multistage probabilistic sampling. Of the 543 participants, 478 had complete data on RNA sequencing (RNA-seq) from nasal epithelial samples, asthma (defined as physician-diagnosed asthma and current wheeze), body mass index (BMI) z-scores32, and relevant covariates. Atopy was defined as a positive IgE (≥ 0.35 IU/mL) to ≥1 of 5 common aeroallergens in Puerto Rico: dust mite (Der p 1), cockroach (Bla g 2), cat dander (Fel d 1), dog dander (Can f 1), and mouse urinary protein (Mus m 1). Of these 478 participants, 235 had asthma and were included in the current analysis of OOA. The study was approved by the Institutional Review Boards (IRBs) of the University of Puerto Rico (San Juan, PR) and the University of Pittsburgh (Pittsburgh, PA). Written parental consent and child assent were obtained for participants under 18 years old, and written consent was obtained from participants 18 years and older.
The Vitamin D Kids Asthma Study (VDKA)
VDKA was a randomized, double-blind, placebo-controlled clinical trial of vitamin D3 supplementation to prevent severe asthma exacerbations in children ages 6 to 16 years33. Children with mild persistent asthma, ≥1 severe asthma exacerbation in the previous year, and a serum vitamin D < 30 ng/ml were recruited from seven U.S. sites. Of the 115 children randomized at the Pittsburgh site, 66 had complete data for RNA-seq from nasal epithelial samples (collected prior to randomization and processed using the same protocol as in EVA-PR), BMI z-scores, and relevant covariates. Atopy was defined as a total IgE ≥ 100 IU/mL or a positive IgE (≥ 0.35 IU/mL) to either dust mite (Der p 1) or cockroach (Bla g 2). The study was approved by the IRBs of all participating institutions, and the RNA-Seq ancillary study was approved by the IRB of the University of Pittsburgh. Written parental consent and assent were obtained from all participants.
RNA-Seq and data pre-processing
RNA-Seq and data preprocessing for both studies (EVA-PR and VDKA) were performed following previously described protocols30,34. In brief, RNA was extracted from nasal epithelial samples collected from the inferior turbinate. RNA-Seq was performed at the Genomics Research Core of the University of Pittsburgh, using 350 ng high-quality RNA (RNA Integrity Number [RIN] >7). Library preparation was done using TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold High Throughput Kit (Illumina), which removes both cytoplasmic and mitochondrial ribosomal RNA (rRNA), according to the manufacturer’s protocol. Libraries were run on the NextSeq 500 with NextSeq® 500/550 High Output Kit v2 (Illumina), using paired-end reads at 75 cycles and 80M reads per sample.
Quality control for raw RNA sequencing FASTQ files was conducted using FastQC35 and summarized using MultiQC36. 3′ adapters and low-quality reads were trimmed with Trim Galore!37 and Cutadapt38. Trimmed reads were aligned to the latest human reference genome at the time of the studies (hg19 for EVA-PR and hg38 for VDKA; reads mapped to hg19 and hg38 are highly concordant39) using STAR40, and subsequently annotated in the Illumina iGenomes database using RSEM41. Samples with low alignment rate were not included in the analysis. After preprocessing and QC, genes with low expression levels (mean count < 2) were removed from downstream analyses, and a total of 18,321 and 19,082 genes were retained for the analyses in EVA-PR and VDKA, respectively. In EVA-PR, genome-wide genotypes were assessed as previously described42, and principal components (PCs) were calculated from genotypes to adjust for population stratification. In VDKA, where genome-wide genotypes were not available, genotypes were imputed from RNA-seq reads using HaplotypeCaller in GATK43, and PCs were calculated to adjust for population stratification.
Statistical Analysis
Overweight or obesity was defined as a BMI z-score ≥ 85th percentile, and normal weight was defined as a BMI z-score < 85th percentile, with percentiles determined using CDC charts32. We compared the transcriptomic profiles of youth with OOA to those of youth with NWA.
Differentially expressed genes (DEGs) were identified based on a raw count table, using multivariable negative binomial regression framework in DESeq244. In EVA-PR, the multivariable model was adjusted for age, sex, sequencing plate number, sample enrichment protocol (CD326+ enriched or not, see Online Supplement), and the first five PCs derived from genotype data. In VDKA, all samples were sequenced in one batch and the multivariable analysis was adjusted for age, sex, and the first five PCs derived from imputed genotypic data. Effect size (the log2-fold change [L2FC] in gene expression) and P-value for each gene were calculated based on these multivariable models. To reduce the impact of low abundance genes and improve estimation stability of effect sizes, L2FC were estimated using a zero-mean normal prior on the non-intercept coefficients. L2FC were transformed to fold change and shown in the tables. A transcriptome-wide meta-analysis was then conducted to combine the results from both cohorts using the weighted sum of z-scores method, a fixed effect model approach45. The Benjamini-Hochberg (BH) false discovery rate (FDR) method was applied to adjust for multiple testing, and significance was defined as an FDR-adjusted P-value < 0.05 in the meta-analysis, provided that the direction of the association was the same in EVA-PR and VDKA.
Functional Enrichment Analysis
Gene Ontology (GO) enrichment analysis was performed to identify over-represented biological process among DEGs in the meta-analysis46,47. This analysis was conducted using Fisher’s exact test in PANTHER48, with FDR adjustment for multiple testing. We then performed an upstream regulatory analysis of DEGs using Ingenuity Pathway Analysis (IPA)49. Finally, Gene Set Enrichment Analysis (GSEA)50 was performed for all 17,770 genes (representing the overlap between the 18,321 genes included in EVA-PR and the 19,082 genes included in VDKA) in the meta-analysis, using function gseGO() in R package clusterProfiler51. Genes were ranked by L2FC from the meta-analysis.
Gene Network Analysis:
see Online Supplement.
RESULTS
The main characteristics of the participants in the two study cohorts are shown in Table 1. In both EVA-PR and VDKA, subjects with OOA had a higher FVC %predicted but a slightly lower FEV1/FVC ratio than those with NWA, consistent with prior studies52,53. In VDKA, subjects with OOA were more likely to be male than those with NWA. There were no significant differences in age, atopy status, the frequency of severe exacerbations, or medication use between OOA and NWA in either cohort.
Table 1
Characteristics of participants in the EVA-PR (Epigenetic Variation and Childhood Asthma) and VDKA (Vitamin D Kids Asthma) studies, according to presence of overweight or obese asthma (OOA) vs. normal weight asthma (NWA)
| EVA-PR† | VDKA‡ | |||
|---|---|---|---|---|
| OOA | NWA | OOA | NWA | |
|
| ||||
| Subjects | 107 | 128 | 37 | 29 |
| Age years | 15.0 [13.0, 17.5] | 15.0 [13.0, 17.0] | 11.0 [9.00, 12.0] | 10.0 [8.00, 11.0] |
| Female | 49 (45.8%) | 52 (40.6%) | 13 (35.1%) | 14 (48.3%) |
| Race | ||||
 Puerto Rican Puerto Rican | 107 (100.0%) | 128 (100.0%) | ||
 Caucasian Caucasian | 7 (18.9%) | 11 (37.9%) | ||
 African American African American | 22 (59.5%) | 16 (55.2%) | ||
 Other Other | 8 (21.6%) | 2 (6.9%) | ||
| BMI z-score | 1.75 [1.38, 2.18]*** | 0.17 [−0.39, 0.61] | 1.83 [1.36, 2.31]*** | −0.12 [−0.68, 0.64] |
| FEV1 % predicted | 98.2 [87.9, 105] | 95.3 [86.8, 102] | 91.6 [82.8, 99.7] | 90.8 [76.2, 99.6] |
| FVC % predicted | 104 [95.9, 112]* | 98.3 [92.9, 108] | 107 [92.8, 114]* | 98.6 [87.2, 113] |
| FEV1/FVC ratio | 0.84 [0.78, 0.87]* | 0.84 [0.79, 0.90] | 0.77 [0.72, 0.82] | 0.82 [0.77, 0.86] |
| Atopy | 77 (72.0%) | 88 (68.8%) | 33 (89.2%) | 23 (79.3%) |
| Severe exacerbation§ | 22 (20.6%) | 17 (13.3%) | 14 (37.8%) | 13 (44.8%) |
| Inhaled steroid | 27 (25.2%) | 35 (27.3%) | N/A¶ | N/A¶ |
| Nasal steroid | 2 (1.9%) | 7 (5.5%) | N/A¶ | N/A¶ |
| Leukotriene | 25 (23.4%) | 29 (22.7%) | N/A¶ | N/A¶ |
Data are presented as number (percentage) for categorical variables or median [Q1, Q3] for continuous variables.
TWAS of overweight or obesity-related asthma (OOA)
The TWAS revealed 7 genes with FDR-adjusted P < 0.05 for OOA in EVA-PR and 50 genes with FDR-adjusted P-value < 0.05 for OOA in VDKA (Table S1). The transcriptome-wide meta-analysis of both cohorts identified 29 DEGs (FDR-adjusted P < 0.05 in the meta-analysis, with the same direction of association in both cohorts) in OOA (Figure 1, Table 2), including CXCL11, CXCL10, CXCL9, CCL8, ISG15, GBP5, GBP1, SOCS1, GZMB, IFI35, and ISG20.
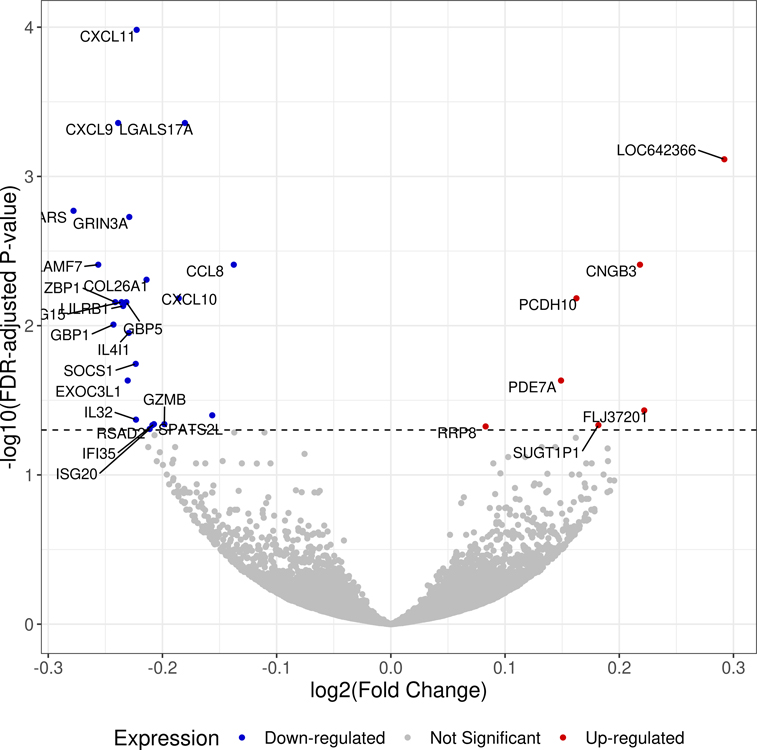
Volcano plot showing the log2 (fold change) and −log10 (FDR-adjusted P-value) of all the genes in the transcriptome-wide meta-analysis of the two study cohorts.
Footnote: Genes that are differentially expressed between overweight or obese subjects with asthma and subjects with asthma of normal weight are colored in red (up-regulated) and blue (down-regulated). Statistical significance for differential expression is defined as a false discovery rate (FDR)-adjusted P-value < 0.05.
Table 2
Top differentially expressed genes identified in the meta-analysis
| Gene Symbol | EVA-PR | VDKA | Meta-Analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Average Expression* | Fold-Change | P-value | Average Expression* | Fold-Change | P-value | Fold-Change | P-value | FDR P-value | |
|
| |||||||||
| CXCL11 | 20.26 | 0.88 | 9.75E-06 | 17.62 | 0.61 | 5.27E-08 | 0.86 | 5.86E-09 | 1.04E-04 |
| CXCL9 | 22.31 | 0.88 | 3.70E-05 | 27.07 | 0.63 | 1.98E-06 | 0.85 | 6.55E-08 | 4.38E-04 |
| LGALS17A | 1.13 | 0.91 | 4.54E-05 | 2.69 | 0.65 | 1.47E-07 | 0.88 | 7.40E-08 | 4.38E-04 |
| LOC642366 | 0.66 | 1.21 | 7.46E-06 | 0.32 | 1.35 | 3.98E-03 | 1.22 | 1.73E-07 | 7.68E-04 |
| WARS | 277.20 | 0.86 | 3.09E-04 | 331.55 | 0.61 | 3.99E-06 | 0.82 | 4.78E-07 | 1.70E-03 |
| GRIN3A † | 0.06 | 0.87 | 1.08E-04 | 0.01 | 0.78 | 6.74E-04 | 0.85 | 6.31E-07 | 1.87E-03 |
| CCL8 | 1.19 | 0.92 | 2.50E-05 | 0.22 | 0.79 | 3.79E-03 | 0.91 | 1.65E-06 | 3.90E-03 |
| CNGB3 † | 0.05 | 1.16 | 1.24E-05 | 0.05 | 1.21 | 4.96E-02 | 1.16 | 1.82E-06 | 3.90E-03 |
| SLAMF7 | 7.09 | 0.83 | 1.31E-05 | 4.81 | 0.85 | 5.61E-02 | 0.84 | 1.98E-06 | 3.90E-03 |
| COL26A1 † | 0.09 | 0.86 | 6.47E-06 | 0.07 | 0.88 | 1.96E-01 | 0.86 | 2.77E-06 | 4.92E-03 |
| CXCL10 | 80.23 | 0.91 | 7.83E-04 | 100.95 | 0.65 | 3.49E-06 | 0.88 | 4.43E-06 | 6.55E-03 |
| ZBP1 | 2.73 | 0.88 | 1.11E-03 | 2.66 | 0.64 | 2.12E-05 | 0.85 | 5.74E-06 | 6.96E-03 |
| ISG15 | 121.24 | 0.88 | 9.47E-04 | 154.93 | 0.64 | 2.85E-05 | 0.85 | 5.79E-06 | 6.96E-03 |
| GBP5 | 24.31 | 0.88 | 1.18E-03 | 15.33 | 0.64 | 1.40E-05 | 0.85 | 5.87E-06 | 6.96E-03 |
| LILRB1 | 2.50 | 0.84 | 3.91E-06 | 0.44 | 0.95 | 6.17E-01 | 0.85 | 6.64E-06 | 7.38E-03 |
| GBP1 | 73.30 | 0.89 | 6.41E-03 | 75.43 | 0.57 | 1.54E-07 | 0.85 | 9.41E-06 | 9.84E-03 |
| IL4I1 | 4.51 | 0.86 | 5.68E-05 | 0.94 | 0.83 | 7.57E-02 | 0.85 | 1.13E-05 | 1.12E-02 |
| SOCS1 | 6.66 | 0.88 | 1.09E-03 | 5.11 | 0.69 | 4.32E-04 | 0.86 | 1.93E-05 | 1.80E-02 |
| PDE7A | 9.24 | 1.08 | 6.46E-03 | 6.39 | 1.23 | 9.99E-05 | 1.11 | 2.74E-05 | 2.33E-02 |
| EXOC3L1 † | 0.28 | 0.89 | 3.48E-03 | 0.24 | 0.67 | 8.01E-05 | 0.85 | 2.76E-05 | 2.33E-02 |
| FLJ37201 | 0.84 | 1.15 | 5.17E-04 | 0.21 | 1.25 | 2.28E-02 | 1.17 | 4.58E-05 | 3.70E-02 |
| SPATS2L | 42.34 | 0.93 | 1.92E-02 | 55.74 | 0.77 | 1.45E-05 | 0.90 | 5.16E-05 | 3.99E-02 |
| IL32 | 11.54 | 0.87 | 9.07E-04 | 15.66 | 0.76 | 1.06E-02 | 0.86 | 5.76E-05 | 4.26E-02 |
| GZMB | 12.53 | 0.88 | 6.54E-04 | 5.61 | 0.78 | 1.78E-02 | 0.87 | 6.45E-05 | 4.57E-02 |
| IFI35 | 40.98 | 0.88 | 1.23E-03 | 66.37 | 0.79 | 9.74E-03 | 0.87 | 6.69E-05 | 4.57E-02 |
| RSAD2 | 21.09 | 0.90 | 6.29E-03 | 15.46 | 0.65 | 4.57E-05 | 0.87 | 7.06E-05 | 4.64E-02 |
| SUGT1P1 | 3.99 | 1.09 | 1.32E-02 | 2.26 | 1.33 | 7.40E-05 | 1.13 | 7.31E-05 | 4.64E-02 |
| RRP8 | 17.92 | 1.04 | 1.08E-02 | 14.48 | 1.14 | 9.91E-05 | 1.06 | 7.72E-05 | 4.73E-02 |
| ISG20 | 41.97 | 0.89 | 2.98E-03 | 61.95 | 0.77 | 2.95E-03 | 0.86 | 8.31E-05 | 4.92E-02 |
Since EVA-PR also included 243 children and adolescents without asthma, we examined the association between these DEGs and overweight/obesity in participants without asthma. Of the 29 DEGs identified in OOA, only five (CXCL11, CCL8, LILRB1, SOCS1, and GZMB) showed nominal associations (P < 0.01) in the same direction in children without asthma, and none were significant after FDR adjustment (P > 0.10 in all instances) (Table S2). Furthermore, and to evaluate the role of atopy in our analysis, we built models additionally adjusting for atopy status, obtaining similar results to those from our original models (Table S3).
Functional enrichment analysis
We found 71 over-represented Gene Ontology (GO) biological processes (FDR-adjusted P< 0.05) for the 29 DEGs associated with OOA. The top 10 over-represented processes, hierarchically clustered and sorted by fold-enrichment, included interferon-gamma production and type-I interferon signaling pathway, T cell and neutrophil chemotaxis, and chemokine-mediated signaling pathways (Table 3). Moreover, upstream regulatory analysis based on the 29 DEGs predicted significant inhibition of interferon pathways in OOA (Table S4). Top inhibited upstream regulators in OOA include IRF7 (P-value = 2.53 × 10−19) (Figure S1) and IFNG (P-value = 7.16 × 10−12). These were confirmed by direct analysis of our transcriptome-wide data, which showed that the expression of IRF7 and IFNG were significantly down-regulated in OOA in both EVA-PR (IRF7: P=0.03; IFNG: P= 0.05) and VDKA (IRF7: P= 0.04; IFNG: P= 0.006). DEGs including CXCL9, CXCL10, CCL8, ISG15, GBP5, GBP1, IL4I1, SOCS1, IFI35, RSAD2, and ISG20, were predicted to be regulated by both IRF7 and IFNG.
Table 3
Top gene ontology (GO) biological processes over-represented by the top DEGs in the meta-analysis
| GO Biological Process | Fold Enrichment | P-value | FDR-adjusted P-value | Gene Symbol |
|---|---|---|---|---|
|
| ||||
| Negative regulation of CD8-positive, alpha-beta T cell activation | > 100 | 4.26E-05 | 1.32E-02 | LILRB1, SOCS1 |
| Interferon-gamma production | > 100 | 8.35E-05 | 2.31E-02 | ISG15, LILRB1 |
| T cell chemotaxis | > 100 | 1.81E-04 | 4.21E-02 | CXCL10, CXCL11 |
| Positive regulation of release of sequestered calcium ion into cytosol | 56.58 | 2.44E-05 | 8.74E-03 | CXCL10, CXCL11, CXCL9 |
| Type I interferon signaling pathway | 46.6 | 1.93E-06 | 1.38E-03 | ISG15, IFI35, RSAD2, ISG20 |
| Negative regulation of viral genome replication | 44.01 | 4.98E-05 | 1.48E-02 | ISG15, RSAD2, ISG20 |
| Neutrophil chemotaxis | 41.15 | 3.09E-06 | 1.81E-03 | CXCL10, CXCL11, CCL8, CXCL9 |
| Chemokine-mediated signaling pathway | 40.11 | 3.41E-06 | 1.85E-03 | CXCL10, CXCL11, CCL8, CXCL9 |
| Killing of cells of other organisms | 37.72 | 4.31E-06 | 2.19E-03 | CXCL10, CXCL11, CCL8, CXCL9 |
| Antimicrobial humoral immune response mediated by antimicrobial peptide | 30.76 | 9.38E-06 | 3.70E-03 | CXCL10, CXCL11, CCL8, CXCL9 |
To overcome the limitations of the analysis based on manually selected genes by P-value or fold-change, we also performed a Gene Set Enrichment Analysis (GSEA) using the unfiltered, whole-genome, ranked gene list (including all 17,770 genes in the meta-analysis), finding 1,412 enriched GO biological processes at FDR-adjusted P <0.05. The top 20 up-regulated and down-regulated pathways, by normalized enrichment score, are listed in Figure 2. Most up-regulated pathways in OOA were related to ciliary structure or function, while most down-regulated pathways in OOA were related to interferon signaling, innate immune responses, and adaptive immune responses pathways.
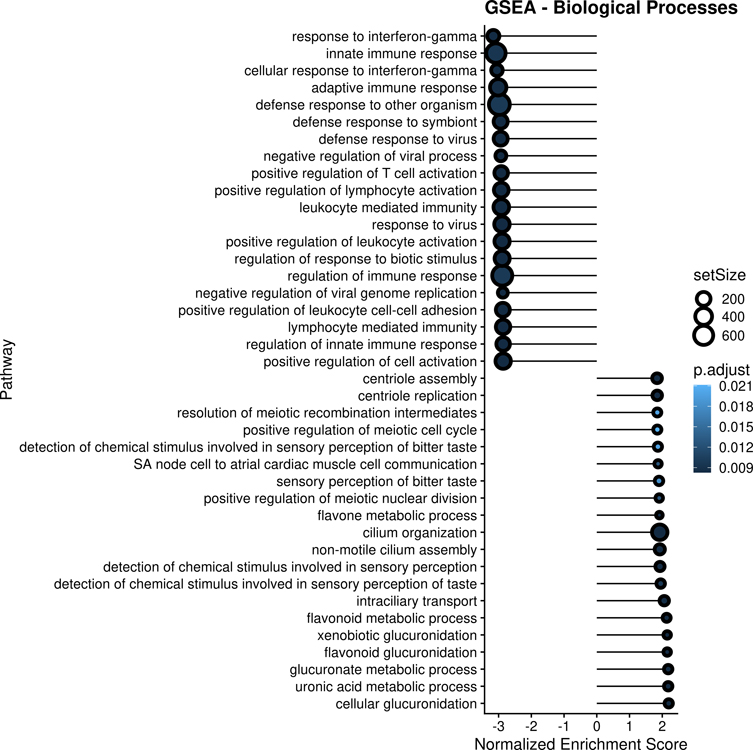
Gene Set Enrichment Analysis (GSEA) of the 17,770 genes included in the meta-analysis.
Footnote: Top 20 up-regulated and top 20 down-regulated significantly enriched Gene Ontology (GO) biological processes are shown. Top pathways are ordered by Normalized Enrichment Score (NES). The size of each circles represents the number of genes included in the gene set for each pathway. The darkness of each circle represents the level of statistical significance (false discovery rate [FDR]-adjusted P-value) for each pathway.
Gene Network analysis
Gene co-expression network analysis in EVA-PR showed differences between OOA and NWA (Kolmogorov-Smirnov P=0.046), indicating that the way genes are connected (co-expressed) differ between the two groups. We identified “hub” genes unique to OOA (GBP5), hub genes unique to NWA (ISG15, SPATS2L, and IFI35), and hub genes shared by both (SOCS1). In addition, we found 22 gene pairs whose co-expression patterns were significantly different by overweight/obesity status (Table S5). For instance, the co-expression between CXCL9 and CXCL10 was markedly higher in OOA (partial correlation= 0.53, P=1.93 × 10−11) than in NWA (partial correlation=0.36, P=6.78 × 10−7) with a significant difference between the two groups (P=7.09 × 10−3). Some gene pairs were co-expressed in OOA only (e.g., PDE7A and IL32, OOA P=1.56 × 10−4, NWA P=0.98) whereas others were co-expressed in NWA only (e.g., IL32 and IFI35, OOA P=0.13, NWA P=9.96 × 10−3).
Among the 29 DEGs in OOA, 25 genes could be mapped to corresponding proteins in the STRING database and were thus included in a protein-protein interaction analysis: 53 interactions were identified among the 25 proteins (Figure S2), showing significant enrichment (P <1.0 × 10−16) compared with a random set of proteins of similar size. STRING literature analysis also revealed significant enrichment (Table S6), with many studies related to innate and adaptive immune processes.
DISCUSSION
To our knowledge, this is the first TWAS of overweight/obesity-related childhood asthma in nasal airway epithelium. We report 29 genes whose expression differs between overweight/obese children with asthma and children with asthma of normal weight. None of these DEGs were significantly associated with overweight/obesity in children without asthma.
Since both obesity and asthma have significant heritability23,54,55, there have been several studies of the genomics of obesity-related asthma56. While studies looking at shared polymorphisms (e.g., candidate-gene or genome-wide association studies) have not yielded as many significant results as expected57,58, other approaches have led to more promising results. SNP microarray studies identified a 0.45-Mb genomic inversion on chromosome 16p11.2 associated with obesity-related asthma24. Smaller studies using PBMCs have identified genes whose expression25 or DNA methylation patterns differed between asthmatic children with or without obesity26. More recently, Rastogi et al. integrated epigenomics and transcriptomics in CD4+ T cells from asthmatic children with or without obesity27,28. Our study builds on those prior reports by examining the transcriptomics of OOA in airway epithelium. Although there are no overlapping DEGs between our study and those in CD4+ T cells, CDC42 and Rho-GTPase pathways (reported to be enriched in CD4+ T cells from obese subjects with asthma27,28) interact with the DEGs and pathways in the current analysis. CDC42 is a member of the Rho GTPase family59, and both CDC42 and Rho-GTPase pathways are involved in chemokine signaling pathways60,61.
We identified several DEGs previously implicated in obesity and/or asthma, as well as several genes related to non-atopic, interferon-related immune pathways. CXCL9, CXCL10, and CXCL11 code for C-X-C motif chemokine ligands involved in Th1-type immunoregulatory and inflammatory responses. Their shared receptor, CXCR3, and CCL8 (differentially expressed in our analysis) regulate interferon-mediated leukocyte chemotactic migration. In particular, the CXCL10/CXCR3 axis mediates the migration of mast cells to airway smooth muscle in subjects with asthma62. CD4+ T cell expression of CXCR3 is lower in obese patients with severe asthma63, while leptin enhances production of CXCL10 by lung fibroblasts63. In a murine model of genetic susceptibility to obesity, ozone exposure led to airway inflammation, as well as to up-regulation of CXCL11 and other inflammatory biomarkers in visceral adipose tissue64.
Although CXCL9, CXCL10, and CXCL11 were down-regulated in nasal epithelium from youth with OOA in the current study, others have reported up-regulation of these genes in adipose tissue from obese individuals65. This highlights the importance of studying different tissues relevant to a disease process; in obese subjects, cytokine gene expression may be strongly modified by asthma and differ between adipose tissue and respiratory epithelium. Moreover, negative feedback mechanisms may exist across different tissues. In a murine model of autoimmune uveitis, for instance, type I interferons can induce splenic sequestration of CD4+ T cells, with reduced migration into the eyes; this results in lower expression of CXCR3, CXCL9, CXCL10, and CXCL11 in the eyes, but higher expression in spleen66. We surmise that up-regulated CXCL9, CXCL10, and CXCL11 from adipose tissue could induce a low-grade systemic interferon response; this, combined with interferon-mediated responses related to asthma67, could elicit inhibited expression of CXCL9, CXCL10, and CXCL11 in the airways of OOA –which could in turn contribute to an impaired response to respiratory viral infections in patients with OOA.
Other DEGs in our analysis, such as SOCS1, IL32, ZBP1, and GBP5, may plausibly influence the pathogenesis of OOA. SOCS1 plays an important role in the attenuation of cytokine signaling -including IL-6 and leukemia inhibitory factor (LIF) pathways- and up-regulation of SOCS1 may lead to interferon deficiency in asthma68. IL-32 can be secreted by NK cells and monocytes, and in turn it can induce IL-6, IL-8, and TNF-alpha production69. IL32 expression in the liver has been reported as a potential biomarker of non-alcoholic fatty liver disease70. Moreover, IL32 transgenic mice fed a normal diet develop greater accumulation of white adipose tissue, as well as adipokine profiles similar to those in metabolic syndrome71. ZBP1 affects innate immune responses by inducing type-I interferon production in response to Z-RNA structures produced by different viruses including influenza A72. GBP5, the main “hub” gene in our analysis of OOA, can activate the NLRP3 inflammasome73. Obese asthmatic adults have increased sputum expression of NLRP374,75, while murine models of obesity-related asthma have shown NLRP3 inflammasome-dependent airway inflammation and hyperreactivity76,77. IRF7, the predicted upstream regulator of 12 out of 29 DEGs, is involved in adipocyte hypertrophy78, metabolic abnormalities79, and susceptibility to influenza80. IRF7 is also down-regulated in influenza-infected mice fed a high-fat diet, compared with their low-fat counterparts81.
We recognize several study limitations. First, we cannot examine temporal relationships between transcriptomic profiles and the development of OOA in this cross-sectional analysis. Second, VDKA differed from EVA-PR with regard to race/ethnicity, asthma severity, and sample size. While EVA-PR included Puerto Ricans and children with various degrees of asthma severity, VDKA was a multi-ethnic study of children with mild persistent asthma and at least one severe asthma exacerbation in the prior year. Such differences and the small sample size of VDKA may explain why only 1 of our 7 genes with FDR-adjusted P< 0.05 in EVA-PR (CXCL11) was fully replicated in the VDKA study. Despite such differences, our meta-analysis of TWAS in nasal epithelial samples from the two cohorts identified biologically plausible genes and pathways for OOA in youth. Third, participants in VDKA had low vitamin D levels. However, 66.2% of the VDKA participants included in this analysis had a serum vitamin D ≥20 ng/ml, which has not been consistently associated with asthma or other disease outcomes. Moreover, a differential expression analysis additionally adjusting for serum vitamin D levels (whether or not higher than 20 ng/ml) yielded similar results (Table S7). Finally, we lacked adequate data on some potential confounders or modifiers of the estimated effects of OOA on gene expression, such as diet.
In summary, we identified 29 differentially expressed genes in nasal airway epithelium from overweight or obese youth with asthma, including pro-inflammatory genes that are differentially expressed in adipose tissue of obese subjects (e.g., CXCL11, CXCL10, and CXCL9) and several novel genes and pathways that warrant further study for potential development of biomarkers and new therapies.
Financial support
This work was supported by grants HL079966, HL117191, MD011764, and HL119952 (to J.C.C.), and U54 MD007587 (to the University of Puerto Rico) from the U.S. National Institutes of Health (NIH). Dr. Forno’s contribution was supported by NIH grant HL149693. Dr. Chen’s contribution was supported by NIH grant HL150431.
Footnotes
Conflicts of interest
Dr. Celedón has received research materials from Pharmavite (vitamin D and placebo capsules) and GSK and Merck (inhaled steroids) in order to provide medications free of cost to participants in VDKA and another NIH-funded study unrelated to the current work. The other authors report no conflicts of interest.
Material in the electronic repository: online methods, data sharing statement, tables, and figures.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1111/pai.13776
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9047012
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/127347353
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1111/pai.13776
Article citations
Body Weight and Allergic Asthma: A Narrative Review.
J Clin Med, 13(16):4801, 15 Aug 2024
Cited by: 0 articles | PMID: 39200943 | PMCID: PMC11355285
Review Free full text in Europe PMC
The functional role of CST1 and CCL26 in asthma development.
Immun Inflamm Dis, 12(1):e1162, 01 Jan 2024
Cited by: 1 article | PMID: 38270326 | PMCID: PMC10797655
Nasal epithelial gene expression and total IgE in children and adolescents with asthma.
J Allergy Clin Immunol, 153(1):122-131, 22 Sep 2023
Cited by: 0 articles | PMID: 37742934
Mechanistic Links Between Obesity and Airway Pathobiology Inform Therapies for Obesity-Related Asthma.
Paediatr Drugs, 25(3):283-299, 19 Jan 2023
Cited by: 4 articles | PMID: 36656428
Review
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Cis- and trans-eQTM analysis reveals novel epigenetic and transcriptomic immune markers of atopic asthma in airway epithelium.
J Allergy Clin Immunol, 152(4):887-898, 02 Jun 2023
Cited by: 2 articles | PMID: 37271320 | PMCID: PMC10592527
Nasal epithelial gene expression and total IgE in children and adolescents with asthma.
J Allergy Clin Immunol, 153(1):122-131, 22 Sep 2023
Cited by: 0 articles | PMID: 37742934
Editorial comments on "Differential gene expression in nasal airway epithelium from overweight or obese youth with asthma".
Pediatr Allergy Immunol, 33(7):e13827, 01 Jul 2022
Cited by: 0 articles | PMID: 35871458
Long-Term PM<sub>2.5</sub> Exposure and Upregulation of <i>CLCA1</i> Expression in Nasal Epithelium from Youth with Asthma.
Ann Am Thorac Soc, 10 Jul 2024
Cited by: 0 articles | PMID: 38986136
Funding
Funders who supported this work.
NHLBI NIH HHS (6)
Grant ID: R21 HL150431
Grant ID: U01 HL119952
Grant ID: R01 HL079966
Grant ID: R01 HL152475
Grant ID: R01 HL149693
Grant ID: R01 HL117191
NIMHD NIH HHS (2)
Grant ID: R01 MD011764
Grant ID: U54 MD007587




