Abstract
Free full text

Chloramphenicol-Sensitive Escherichia coli Strain Expressing the Chloramphenicol Acetyltransferase (cat) Gene
Abstract
An Escherichia coli strain (strain CM2555) bearing the chloramphenicol acetyltransferase (cat) gene was found to be sensitive to chloramphenicol. We demonstrate that the cat gene is efficiently expressed in strain CM2555. Our results suggest that decreased levels of acetyl coenzyme A in cat-expressing CM2555 cells in the presence of chloramphenicol may cause the bacterium to be sensitive to this antibiotic.
One of the best-characterized bacterial antibiotic resistance mechanisms is the synthesis of chloramphenicol acetyltransferase (CAT) (3, 9, 12, 23). Production of this enzyme, encoded by the cat gene, is the most common means by which bacteria become resistant to chloramphenicol (13, 22), a small bacteriostatic antibiotic that interacts with a peptidyl transferase center (16). CAT catalyzes transfer of the acetyl moiety from acetyl coenzyme A (acetyl-CoA) to a chloramphenicol molecule (8, 12). Thus modified, chloramphenicol no longer binds to the ribosomes and protein synthesis proceeds (12).
Recently, an Escherichia coli strain, strain CM2555, was identified in which no transformants were selected when chloramphenicol was used as the selective agent during attempts to introduce plasmids carrying the cat gene (21). cat-bearing strain CM2555 was found to be sensitive to chloramphenicol (21). The aim of the work described here was to investigate the mechanism of this phenomemon.
The bacterial strains and plasmids used in this work are listed in Table Table1.1.
TABLE 1
E. coli strains and plasmids used in the study
| Strain or plasmid | Genotype or characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
 CM732 CM732 | met46 trp-6 his-4 galK2 lacY or lacZ4 mtl-1 ara-9 tsx-3 ton-1 rpsL8 or rpsL9 supE44 ilvE12 | 4 |
 CM2555 CM2555 | Same as CM732, but dnaA508 ilv+ | 4 |
 CM732-508 CM732-508 | Same as CM732, but dnaA508 zid-3162::Tn10kan | This worka |
 CM2555 dnaA+ CM2555 dnaA+ | Same as CM2555, but dnaA+ | This worka |
 CM2555-72 CM2555-72 | Same as CM255, but dnaA+zid-3162::Tn10kan | This worka |
 MG1655 MG1655 | Wild type | 5 |
 MG1655-508 MG1655-508 | Same as MG1655, but dnaA508zid-3162::Tn10kan | This worka |
 CAG18558 CAG18558 | zid-3162::Tn10kan | 15 |
| Plasmids | ||
 pACYC184 pACYC184 | Replication origin p15A, Cmr, Tetr | 2 |
 pBR328 pBR328 | Replication origin pMB1, Ampr Cmr Tetr | 1 |
 pJKP1 pJKP1 | Replication origin pMB1, plac-cat, Ampr | 7 |
 pJMH1 pJMH1 | Replication origin pSC101, lacIq Kanr | 19 |
 pUC19 pUC19 | Replication origin pMB1, Ampr | 17 |
For Western blotting, after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) with 12.5% polyacrylamide gels, or PAGE the proteins were transferred onto a nitrocellulose membrane (Bio-Rad) with a semidry blot apparatus (Schleicher & Schuell). The CAT protein was visualized with a specific anti-CAT serum (Sigma Aldrich) with alkaline phosphatase-conjugated secondary antibodies as described previously (20). The relative amount of CAT was estimated by densitometry.
Preparation of crude cellular extracts and spectrophotometric measurements of CAT biochemical activity were performed as described previously (11). The intracellular concentration of acetyl-CoA was determined spectrophotometrically as described earlier (10).
The efficiency of protein synthesis was estimated by measurement of the level of incorporation of radioactive precursors into trichloroacetic acid-precipitable material as described previously (18), but 14C-labeled amino acids (UVVVR) were added to bacterial cultures to a final concentration of 2.5 μCi/ml.
E. coli CM2555 has a genetic defect that leads to its chloramphenicol sensitivity in the presence of cat.
Strain CM2555 has been described as one of the series of ilv+ dnaA mutants (CM2555 bears the dnaA508 allele) otherwise isogenic to strain CM732 (4). However, using a set of bacterial strains and plasmids (Table (Table1),1), we found that the chloramphenicol sensitivity of strain CM2555 bearing the cat gene is not caused by the dnaA508 allele or the combination of ilv+ and dnaA508 alleles (data not shown). Thus, we conclude that strain CM2555 must contain another, previously unidentified mutation(s) which makes it sensitive to chloramphenicol, despite the presence of the cat gene.
Expression of cat gene in strain CM2555.
Densitometric analyses of electropherograms after SDS-PAGE and of Western blots with anti-CAT serum revealed that CM2555/pBR328 produces at least three times more CAT than the parental strain, CM732/pBR328 (data not shown). To test whether an increased level of CAT plays a role in the mechanisms of the chloramphenicol sensitivity of CM2555, we used plasmid pJKP1 bearing the cat gene under control of an inducible promoter, plac, together with another plasmid, pJMH1, harboring the lacIq allele. Addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM resulted in CAT levels (in both CM2555 and CM732 hosts) that corresponded to about 90% of that found in CM732/pBR328 (data not shown). Contrary to the results obtained in control experiments, the increased efficiency of cat expression correlated with the faster growth of strain CM2555/pJKP1/pJMH1 in a chloramphenicol-containing medium only up to IPTG concentrations of 0.1 mM, and a further increase in the efficiency of cat expression resulted in the inhibition of bacterial growth (Fig. (Fig.1).1). Therefore, we suggest that the efficiency of expression of the cat gene may be important for the chloramphenicol sensitivity of strain CM2555.
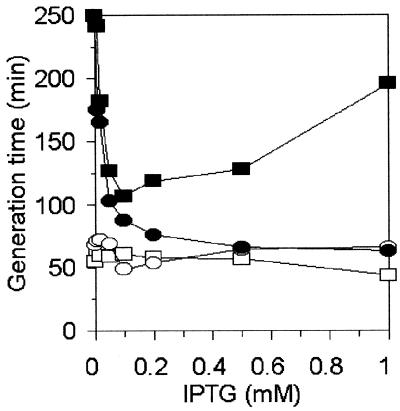
Effect of intracellular amount of CAT on bacterial growth in the presence of chloramphenicol. E. coli CM732 and CM2555 harboring pJKP1 and pJMH1 were grown in Luria-Bertani liquid medium. cat gene expression from the plac-cat fusion was induced overnight, and then in the course of the experiment expression was maintained by the presence of various amounts of IPTG. Chloramphenicol was added when the optical density at 575 nm was 0.1. Doubling times were determined by monitoring bacterial growth (optical density at 575 nm) in the presence and absence of chloramphenicol (34 μg/ml). Symbols: open circles, CM732/pJKP1/pJMH1 grown without chloramphenicol; closed circles, CM732/pJKP1/pJMH1 grown in the presence of chloramphenicol; open rectangles, CM2555/pJKP1/pJMH1 grown without chloramphenicol; closed rectangles, CM2555/pJKP1/pJMH1 grown in the presence of chloramphenicol.
Activity of CAT protein in strain CM2555.
We measured the acetylation of chloramphenicol and calculated that the total CAT activity per 1 mg of cellular proteins in strain CM2555/pBR328 was higher than that in analogous samples prepared from the parental strain, CM732/pBR328 (1.20 CAT activity units in CM2555/pBR328 versus 0.73 CAT activity units in CM732/pBR328). Moreover, CAT reveals biological activity in both CM2555/pBR328 and CM732/pBR328, as the addition of chloramphenicol (up to 34 μg/ml) to cultures of these strains had no significant effect on protein synthesis efficiency (data not shown).
Sensitivity to chloramphenicol of strain CM2555 expressing cat results from decreased levels of acetyl-CoA.
Since CAT catalyzes the transfer of the acetyl group from acetyl-CoA to a chloramphenicol molecule, we measured the levels of acetyl-CoA in strain CM2555 and strain CM732 bearing plasmid pBR328 in the absence and presence of chloramphenicol. We found that the residual level of acetyl-CoA, without the addition of the antibiotic, was slightly lower in CM2555/pBR328 relative to that in CM732/pBR328 (the level of acetyl-CoA in CM2555/pBR328 was about 90% of that found in CM732/pBR328). However, after the addition of chloramphenicol, the level of acetyl-CoA was roughly constant in CM732/pBR328, while it gradually decreased in CM2555/pBR328 (Fig. (Fig.2A2A and B).
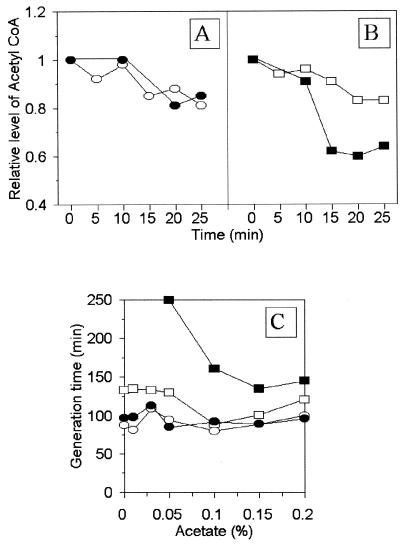
Intracellular amount of acetyl-CoA as affected by chloramphenicol in E. coli CM732/pBR328 (A) and CM2555/pBR328 (B) and the effect of sodium acetate on bacterial growth in the presence of chloramphenicol (C). In the experiments whose results are depicted in panels A and B, strains harboring pBR328 were grown in Luria-Bertani liquid medium. Chloramphenicol (34 μg/ml) was added when the optical density at 575 nm was 0.1 (time zero). Samples were withdrawn every 5 min after addition of the antibiotic and were then assayed for acetyl-CoA. For each strain, the amount of acetyl-CoA at time zero was taken to be 1, and the relative concentration was calculated. In the experiments whose results are depicted in panel C, E. coli CM732 and CM2555 harboring pBR328 were grown in Luria-Bertani liquid medium supplemented with different amounts of sodium acetate. Chloramphenicol was added when the optical density at 575 nm was 0.1. The doubling times of the cultures were determined by monitoring bacterial growth (optical density at 575 nm) in the presence and absence of chloramphenicol (34 μg/ml). The values presented are means from three experiments. In all cases the standard deviation was below 10%. Symbols: open circles, CM732/pBR328 grown without chloramphenicol; closed circles, CM732/pBR328 grown in the presence of chloramphenicol; open rectangles, CM2555/pBR328 grown without chloramphenicol; closed rectangles, CM2555/pBR328 grown in the presence of chloramphenicol.
To test whether a decrease in the level of acetyl-CoA in strain CM2555/pBR328 is responsible for the sensitivity of this strain to chloramphenicol, we measured the growth rates of bacterial cultures in the presence of sodium acetate. Sodium acetate can be used as an alternative source of acetyl-CoA production in cells (6). It was reported previously (12) that cat-expressing E. coli mutants with mutations in aceE or aceF, which cannot make acetyl-CoA from pyruvate but which can make acetyl-CoA from sodium acetate, are resistant to chloramphenicol only when sodium acetate is supplied. Thus, if the hypothesis stated above were true, we should have observed improved growth of CM2555/pBR328 in the presence of sodium acetate. Indeed, we found that the presence of sodium acetate dramatically improved the growth of CM2555/pBR328 in the medium containing chloramphenicol (Fig. (Fig.2C).2C). We also found that strain CM2555 could be transformed by different plasmids bearing the cat gene when selection was performed on plates containing both chloramphenicol (34 μg/ml) and 0.15% sodium acetate (data not shown). Neither the pBR328 copy number nor the amount of CAT in CM2555 cells was affected by the presence of sodium acetate in the medium (data not shown). These results strongly support the hypothesis that decreased levels of acetyl-CoA in cat-expressing CM2555 cells in the presence of chloramphenicol result in the sensitivity of the bacterium to this antibiotic.
Acknowledgments
We are very grateful to S. Barañska and A. Szalewska-Palasz for assistance at the stage of preliminary experiments and to J. Davies for discussions and support at the beginning of this project.
This work was supported by the Polish State Committee for Scientific Research (project 6 P04A 060 18). G.W. also acknowledges financial support from the Foundation for Polish Science (subsidy 14/2000).
REFERENCES
Articles from Antimicrobial Agents and Chemotherapy are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aac.45.12.3610-3612.2001
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc90880?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/116782539
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/aac.45.12.3610-3612.2001
Article citations
Molecular mechanisms of re-emerging chloramphenicol susceptibility in extended-spectrum beta-lactamase-producing Enterobacterales.
Nat Commun, 15(1):9019, 18 Oct 2024
Cited by: 1 article | PMID: 39424629 | PMCID: PMC11489765
Prevalence and antimicrobial susceptibility profile of bacteria isolated from the hands of housemaids in Jimma City, Ethiopia.
Front Public Health, 11:1301685, 29 Jan 2024
Cited by: 0 articles | PMID: 38348381 | PMCID: PMC10859430
Molecular insights into novel environmental strains of Klebsiella quasipneumoniae harboring different antimicrobial-resistance genes.
Front Public Health, 10:1068888, 12 Jan 2023
Cited by: 1 article | PMID: 36711372 | PMCID: PMC9878601
Strategies for Enzymatic Inactivation of the Veterinary Antibiotic Florfenicol.
Antibiotics (Basel), 11(4):443, 25 Mar 2022
Cited by: 1 article | PMID: 35453195 | PMCID: PMC9029715
Analysis of isolates from Bangladesh highlights multiple ways to carry resistance genes in Salmonella Typhi.
BMC Genomics, 20(1):530, 28 Jun 2019
Cited by: 12 articles | PMID: 31253105 | PMCID: PMC6599262
Go to all (16) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The acrAB locus is involved in modulating intracellular acetyl coenzyme A levels in a strain of Escherichia coli CM2555 expressing the chloramphenicol acetyltransferase (cat) gene.
Arch Microbiol, 180(5):362-366, 03 Sep 2003
Cited by: 3 articles | PMID: 14614545
Inactivation of the acrA gene is partially responsible for chloramphenicol sensitivity of Escherichia coli CM2555 strain expressing the chloramphenicol acetyltransferase gene.
Microb Drug Resist, 8(3):179-185, 01 Jan 2002
Cited by: 3 articles | PMID: 12363006
The low level expression of chloramphenicol acetyltransferase (CAT) mRNA in Escherichia coli is not dependent on either Shine-Dalgarno or the downstream boxes in the CAT gene.
Microbiol Res, 153(2):173-178, 01 Aug 1998
Cited by: 4 articles | PMID: 9760750
Ribosomal RNA and the site specificity of chloramphenicol-dependent ribosome stalling in cat gene leaders.
Ann N Y Acad Sci, 646:31-34, 01 Dec 1991
Cited by: 1 article | PMID: 1809198
Review




