Abstract
Free full text

Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design
Abstract
Genetic information is translated into proteins by the ribosome. Structural studies of the ribosome and of its complexes with factors and inhibitors have provided invaluable information on the mechanism of protein synthesis. Ribosome inhibitors are among the most successful antimicrobial drugs and constitute more than half of all medicines used to treat infections. However, bacterial infections are becoming increasingly difficult to treat because the microbes have adapted to become resistant to the most effective antibiotics, creating a major public health care threat. This has spurred a renewed interest in structure-function studies of protein synthesis inhibitors and, in few cases, compounds have been developed into potent therapeutic agents against drug-resistant pathogens. In this review, we describe the modes of action of many ribosome-targeting antibiotics, highlight the major resistance mechanisms developed by pathogenic bacteria, and discuss recent advances in structure-assisted design of new molecules.
1. INTRODUCTION
Protein synthesis is an essential process in all cells. The ribosome performs this task by translating the genetic information encoded in messenger RNAs (mRNAs) into proteins (1, 2). The bacterial ribosome is composed of three RNA chains (16S, 23S, and 5S) and more than 50 proteins assembled into two individual subunits, the small 30S and the large 50S subunits, which join together to form the 70S ribosome (3). For translation to proceed efficiently, many protein factors are needed and interact sequentially with the ribosome. Through highly regulated and choreographed steps, translation factors guide the ribosome during the protein synthesis cycle, which can be divided into four main steps: initiation, elongation, termination, and ribosome recycling (3).
Protein synthesis is initiated on the 30S subunit with the help of initiation factors which recruit the initiator formyl-methionine transfer RNA (fMet-tRNAifMet) in the ribosomal peptidyl (P) site [reviewed in (4)]. The 50S subunit associates with the 30S, forming the 70S initiation complex that is primed for the elongation phase of protein synthesis. The first codon of the gene open reading frame located in the aminoacyl (A) site of the ribosome is decoded by the ternary complex, composed of aminoacyl-tRNA (aa-tRNA), elongation factor Tu (EF-Tu), and GTP. Decoding of the A-site codon by a cognate aa-tRNA triggers GTP hydrolysis on EF-Tu and release of aa-tRNA into the A site of the ribosome. The 3’-CCA termini of aa-tRNA is accommodated into the peptidyl transferase center (PTC) of the 50S subunit, and the peptidyl transferase reaction occurs spontaneously extending the nascent peptide chain by one amino acid residue. Translocation of mRNA and tRNAs is catalyzed by elongation factor G (EF-G) and GTP. Elongation is repeated until a stop codon enters the A site. Release factors (RF1 and RF2) recognize the stop codon and promote hydrolysis of the peptidyl-tRNA in the P site, releasing the newly synthesized protein from the ribosome. The 70S ribosome is then recycled into individual subunits by the concerted action of EF-G and RRF.
During the past two decades, the structural details of key ribosome complexes have been elucidated with the use of X-ray crystallography and cryo-electron microscopy (cryo-EM) reconstructions, providing important insights into the process of protein synthesis and its inhibition. The first high-resolution views of the ribosomal subunits (5–8) provided a wealth of structural information and laid the basis for understanding the mechanisms of action of many ribosome-targeting antibiotics (Figure 1a,,b)b) (9–27). Subsequently, multiple crystal structures of the 70S ribosome were obtained (28–30), which opened further possibilities for understanding the mechanism of protein synthesis and the action of ribosome-targeting inhibitors.
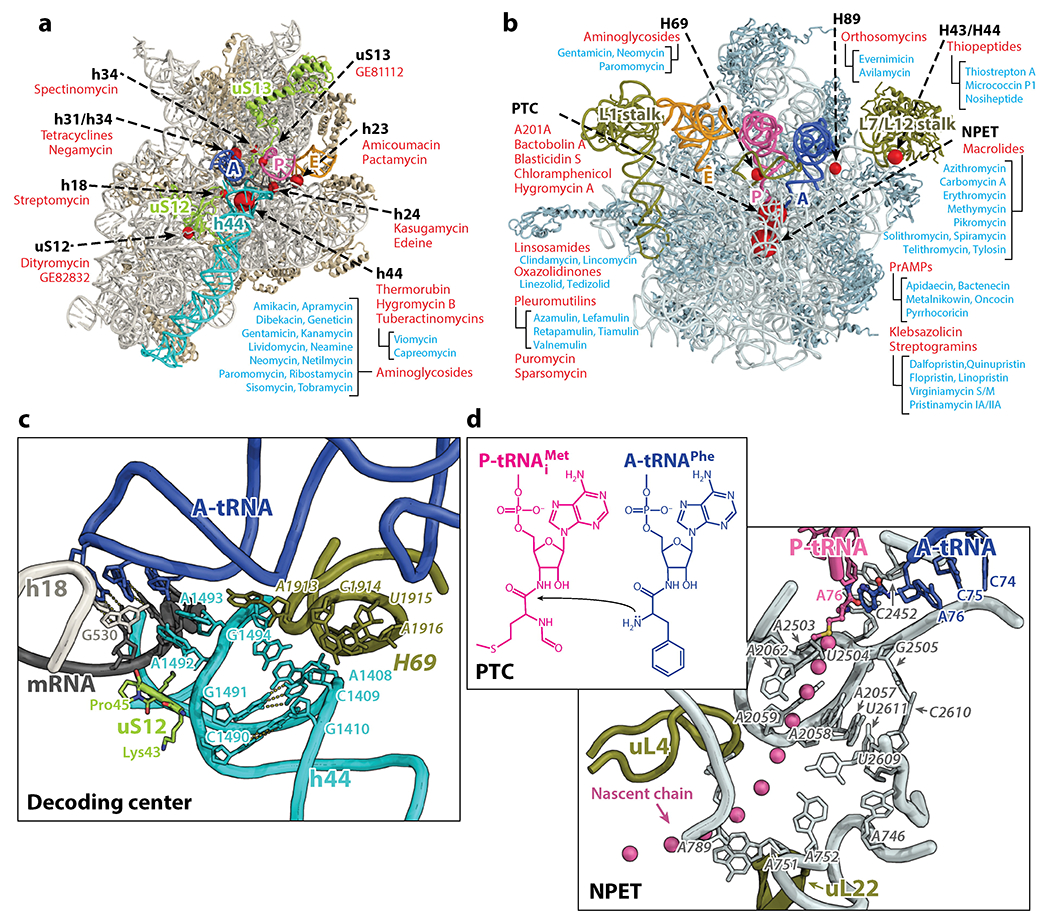
(a, b) Major antibiotic binding sites on the 30S and 50S subunit, respectively. All binding sites are shown as red spheres of variable sizes with the larger ones indicating more drugs discovered at that site. The names of representative antibiotics (or classes) bound to each site are listed. (c) A detailed view of the decoding center (DC) on the 30S subunit. (d) A detailed view of the peptidyl transferase center (PTC) and the nascent peptide exit tunnel (NPET) on the 50S subunit. E. coli numbering is used throughout the text and figures. Abbreviations: A-tRNA, A-site tRNA; mRNA, messenger RNA; PrAMPs, proline-rich antimicrobial peptides; P-tRNA, P-site tRNA.
This review seeks to provide a one-stop resource for those interested in the mechanisms of action and resistance of ribosome-targeting inhibitors. Because antibiotics inhibit protein synthesis by targeting specific functional centers of the ribosome, we discuss inhibitors based on their binding sites in the ribosome. We describe their modes of action, as well as prominent examples of the resistance mechanisms that have developed among pathogens. Finally, we discuss approaches currently being used to identify new antibiotics.
2. ANTIBIOTICS TARGETING THE SMALL SUBUNIT OF THE RIBOSOME
2.1. Antibiotics that Bind in the Decoding Center
The decoding center (DC) is the region comprising the A site on the 30S subunit that monitors the correct base pairing between the mRNA codon and the anticodon of the aa-tRNA. This part of the ribosome consists of regions of the 30S head, shoulder and the top of helix 44 (h44) of 16S rRNA. Biochemical (31–33) and structural studies (25, 34) have shown that conserved nucleotides A1492, A1493 (h44), and G530 (h18) play a critical role in the decoding process. Binding of cognate tRNA in the A site makes nucleotides A1492 and A1493 flip out of h44 and form A-minor interactions that monitor the geometry of the first two codon-anticodon base pairs, ensuring that only Watson-Crick pairs are accepted (34). The base pair geometry in the third position of the codon is monitored less strictly by ribosomal protein uS12 (Pro45) and nucleotide G530, allowing wobble base pairs in this position (Figure 1c) (34).
Antibiotics belonging to several different chemical classes bind in the DC, and some of them are used to treat a broad range of infections. Although tetracyclines (Figure 2a) and negamycin (Figure 2b) bind to overlapping sites in DC, their mechanisms for inhibiting protein synthesis are quite different. Tetracyclines inhibit translation by preventing binding of aa-tRNA to the A site (35), while the smaller molecule negamycin stabilizes tRNA binding in the A site, inhibiting translocation and stimulating miscoding (36).
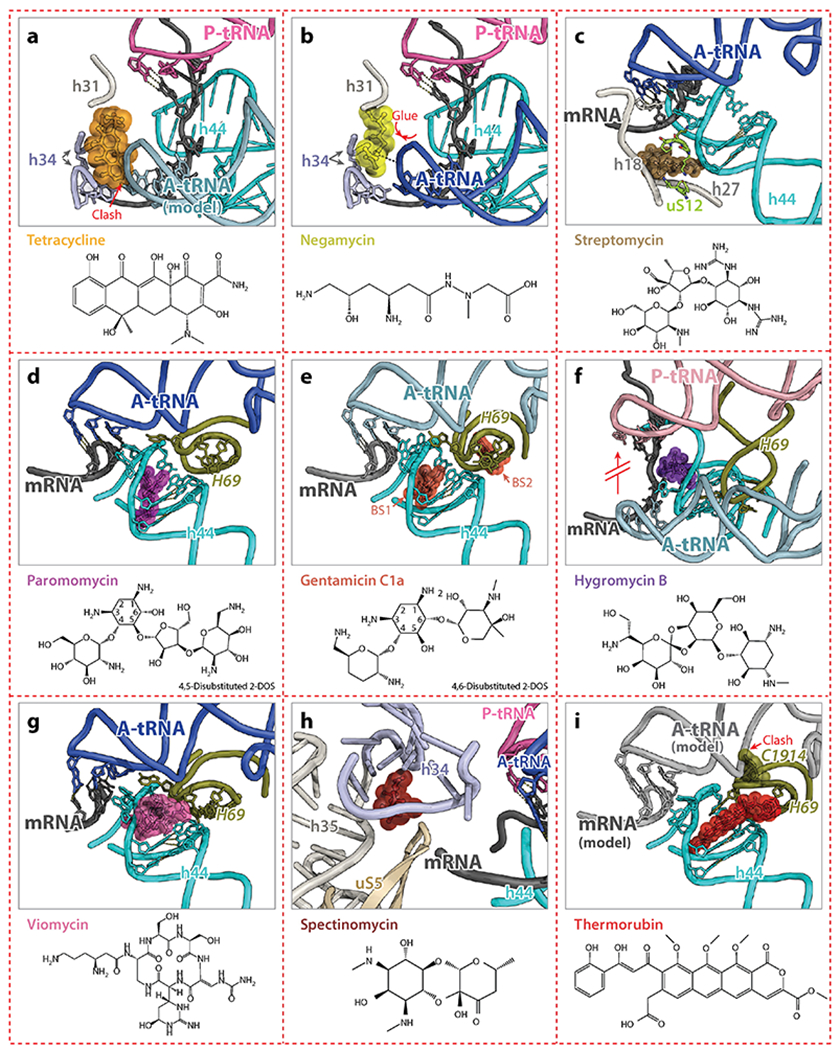
All drugs are shown as spheres with their chemical structures provided on the bottom of each panel. Major drug-interacting elements on the ribosome are indicated. (a) Tetracycline (PDB identification code 4V9B) (35) collides with the A-site tRNA, thereby preventing binding of aa-tRNA to the A site. (b) Negamycin (PDB code 4W2I) (36) binds to an overlapping position with that of tetracycline, but stabilizes the A-site tRNA and inhibits translocation. (c) Streptomycin (PDB code 4DR5) (37) binds outside h44 and interacts with h18, h27, and the ribosomal protein uS12. (d, e) Paromomycin (PDB code 4V51) (28) and gentamicin C1a (PDB code 4V53) (39) bind inside h44 and induce a flipped-out conformation of nucleotides A1492 and A1493, thereby increasing miscoding. Gentamicin has two binding sites on the ribosome: one involves h44 (BS1) and the other involves H69 (BS2). (f) Hygromycin B (PDB code 4V64) (38) binds to the top of h44, preventing tRNA translocation by interacting and stabilizing mRNA. (g) Viomycin (PDB code 4V7C) (41), a drug from tuberactinomycins class, interacts with both h44 and H69 and stabilizes a ratcheted state of the ribosome. (h) Spectinomycin (PDB code 1FJG) (25) binds to h34 located at the 30S neck and interferes with the dynamics of the 30S head domain. (i) Thermorubin (PDB code 4V8A) (44) binds to h44 and extends to H69, causing C1914 to flip out and to collide with the A-site tRNA.
The region of the 30S subunit consisting of h44 and its immediate vicinity is targeted by several classes of antibiotics. Streptomycin interacts with h44, h27, h18 and ribosomal protein uS12, inducing conformational changes of nucleotides A1492 and A1493 (Figure 2c) (25, 37). Aminoglycosides, such as paromomycin and gentamicin, bind at the top of h44 and induce conformational rearrangements of A1492 and A1493 similar to those induced by streptomycin (Figure 2d,,e)e) (25). These drugs favor the flipped-out conformation of A1492 and A1493, and thereby increase miscoding by stimulating the binding of near- and non-cognate tRNAs in the A site. Hygromycin B, another aminoglycoside antibiotic, which, however, does not stimulate miscoding, increases the affinity of tRNA for the A site, and similarly to negamycin, inhibits tRNA translocation (Figure 2f) (38). Some aminoglycosides, such as gentamicin and neomycin, have a second binding site on the ribosome. They bind to H69 on the 50S subunit (Figure 2e) and may inhibit ribosome recycling mediated by RRF (39).
Circular peptide antibiotics belonging to the tuberactinomycin class, such as viomycin and capreomycin IA, bind to a site that overlaps with that of hygromycin B and paromomycin (Figure 2g) (40, 41). Tuberactinomycins inhibit protein synthesis by interfering with the conformational dynamics of the ribosome. Although viomycin binds also at the top of h44, it stabilizes the ratcheted conformation of the ribosome (42). The chemically unrelated antibiotic spectinomycin is known to affect ribosome dynamics by interacting with h34 of the 16S rRNA and limiting rotation of the 30S head domain required for tRNA translocation (Figure 2h) (43).
Thermorubin is chemically related to tetracyclines. However, it binds to the ribosome in a completely different site, namely at the interface between the small and the large subunits where h44 and H69 form the inter-subunit bridge B2a. Thermorubin induces rearrangement of two bases in H69 of the 23S rRNA (44), leaving the displaced nucleotide C1914 in H69 in a position where it would collide with an A-site tRNA (Figure 2i), which explains its inhibitory effect on the initiation and elongation steps of protein synthesis.
2.2. Antibiotics that Bind to the P and E Sites in the 30S Subunit
The P and exit (E) sites in the 30S subunit refer to the locations occupied by the anticodon loops of the peptidyl- and deacyl-tRNAs, respectively, during translation. The E site is the third and final binding site for a tRNA before it dissociates from the ribosome. Antibiotics that bind to the P and E sites in the 30S subunit are generally referred to as translation initiation inhibitors, but some of them also affect tRNA and mRNA translocation. These antibiotics include kasugamycin, pactamycin, edeine, GE81112, and amicoumacin A. The binding site of kasugamycin spans both the P and E sites in a position that overlaps with the mRNA (Figure 3a) (45, 46). The position of the bound drug is incompatible with the canonical location of mRNA, preventing formation of the 30S initiation complex. Similarly, pactamycin also disturbs the path of the mRNA and prevents the formation of the 30S initiation complex by binding to the E site of the small subunit (Figure 3b) (26, 47). Amicoumacin A is another potent protein synthesis inhibitor that binds to the E site of the small subunit. The crystal structure of the 70S ribosome carrying mRNA and tRNAs and with amicoumacin A bound revealed that this antibiotic simultaneously interacts with conserved nucleotides of the 16S rRNA in the E site and with the mRNA phosphate backbone, thereby tethering mRNA to the ribosome and preventing its translocation (Figure 3c) (47).
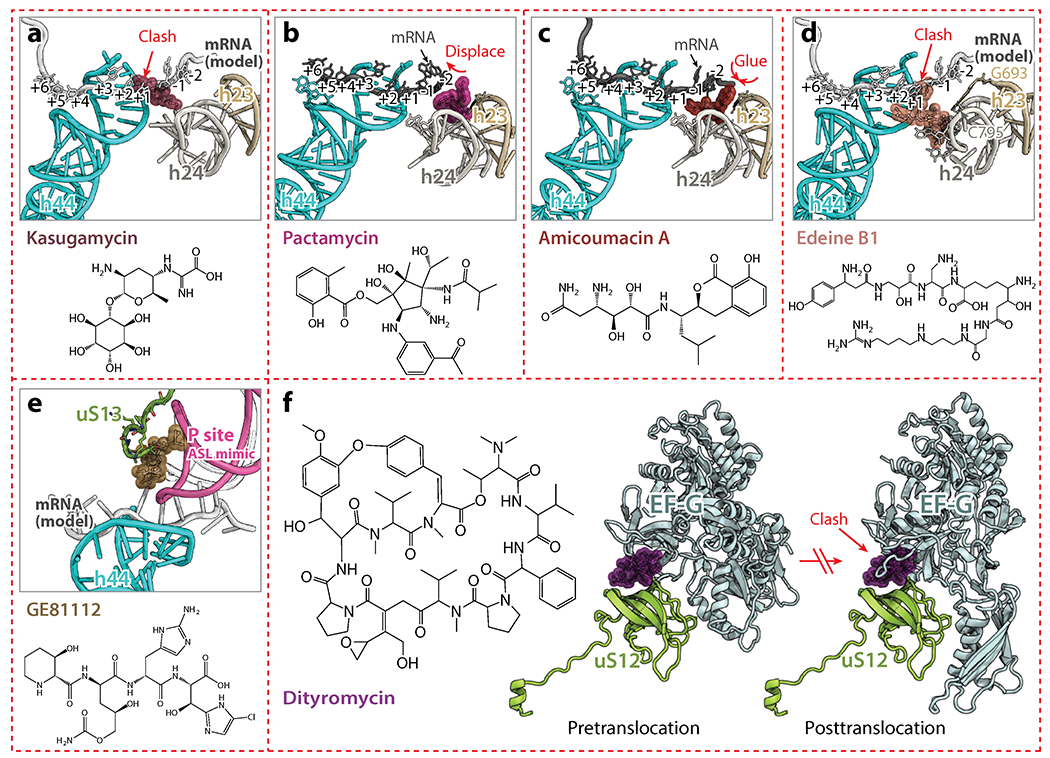
(a) Kasugamycin (PDB identification code 2HHH) (46) binds against h24 and overlaps with mRNA, interfering with translation initiation. (b, c) Pactamycin (PDB code 4W2H) and amicoumacin A (PDB code 4W2F) (47) bind to the E site. While pactamycin displaces the mRNA from its usual path in the E site, amicoumacin A tethers the mRNA to the ribosome. (d) Edeine B1 (PDB code 1I95) (24) binds to the P site and prevents translation initiation. (e) GE81112 (PDB code 5IWA) (48) binds to the P site of the small subunit and interacts with the C-terminus of uS13. It stabilizes the anticodon-stem loop of fMet-tRNAifMet in a distorted conformation, thereby interfering with translation initiation. (f) Dityromycin (PDB code 4WQU) (53) interacts solely with uS12 and prevents conformational changes in EF-G required for tRNA translocation. Abbreviations: ASL, anticodon-stem loop; clash, unnatural overlap of any nonbonding atoms in a structure; EF-G, elongation factor G; fMet-tRNAifMet, initiator formyl-methionine transfer RNA; mRNA, messenger RNA; PDB, Protein Data Bank.
The cationic peptide antibiotic edeine binds to the P site of the small subunit and induces base pair formation between C795 and G693 in the 16S rRNA (24), thereby preventing the binding of fMet-tRNAifMet to the small subunit (Figure 3d). Conversely, the tetrapeptide antibiotic GE81112, which also binds to the P site of the small subunit, prevents proper P site decoding by stabilizing the anticodon-stem loop of fMet-tRNAifMet in a distorted conformation (Figure 3e) (48). This results in a stalling of the initiation step of protein synthesis caused by a reduction in the association rate for the 50S subunit (48).
While ribosome protein uS13 contributes significantly to the binding of GE81112 (Figure 3e), ribosomal protein uS12 in the 30S subunit is also an important component of the ribosome. In an effort to overcome the shortage of new antibiotics, natural compound library screens are now being performed to detect bioactive molecules that could be further improved by rational drug design, fragment-based drug discovery and computational chemistry. Such screens have led to the identification of GE82832, a peptide antibiotic produced by Streptosporangium cinnabarinum (strain GE82832) that inhibits tRNA translocation by interacting with the 30S subunit (49). Further analysis revealed that GE82832 is related to a poorly characterized antibiotic called dityromycin that was discovered decades ago (50). Characterization of both antibiotics has shown that they are structurally and functionally related, and inhibit EF-G-dependent tRNA translocation on the ribosome (51). The crystal structure of the 70S ribosome in complex with dityromycin and GE82832 showed that both antibiotics bind exclusively to ribosomal protein uS12 (52). To our knowledge, dityromycin and GE82832 are the only two ribosome-targeting antibiotics that bind solely to a ribosomal protein in the bacterial ribosome. The mechanism by which dityromycin and GE82832 interfere with tRNA and mRNA translocation has recently been elucidated using a crystal structure of EF-G bound to a dityromycin-70S ribosome complex (Figure 3f) (53). The binding of dityromycin to protein uS12 traps EF-G in a compact conformation on the ribosome, inhibiting EF-G-mediated tRNA translocation.
3. ANTIBIOTICS TARGETING THE LARGE SUBUNIT OF THE RIBOSOME
3.1. Antibiotics that Bind the Peptidyl Transferase Center
The majority of antibiotics that bind to the 50S subunit cluster at the PTC, where the peptide bond formation occurs, and within the nascent peptide exit tunnel (NPET) (Figure 1d). Several of them inhibit protein synthesis by competing with the amino acid side chains of incoming aa-tRNAs for binding to the ribosome A-site crevice, the wedge-shaped gap formed by the bases of nucleotides U2504, A2451, and C2452 of the 23S rRNA (54). Chloramphenicol is one such antibiotic which inhibits translation in a wide range of gram-positive and gram-negative bacteria as well as in mitochondria, but not in the cytoplasm of eukaryotic cells. It binds to the A-site crevice of the 50S subunit, and its nitrobenzyl ring forms a π-stacking interaction with the base of C2452 of the 23S rRNA. The binding site of the aromatic ring of chloramphenicol overlaps with that of the amino acid side chain of the incoming aa-tRNA, effectively preventing it from binding (Figure 4a). Because all aa-tRNAs make key interactions with the ribosome in the A site, chloramphenicol was viewed as a universal inhibitor of peptide bond formation. However, this commonly accepted model of chloramphenicol action fails to explain several experimental observations such as the differential inhibition of translation of specific mRNA templates (55). There is increasing evidence that chloramphenicol acts in a context-specific way and that its activity critically depends on the nature of specific amino acids in the nascent chain and the identity of the residue entering the A site (55).
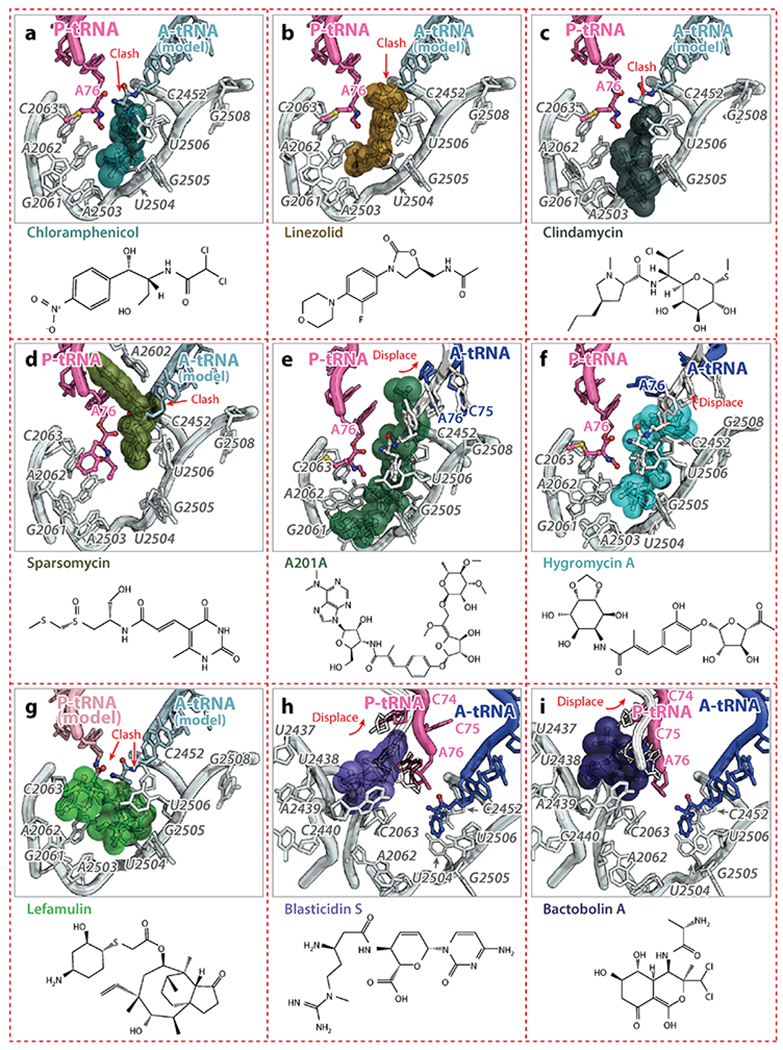
(a, b, c) Chloramphenicol (PDB identification code 4V7W) (91), linezolid (PDB code 4WFA) (57), and clindamycin (PDB code 4V7V) (59) collide with the CCA-end of the A-site tRNA and prevent its accommodation. (d) Sparsomycin (PDB code 1VQ9) (62) collides with the CCA-end of the A-site tRNA and stabilizes the P-site tRNA. (e, f) A201A (PDB code 4Z3S) and hygromycin A (PDB code 5DOY) (67) induce an unusual conformation of the CCA-end of the A-site tRNA, which is not competent of peptide bond formation. (g) Lefamulin (PDB code 5HL7) (10) occupies the PTC and interferes with the binding of both the A- and P-site tRNAs. (h, i) Blasticidin S (PDB code 4V9Q) (77) and bactobolin A (PDB code 4WT8) (81) distort the 3’-terminus of the P-site tRNA, inhibiting peptidyl-tRNA hydrolysis mediated by release factors.
The binding of chloramphenicol to the A-site crevice strikingly resembles that of another drug, anisomycin, which is effective only against archaeal and eukaryotic, but not eubacterial ribosomes. The specificity of chloramphenicol for the bacterial ribosome versus that of anisomycin for the archaeal Haloarcula marismortui 50S (16) can be explained by a steric collision that would occur between anisomysin and U2504 (that pairs with C2452) in the bacterial ribosome. The reorientation of U2504 and G2505 observed in the H. marismortui 50S subunit widens the A-site cleft (14), providing a rationale for the specificity of anisomycin for the archaeal ribosome.
The antibiotic linezolid, a member of the oxazolidinone family, is active against all major pathogenic Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium, and penicillin-resistant Streptococcus pneumoniae (56). Similar to chloramphenicol, linezolid binds to the ribosome in the A-site cleft and, based on the crystal structures of the H. marismortui, S. aureus and Deinococcus radiodurans 50S subunit, the oxazolidinone ring of linezolid forms a π-stacking interaction with the base of U2504, fitting perfectly into the A-site cleft of the PTC (Figure 4b) (15, 57, 58). Also, the fluorophenyl moiety of linezolid stacks onto the base of C2452, the way nitrobenzyl ring of chloramphenicol does, a structural feature observed for several classes of antibiotics. The remaining part of linezolid would collide with the CCA-end of aa-tRNA and, presumably, interfere with its accommodation.
Clindamycin, a semisynthetic derivative of the lincosamide family of compounds, is widely used clinically to treat Gram-positive bacterial infections. In the crystal structures of clindamycin with the H. marismortui 50S subunit (18) and the Escherichia coli 70S ribosome (59), the pyrrolidinyl propyl group of clindamycin, and likely of other lincosamides, occupies the same space as the O-methyl tyrosine aromatic ring of CC-puromycin (60) and the phenylalanine residue of the Phe-tRNAPhe bound in the A site (Figure 4c) (61). Similarly, the sulfur-containing tail of sparsomycin forms hydrophobic interactions with the A-site crevice (22, 60, 62), illustrating the similarity of the strategies used by small molecules to target the PTC region of the ribosome. The remaining moieties of sparsomycin, the pseudouridine group and a part of its tail, are sandwiched between the CCA-end of the P-site tRNA and the base of A2602 (Figure 4d) (22, 60, 62). In this position, sparsomycin stabilizes the binding of the P-site tRNA, which explains why it induces translocation of an A-site tRNA to the P site (63).
Antibiotic A201A, which is structurally similar to puromycin, is active against Gram-positive bacteria and most Gram-negative anaerobic species (64). Hygromycin A shares its dihydroxyphenyl and hexofuranose rings with A201A and has broad-spectrum activity against Gram-positive and, to a lesser extent, Gram-negative bacteria (65, 66). The structures of hygromycin A and A201A bound to the ribosome show that their dihydroxyphenyl moieties localize in the A-site crevice, and stack on the base of C2452 (67). These structures and single-molecule fluorescence resonance energy transfer experiments have revealed that hygromycin A and A201A sterically occlude the binding of the CCA-end of the A-site tRNA, leading to the accumulation of partially accommodated aa-tRNAs on ribosomes which prohibits peptide bond formation (Figure 4e,,f)f) (67).
Streptogramin A (SA) is another class of antibiotics, whose binding site spans not only the A-site cleft, as in the previous examples, but also encroaches into the P site. Madumycin II, the simplest alanine-containing SA antibiotic, inhibits the ribosome prior to the first cycle of peptide bond formation (68). It allows binding of the tRNAs to the A and P sites, but prevents correct positioning of their CCA-ends into the PTC, thus making peptide bond formation impossible. Also, binding of madumycin II induces rearrangement of nucleotides U2506 and U2585 of the 23S rRNA resulting in the formation of the U2506•G2583 wobble pair that has been attributed to a catalytically inactive state of the PTC (68). Virginiamycin M is another SA antibiotic that binds in the PTC, causes rearrangements of nucleotides A2062 and U2585 (22) and inhibits binding of A- and P-site substrates (69, 70). In this case, the oxazole ring of virginiamycin M reaches into the A-site cleft, where it establishes hydrophobic interactions (22).
Pleuromutilins, such as lefamulin, are a class of antibiotics that are active against drug-resistant pathogens. They inhibit protein synthesis by binding to the PTC and interfering with the coordination of both A- and P-site tRNAs. In the crystal structure of the S. aureus 50S subunit (10), the tricyclic mutilin core moiety of lefamulin forms hydrophobic stacking interactions with nucleotides U2504 and C2452, anchoring the drug to the ribosome and blocking the A site (Figure 4g). The C14 extension on the tricyclic core of lefamulin points into the P site, thereby hindering tRNA positioning at the P and A sites.
Blasticidin S, produced by some species of Streptomyces, is a nucleoside analog consisting of a cytosine linked to a pyranose sugar ring and attached to an N-methyl-guanidine tail. It is a potent inhibitor of protein synthesis in bacterial and eukaryotic cells (71, 72). The crystal structure of the H. marismortui 50S subunit bound to blasticidin S revealed that it occupies the P site of the large subunit where it would presumably compete with the binding of the P-site tRNA (22). However, observations that blasticidin S enhances binding of aa-tRNA-mimics to the P site (73) and decreases binding of the same tRNA mimics to the A site (73, 74), combined with the fact that blasticidin S competes for binding with sparsomycin (75) and puromycin (76), two antibiotics that bind the hydrophobic A-site cleft, raised doubts that the 50S crystal structure fully explained its mode of action. The crystal structure of blasticidin S in complex with the Thermus thermophilus 70S ribosome with tRNAs shows that antibiotic molecule forms a Watson-Crick-like interaction with nucleotide G2251 in the P-loop of the 23S rRNA and distorts the 3’-termini of the P-site tRNA by intercalating between nucleotides C74 and A76 of the P-site tRNA, thereby displacing C75 from its normal position in the PTC (Figure 4h) (77). Biochemical experiments show that distortion of the P-site tRNA by blasticidin S inhibits peptidyl-tRNA hydrolysis promoted by release factors, rather than peptide bond formation (77). Furthermore, blasticidin S and chloramphenicol were recently reported to inhibit subunit dissociation mediated by EF-G and RRF or by low magnesium concentrations (78). This highlights the often promiscuous and collateral effects of antibiotics binding to functional centers that are critical for various ribosomal activities.
The polyketide-peptide family of molecules produced by Burkholderia thailandensis, like bactobolin A, inhibit protein synthesis (79). Mutations in ribosomal protein uL2 that would disrupt the E236:A2450 contact in the PTC leads to bactobolin A resistance (80, 81), but the ribosome remains susceptible to other known antibiotics, which hinted that bactobolin A binds to a new location on the ribosome. However, the reported structure of the bacterial ribosome bound to bactobolin A (81) shows that its binding site overlaps with that of blasticidin S, and that it distorts the CCA-end of the P-site tRNA toward the A site, suggesting that both antibiotics may interfere with protein synthesis the same way (Figure 4i).
3.2. Antibiotics that Bind in the Nascent Peptide Exit Tunnel
The nascent polypeptide chain remains attached to a tRNA molecule inside the ribosome at all times during translation until it is hydrolyzed by a release factor and released from the ribosome in response to a stop codon. During this process, a part of the synthesized polypeptide chain is located in the NPET – an important structural and functional element of the ribosome (5, 8, 54). The function of the exit tunnel appears to be not only to provide an unobstructed passage through the ribosome for newly synthesized polypeptide chains but also to regulate translation itself. Specific elements of the tunnel monitor the amino acid sequence of the nascent polypeptide chain, and arrest of translation may occur in response to particular drugs or metabolites (82, 83). The NPET is the target for many clinically important antibiotics (23, 27, 54, 59) and its alteration can lead to antibiotic resistance in pathogenic bacteria (84–87).
Macrolide antibiotics represent a large class of polyketide-based compounds composed of a 12-, 14-, 15-, or 16-membered macrocyclic lactone ring decorated with one or several sugars and/or side chains. The natural macrolide erythromycin and its semi-synthetic derivatives, the ketolides, bind near the entrance of the NPET (Figure 5a). By restricting the effective diameter of the tunnel and partially blocking it, macrolides were initially thought to simply obstruct the progression of the nascent polypeptide chain resulting in peptidyl-tRNA drop off and abortion of protein synthesis. As in the case with chloramphenicol and linezolid, that model for the mechanism of macrolide action has had to be amended as new data have become available. Currently, there is compelling evidence that macrolides inhibit translation in a context-specific manner by interfering with polymerization of only certain amino acid sequences (88, 89). Some proteins lack the “problematic” sequences and continue to be synthesized when macrolide antibiotics are bound in the exit tunnel (90).
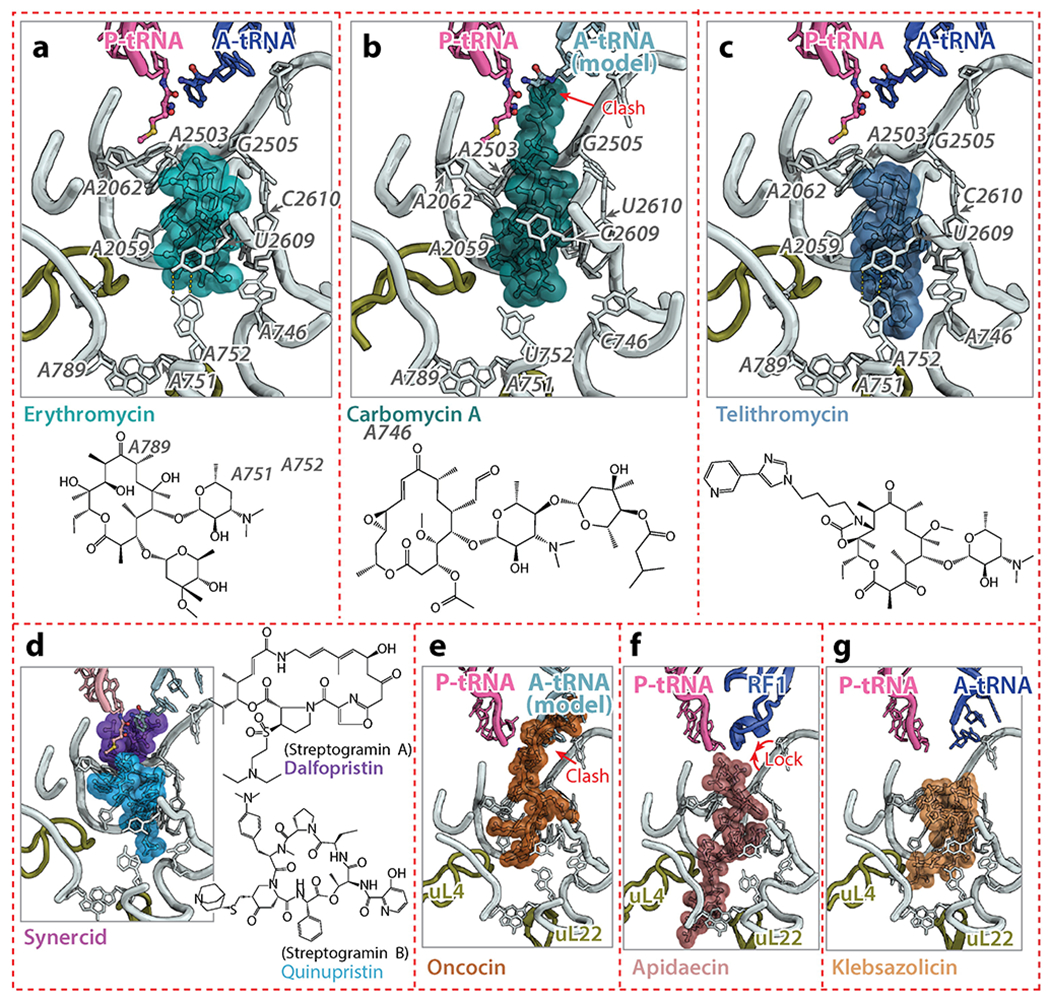
(a, b, c) Erythromycin (PDB identification code 4V7X) (91), carbomycin A (PDB code 1K8A) (23), and telithromycin (PDB code 4V7Z) (91) are macrolides that bind to the NPET. Carbomycin A has a disaccharide extension that reaches into the PTC and collides with the A-site tRNA. Telithromycin has an alkyl-aryl group that extends further into the NPET, increasing its affinity for the ribosome. (d) Synercid is a combination of dalfopristin, a streptogramin A drug, and quinupristin, a streptogramin B drug, with both binding synergistically on the ribosome, occupying the PTC and the NPET. (e, f) Oncocin (PDB code 4Z8C) (115) and apidaecin (PDB code 5O2R) (116) are proline-rich antimicrobial peptides that bind to the 50S subunit. Oncocin prevents the accommodation of the A-site tRNA, whereas apidaecin interferes with translation termination by locking RF1 on the ribosome. (g) Klebsazolicin (PDB code 5W4K) (119) is a ribosomally-synthesized post-translationally modified peptide that obstructs the NPET. Abbreviations: A-tRNA, A-site tRNA; clash, unnatural overlap of any nonbonding atoms in a structure; PDB, Protein Data Bank; PTC, peptidyl transferase center; P-tRNA, P-site tRNA.
The most recent structures of erythromycin in complex with the 70S ribosome from T. thermophilus (91) and E. coli (59) reveal that the desosamine sugar, an essential element of every macrolide, interacts with A2058 and A2059 of the 23S rRNA. This is consistent with probing experiments showing that macrolides protect A2058 and A2059 from chemical modification (92). Methylation of A2058 or its mutation to G renders ribosomes macrolide-resistant (see Section 4.3) (93). Curiously, archaeal and eukaryotic ribosomes have a G in the position equivalent to A2058, making them naturally resistant to macrolides. Although the mutation G2058A in archaea enhances erythromycin binding (18), the same mutation in yeast is not sufficient to render ribosomes susceptible to erythromycin, suggesting that other determinants contribute to resistance in eukaryotes (94).
Larger macrolides, such as 16-membered tylosin, carbomycin A, and spiramycin have a disaccharide extension at the C5 position of their macrolactone ring. This extended tail can reach deeper into the PTC and interfere with the peptide bond formation when these drugs are bound to the ribosome (23). Erythromycin lacks such a tail. These drugs directly inhibit peptide bond formation to varying extents (carbomycin A – 100%, spiramycin – 85%, and tylosin – 60%) (95). The strong inhibition of peptide bond formation by carbomycin A is likely due to the presence of the isobutyrate extension on the C5-disaccharide that collides with the amino acid of aa-tRNAs (Figure 5b) (23).
Ketolides are semi-synthetic derivatives of macrolides in which the sugar in position C3 is replaced with a keto group. Telithromycin is a ketolide that is derived from 14-membered macrolides, and it is representative of a new generation of antimicrobials that has been developed to overcome the problem of macrolide resistance; it is active against erythromycin-resistant pneumococci (96). Telithromycin has an alkyl-aryl arm that extends from positions C11/C12 and forms a stacking interaction with base-pair A752-U2609 of the 23S rRNA (Figure 5c). This additional point of contact increases the affinity of telithromycin for the wild-type ribosome by 10-fold compared to erythromycin (97). However, adverse side effects have limited the clinical use of telithromycin (98) and led to its withdrawal from the market. For that reason, new ketolides are being sought that will fight bacterial infections while having fewer side effects. The fluorine-containing ketolide solithromycin (CEM-101) is a promising drug of this class (99). The structure of solithromycin bound to the E. coli ribosome (100) shows that its binding site is similar to that of telithromycin (59, 91), with differences being confined to the interactions mediated by the alkyl-aryl extension, which appears to anchor solithromycin to the ribosome better than telithromycin (100).
The exit tunnel of the ribosome is also targeted by type B streptogramins (SB), such as pristinamycin IA, quinupristin, and virginiamycin S. While type A streptogramins (SA) contain a 23-membered unsaturated ring with lactone and peptide bonds, SB antibiotics are depsipeptides produced by various species of Streptomyces. Crystal structures of SA (22, 68, 101) and SB (20, 101) antibiotics in complex with the ribosome show that both classes bind to adjacent, but not overlapping, sites on the ribosome, which explains their synergistic action (Figure 5d) (102). As discussed in Section 3.1, SA antibiotics bind at the PTC and prevent proper positioning of the A- and P-site tRNAs, whereas SB antibiotics bind to a site that overlaps with that of macrolides and presumably interfere with the passage of the nascent peptide through the exit tunnel. Interestingly, nucleotide A2062 of the 23S rRNA is sandwiched between quinupristin and the macrocyclic ring of dalfopristin (20, 101), rationalizing why mutation A2062C in the 23S rRNA of Streptococcus pneumoniae leads to SA and SB cross-resistance (103). Several streptogramins that are approved for clinical use, such as Synercid, a mixture of type A streptogramin dalfopristin and type B streptogramin quinupristin (Figure 5d) (101), are effective against MRSA (104).
3.3. Ribosome Inhibition by Antimicrobial Peptides
The cecropins, which were discovered more than 35 years ago, is a group of antibacterial peptides (AMPs) produced by insects to protect them from pathogen infections (105). They are now known to be widely expressed in both animals and plants. Because of their potential as therapeutic agents, we will discuss one sub-class of AMPs, the proline-rich antimicrobial peptides (PrAMPs), which are expressed in both mammals and insects. They have attracted attention because of their wide distribution, and because they kill bacteria by mechanisms that do not involve cell-membrane disruption (106). Oncocin, apidaecin, drosocin, pyrrhocoricin, metalnikowin and bactenecin 7 are PrAMPs known to interact with micromolar affinity with the substrate-binding domain of the chaperone DnaK, leading to protein misfolding and aggregation, and ultimately bacterial death (107, 108). However, DnaK-deficient E. coli strains remain susceptible to PrAMPs, indicating that PrAMPs have other targets inside the cell (109).
The mystery of the primary cellular target of these insect-derived PrAMPs remained unsolved until it was discovered that oncocin and apidaecin bind to the 70S ribosome with nanomolar affinities (109). These results explain why DnaK null mutant E. coli strains remain susceptible to apidaecin and oncocin. Similarly, it was reported that bactenecin 7, which belongs to the cathelicidin family, inhibits protein synthesis through its interaction with the 70S ribosome (110). Biochemical and structural studies of several ribosome complexes with oncocin, metalnikowin, pyrrhocoricin and bactenecin 7 revealed that these PrAMPs bind inside the ribosomal exit tunnel in a reverse orientation compared to that of a nascent polypeptide chain (111–115). The binding location of these PrAMPs in the peptide exit tunnel and their overlap with the PTC allows the initiation of protein synthesis to take place, but prevents the transition to the elongation phase of translation by interfering with the binding of the aa-tRNA in the A site of the 50S subunit (Figure 5e).
While most of the recently studied PrAMPs inhibit translation initiation, apidaecin has recently been shown to interfere with translation termination (116). Apidaecin traps RF1 or RF2 on the ribosome subsequent to the release of the nascent peptide chain. A cryo-EM reconstruction of the E. coli 70S ribosome complexed with RF1 and Api137 (116), a more stable 18-amino acid derivative of the natural apidaecin 1b (117), reveals that Api137 also binds within the peptide exit tunnel of the ribosome and that its C-terminal amino acids interact with 23S rRNA nucleotides in the PTC and with the GGQ motif of RF1 (Figure 5f). Contrary to other PrAMPs, for which structures are available, the orientation of Api137 matches that of a nascent polypeptide chain. Although several inhibitors, such as blasticidin S, bactobolin A, and negamycin, have been reported to interfere with translation termination (77, 81, 118), these antibiotics also hamper other steps of protein synthesis. This makes apidaecin the first inhibitor known to be specific for the termination step of translation. The mode of interaction of these PrAMPs with the exit tunnel of the ribosome, and the fact that they overlap with the binding sites of several other drugs, provide opportunities to develop these peptides into novel therapeutics.
A ribosomally-synthesized post-translationally modified peptide klebsazolicin (KLB), which directly inhibits the ribosome, was recently identified using a genome mining approach (119). Structural analysis of the ribosome-KLB complex reveals that the compound binds in the peptide exit tunnel to a site that overlaps with the binding sites of macrolides, SB, the PrAMPs Onc112 and Bac7, and significantly obstructs the tunnel (Figure 5g) (119). Unlike PrAMPs, which are highly enriched in positively charged amino acids and interact with the negatively charged rRNA backbone, KLB interacts with the ribosome mainly via stacking of heterocycles over rRNA bases. Unlike the PrAMPs, which bind the ribosome in an elongated conformation, KLB binds the exit tunnel in a compact, globular conformation.
3.4. Antibiotics that Interfere with Translation Factors
Thiopeptide antibiotics, such as thiostrepton and micrococcin, interact with the large subunit of the ribosome and interfere with the function of EF-Tu, EF-G and IF2. The crystal structure of the D. radiodurans large subunit bound to thiostrepton and micrococcin (120) revealed that this class of antibiotics targets the GTPase-associated center of the ribosome. They bind in a cleft formed between ribosomal protein uL11 and helices H43 and H44 of the 23S rRNA. In this site, thiopeptides overlap with the location of domain V of EF-G explaining how these compounds interfere with the binding of translation factors to the ribosome (Figure 6a).
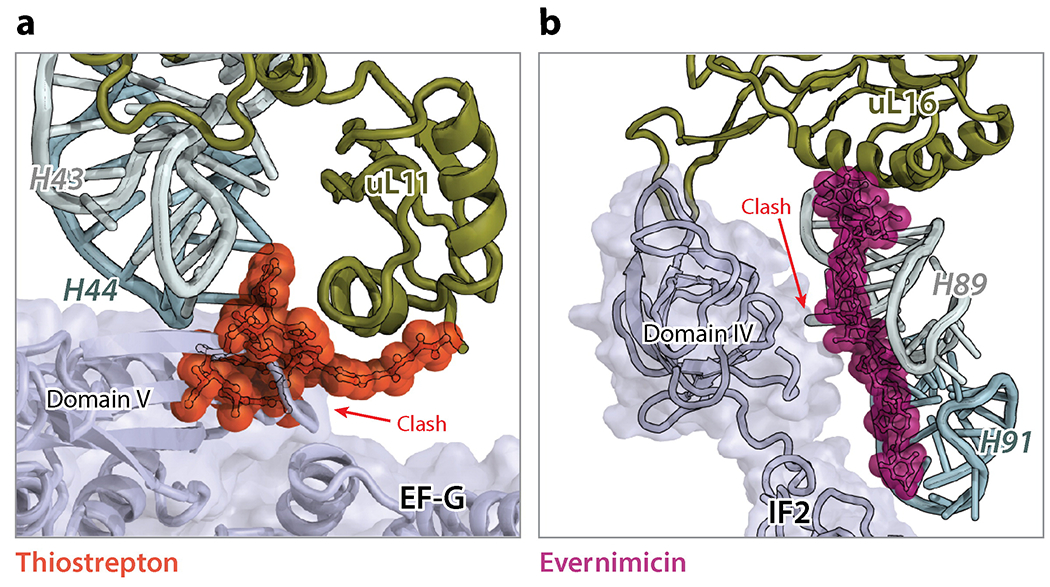
(a) Thiostrepton (PDB identification code 3CF5) (120), a thiopeptide interacting with H43/H44 and ribosomal protein uL11, sterically clashes with domain V of EF-G and inhibits the EF-G-catalyzed tRNA translocation. (b) Evernimicin (PDB code 5KCS) (121) interacts with H89, H91 and protein uL16, and may interfere (clash) with the binding of IF2 during translation initiation.
A new antibiotic binding site on the ribosome has recently been discovered from the structures of the 70S ribosome in complex with the orthosomycin antibiotics evernimicin and avilamycin (9, 121). Both antibiotics adopt elongated conformations spanning the minor grooves of helices H89 and H91 of the 23S rRNA, contacting few arginine residues of ribosomal protein uL16 (Figure 6b). In this location, evernimicin has been reported to inhibit formation of the IF2-dependent 70S initiation complex and to prevent accommodation of the aa-tRNA into the A site on the large subunit (122, 123).
4. MECHANISMS OF ANTIBIOTIC RESISTANCE
4.1. General Mechanisms of Antibiotic Resistance
The development of resistance to antibiotics often rapidly follows their introduction into the clinic. Antibiotics that target the ribosome are no exception. Antibiotic resistance, however, should be considered as an ancient feature, since naturally produced antibiotics have existed in the environment for billions of years and resistance to them was spread among environmental and pathogenic bacteria way before modern use of antibiotics to cure infections (124). Resistance to antibiotics is inherently linked to their production because microorganisms that produce antibiotics must be able to protect themselves against the action of the drugs they make. These intrinsic mechanisms of resistance that exist in nature lay the foundation for the acquired resistance in pathogens under selective pressure caused by the human use of antibiotics. Through horizontal gene transfer (HGT), pathogens can acquire resistance genes on mobile elements, e.g. plasmids, transposons, or integrons, which may have their evolutionary roots in the environment. In addition to HGT, pathogens can also acquire resistance through spontaneous point mutation or collection of such in their chromosomes.
Bacteria can become resistant to an antibiotic in three different ways: (i) by contriving to keep the intracellular concentration of the drug low; (ii) by inactivating the drug chemically; and (iii) by altering its target so that it will no longer interact with the drug. As a first line of defense, bacterial cells can control antibiotic concentrations either by restricting their influx or enhancing their efflux from the cell. Some bacteria are intrinsically resistant to particular antibiotics because the uptake of those drugs is impaired. For example, Gram-negative bacteria are resistant to macrolides because the hydrophobic nature of macrolides prevents them from penetrating their outer membranes. Anaerobic bacteria are naturally resistant to aminoglycosides because the transport of aminoglycosides into the cell is driven by energy produced by oxidative metabolism, which these bacteria cannot perform. In addition, bacteria can develop resistance by increasing the efflux of a particular drug, thereby lowering its concentration inside the cell to a non-toxic level. This can be achieved by acquisition of genes encoding drug-specific efflux pumps or chromosomal mutations resulting in overexpression of a pre-existing efflux pump, such as for example the multidrug efflux pump TolC in E. coli (125). The drug resistance that is conferred by the up-regulation of the efflux pumps is known for almost all ribosome-targeting antibiotics. For some antibiotics, enhanced activity of porins was indicated in drug resistance (126). Once it is inside the cell, an antibiotic can be inactivated by either enzymatic modification (e.g., aminoglycosides), or degradation (e.g., tetracyclines). Similarly, genes encoding these enzymes can be obtained by either HGT or mutation. The third general principle of acquiring drug resistance is to alter the target. Mutations at or near the binding sites diminish binding of antibiotics by physically changing their target. The binding sites for most known ribosome-targeting antibiotics are mostly made of rRNA. Therefore, mutations in the genes encoding rRNAs can confer resistance. However, most bacteria have multiple copies of rRNA operons, and only if the same mutation occurs simultaneously on all copies will the resistance manifests itself. As a result, antibiotic resistance caused by a mutation in rRNA is rare in worst clinical pathogens. Nevertheless, such mutations often occur in pathogens such as Mycoplasma pneumoniae, Mycobacterium tuberculosis, and other bacterial species that have only one or two rRNA operons (93). In some rare cases, antibiotics bind directly to ribosomal proteins or have ribosomal proteins in the vicinity of their binding sites. Resistance to such antibiotics more frequently arises from mutations of ribosomal proteins, whose genes usually exist as a single copy in the genome. Besides mutations, antibiotic binding sites can also be altered by enzymatic modification, which affects all copies of rRNA in the cell and leads to strong resistance. In one known case, resistance to kasugamycin that arises from the loss of an intrinsic modification in the 16S rRNA caused by inactivation of the responsible enzyme (127).
Although the mechanisms of resistance are very diverse for any given antibiotic, one mechanism often prevails for many pathogens. Next, we highlight the major mechanisms of resistance that emerged from the clinical use of some important ribosome-targeting antibiotics.
4.2. Mechanisms of Resistance to Tetracyclines
Tetracycline resistance usually results from the acquisition of mobile genetic elements carrying tetracycline-specific resistance genes that encode for efflux pumps, ribosome protection proteins (RPPs), and drug-degrading enzymes (Figure 7). Tetracycline-specific efflux pumps are most effective against 1st generation tetracyclines. They confer moderate resistance to the 2nd generation tetracyclines, such as doxycycline and minocycline, and little resistance to the 3rd generation tetracyclines (also called glycylcyclines), such as tigecycline (128). Use of RPPs is another way to resist the inhibitory action of tetracyclines. RPPs, represented by TetO and TetM, are paralogues of EF-G (129). They bind to the tetracycline-stalled ribosome and cause dissociation of tetracycline from the ribosome (Figure 7). A cryo-EM reconstruction of TetM bound to the E. coli 70S shows that Pro509 at the tip of domain IV overlaps with the binding position of tetracycline, suggesting a mechanism for dislodging ribosome-bound tetracycline (130). Interestingly, binding of tigecycline to the ribosome is not affected by TetM, even though a more severe clash between TetM and tigecycline is anticipated. Recently, members of the ABC-F subfamily of ATP-binding cassette proteins were shown to protect the ribosome from inhibition by antibiotics that bind near the PTC, such as macrolides, streptogramins, lincosamides, and oxazolidinones (131).
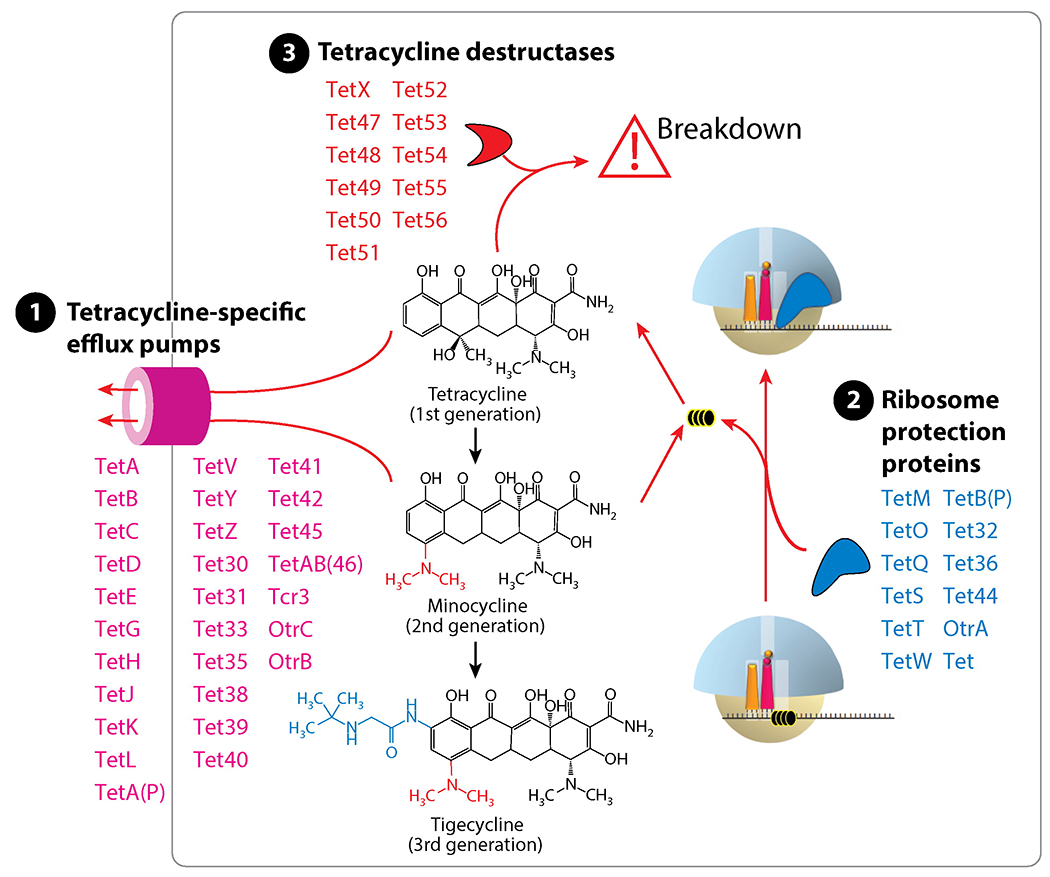
Resistance to tetracyclines is mainly conferred by the acquisition of mobile genes expressing tetracycline-specific efflux pumps, ribosome protection proteins, or tetracycline degrading enzymes (tetracycline destructases).
Enzymatic degradation is a mechanism of tetracycline resistance that is rare in the clinic. A flavin-dependent monooxygenase encoded by the tetX gene modifies tetracycline at position C11a to generate 11a-hydroxyl-tetracycline, which undergoes spontaneous non-enzymatic breakdown (Figure 7). For a long time, TetX remained the only tetracycline-degrading enzyme that had been characterized biochemically, and that had been detected in few human pathogens (132). However, a family of widespread TetX-related genes, termed destructases, was discovered recently (133). Products of these genes utilize various mechanisms to inactivate tetracycline, but not tigecycline (134). The existence of the destructase family is alarmingly worrisome. If disseminated into human pathogens and mutated, these genes would exacerbate the problems caused by tetracycline resistance, and, potentially, threaten the clinical application of tigecycline (135).
4.3. Mechanisms of Resistance to Aminoglycosides
Drug modification by cellular enzymes is the prevalent mechanism of resistance to aminoglycosides. Owing to the exposed amide and hydroxyl groups, aminoglycosides are prone to enzymatic modification. Over 300 enzymes, which can be categorized into three classes, including acetyltransferases, nucleotidyltransferases, or phosphotransferases, modify aminoglycosides at their amino groups or hydroxyl groups (Figure 8a) and cause them to lose ribosome-binding activity (Figure 8b) (136). These enzymes, which likely evolved from aminoglycoside-producing bacteria, now exist in transmissible elements posing a significant clinical threat as they confer resistance to a broad range of aminoglycosides.
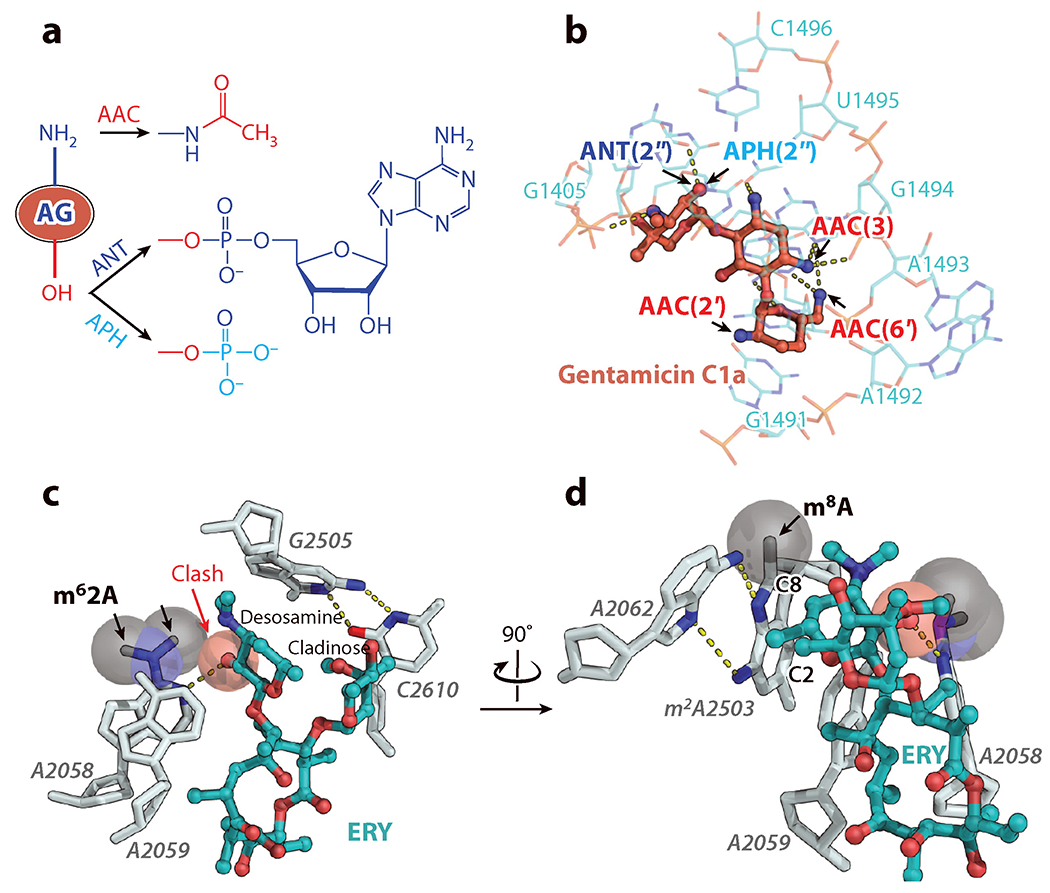
(a) Drug modification at the amino and hydroxyl groups is the main mechanism of resistance to aminoglycosides (AG). (b) Modifications of gentamicin C1a disrupt its binding to h44. (c) Resistance conferred by drug-target modification exemplified by the macrolide erythromycin (ERY). Dimethylation of A2058 at the N6 atom abolishes the binding of erythromycin. (d) Cfr-catalyzed methylation at the C8 position of A2503, which has an intrinsic methylation at the C2 position, confer resistance against a wide range of antibiotics and synergizes with the A2508 modification to extend resistance to macrolides. Abbreviations: AAC, acetyltransferase; ANT, nucleotidyltransferase; APH, phosphotransferase; Cfr, chloramphenicol florphenicol resistance; clash, unnatural overlap of any nonbonding atoms in a structure; m62A, N6, N6-dimethyladenine; m2A and m8A, 2-methyladenine and 8-methyladenine, respectively.
Resistance to aminoglycosides can also be conferred by ribosomal mutations in pathogens that have one or two copies of rRNA operon. For example, the A1408G mutation in h44 of the 16S rRNA has been found in clinical isolates of resistant M. tuberculosis (137). Notably, G1408 normally exist in human cytosolic ribosomes, explaining why aminoglycosides are inactive against human ribosomes and rationalizing their specificity for bacteria. Unlike most aminoglycosides that bind h44, streptomycin interacts with the G530-loop (h18) as well as ribosomal protein uS12 (Figure 2c). Accordingly, the most frequently occurring mechanism of resistance to streptomycin in M. tuberculosis is a mutation of one of the two lysine residues K43 or K88, both of which contact streptomycin bound to the ribosome (138).
In some clinical isolates, methylation of either G1405 or A1408 mediated by specific 16S rRNA methyltransferases confers resistance to many h44-binding aminoglycosides (139). The methylase-encoding genes are usually found on plasmids that probably originated in aminoglycosides-producing actinomycetes. It has also been shown that the loss of the normal G527 methylation caused by mutations in a gene encoding a 7-methylguanosine (m7G) methyltransferase (RsmG/GidB) confers low-level streptomycin resistance (140).
4.4. Mechanisms of Resistance to Macrolides, Lincosamides, Streptogramins B, and Ketolides
Antibiotics, such as macrolides, lincosamides, streptogramins B, and ketolides (collectively termed MLSBK), share overlapping binding positions in the NPET on the 50S subunit. Therefore, resistance developed against one such drug often extends to other MLSBK members. A common mechanism of resistance to MLSBK antibiotics is mono- or di-methylation of A2058 of the 23S rRNA, which is located in the upper part of NPET. These modifications prevent binding of all MLSBK antibiotics to the ribosome (Figure 8c). This type of ribosome modification is catalyzed by a class of plasmid-encoded resistance enzymes called erythromycin resistance methyltransferases. Because A2058 methylation decreases cell fitness (141), erythromycin resistance methyltransferase genes are generally inducible by erythromycin and other macrolides through a well-studied mechanism of translation attenuation (142). Another methyltransferase, Cfr, targets A2503 of the 23S rRNA, and its activity results in the formation of a dimethylated m2m8A2503 residue (Figure 8d) that confers resistance to a broad range of antibiotics, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramins A (67, 143). Interestingly, in the chromosome of the MRSA strain CM05, the ermB and cfr genes exist in a single operon and modification of both A2058 and A2503 synergistically enhances resistance to 16-membered-ring macrolides (144).
Mutations of A2058 or A2059 that confer resistance to MLSBK antibiotics have been found in pathogens having one or two rRNA operons (145). In addition, mutations in ribosomal proteins uL4 and uL22 can reduce susceptibility to one or more MSLBK antibiotics. These proteins have extended loops that form the wall of the peptide exit tunnel, near the binding site of MLSBK antibiotics. Although these loops do not directly contact ribosome-bound drugs, mutations in them can trigger conformational changes of surrounding rRNA that propagate into the binding sites of MSLBK antibiotics (146).
5. IMPLICATIONS FOR DRUG DESIGN
Since the discovery of the first antibiotics, natural products of microbial origin have been recognized as a highly valuable source of lead compounds for the development of new therapeutic agents. The rise of life-threatening infections caused by multidrug-resistant bacteria and the spread of plasmid-mediated resistance against the most effective antibiotics have stimulated the development of new approaches to revive the natural products discovery pipeline and to enrich the arsenal of structural scaffolds suitable for optimization by medicinal chemists. An important criterion that should be applied to the search of new chemical scaffolds is the ability to overcome acquired resistance, which, in the case of the ribosome, occurs mainly through modification of the target sites and, much more rarely, through mutation(s) of rRNA.
Currently, to minimize possible cross-resistance with the existing clinical drugs there is a high demand for new chemical scaffolds which would interact with new ribosomal sites and inhibit translation via novel mechanisms of action. Unfortunately, screening of the small-molecular metabolites produced by cultivatable microorganisms often results in the rediscovery of already known compounds. This problem could be tackled by genome mining, a novel approach that allows one to harness a much greater chemical diversity and can result in the discovery of entirely new molecular scaffolds. Analysis of genomic data makes it possible to identify gene clusters encoding biosynthetic pathways for potential drug candidates, which may otherwise escape attention due to their inactivity under laboratory growth conditions (147). The human microbiome, for example, has been shown to contain a wealth of antibiotic biosynthesis genes, and recently two new promising antibiotics, lactocillin and lugdunin, were isolated from human commensals (148, 149). Ribosomally-synthesized post-translationally modified peptides are among the most abundant antimicrobial agents synthesized by human microbiota (148, 150). Recently, the first member of a new class of protein synthesis inhibitors, klebsazolicin (KLB), which comes from the opportunistic human pathogen Klebsiella pneumoniae, was discovered using this genome-guided approach (119). KLB is the first linear azole-containing peptide for which the mode of binding to its target has been structurally characterized (see section 3.3). KLB is synthesized on the ribosome as a precursor, which next undergoes post-translational modifications by dedicated enzymes encoded in a compact gene cluster (119). Moreover, KLB can be expressed in a surrogate E. coli host, which suggests avenues for future rational drug design: by changing the primary sequence of amino acids in the KLB precursor, we can change properties of the final processed compound.
Most of the major classes of antibiotics were discovered more than 60 years ago. Since then, the primary tool by which new antibiotics have been obtained is semi-synthesis, which is to say chemical modification of natural compounds produced by fermentation. Today, macrolide antibiotics are among the most clinically successful antimicrobial compounds. However, most of the macrolides that are approved for use in clinic have been produced by semi-synthesis, an approach which is inherently limited by the low chemical reactivity of the atoms at most positions of the macrolactone ring. As a result, only a handful of macrolides have been commercialized since the discovery of erythromycin in 1948. Recently, a novel platform has been developed for the fully synthetic preparation of macrolide antibiotics (151). This platform provides both a discovery engine for structurally novel antibiotic candidates that would be difficult or impossible to obtain from erythromycin, as well as a basis for their eventual manufacture. Using simple building blocks and a highly convergent assembly process, more than 300 structurally diverse macrolide antibiotic candidates have been synthesized including the approved drug telithromycin, and the promising clinical candidate solithromycin (151).
In combination with recent advances in computational and structural methods, the determination of ribosomal structures with substrates and inhibitors bound has opened the possibility of developing new antibiotics by structure-based methods. Although it is accepted that structures of antibiotics complexed with the ribosome in a functionally meaningful state provide detailed views of the binding pockets that, in turn, could be used this way, in reality, however, little use has been made of the structures in the beginning of the drug development process. Another extremely important aspect of the structure-based drug design process is the resolution of the structures used to guide it. Only high-resolution ribosome structures (3Å or better) can provide reliable information on the interactions between individual chemical groups that are being changed during optimization. Finally, structural studies seeking to explain mechanisms of action of ribosome-targeting antibiotics should focus on ribosome functional complexes carrying mRNA and acylated-tRNAs, which most closely mimic the states of the ribosome in a living cell. A number of structural models showing how various drugs interact with a vacant bacterial ribosome have provided information that is incomplete or possibly even misleading (22, 152). Often, this is because key interactions critically depend on the presence of ribosome ligands, such as mRNA, tRNAs or translation factors.
6. CONCLUSION
The rise of pathogens that are resistant to antibiotics warrants the search for new drugs capable of inhibiting bacterial growth and curing infections. Many of the antibiotics currently in use target the bacterial ribosome and usually hamper protein synthesis by binding to ribosomal functional centers, such as the decoding center, the peptidyl transferase center, and the peptide exit tunnel. High-resolution structures of the ribosome in complex with factors and inhibitors have already been instrumental in advancing our understanding of the mechanism of protein synthesis and its modes of inhibition. As new ribosome functional complex structures become available, we expect that the incremental but crucially important progress on drug discovery will continue and lead to novel protein synthesis inhibitors that will be part of the next-generation antibiotic arsenal.
ACKNOWLEDGMENTS
We thank Peter B. Moore for critical reading of the manuscript and valuable suggestions. This work was supported by grants from the National Natural Science Foundation of China (No. 31770784) to J.L., the National Institutes of Health (GM022778) to T.A.S., by Illinois State startup funds to Y.S.P., by the Sealy Center for Structural Biology and Molecular Biophysics at the University of Texas Medical Branch (UTMB), and by a Rising Science and Technology Acquisition and Retention (STARs) Program award from the University of Texas System to M.G.G.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
Full text links
Read article at publisher's site: https://doi.org/10.1146/annurev-biochem-062917-011942
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9176271
Citations & impact
Impact metrics
Article citations
DMDA-PatA mediates RNA sequence-selective translation repression by anchoring eIF4A and DDX3 to GNG motifs.
Nat Commun, 15(1):7418, 02 Sep 2024
Cited by: 1 article | PMID: 39223140 | PMCID: PMC11369270
RNA polymerase stalling-derived genome instability underlies ribosomal antibiotic efficacy and resistance evolution.
Nat Commun, 15(1):6579, 03 Aug 2024
Cited by: 0 articles | PMID: 39097616 | PMCID: PMC11297953
Fluorogenic RNA-Based Biosensors of Small Molecules: Current Developments, Uses, and Perspectives.
Biosensors (Basel), 14(8):376, 01 Aug 2024
Cited by: 0 articles | PMID: 39194605 | PMCID: PMC11352751
Review Free full text in Europe PMC
Preserving the efficacy of antibiotics to tackle antibiotic resistance.
Microb Biotechnol, 17(7):e14528, 01 Jul 2024
Cited by: 1 article | PMID: 39016996 | PMCID: PMC11253305
Review Free full text in Europe PMC
Hibernating ribosomes as drug targets?
Front Microbiol, 15:1436579, 29 Jul 2024
Cited by: 0 articles | PMID: 39135874 | PMCID: PMC11317432
Review Free full text in Europe PMC
Go to all (128) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (Showing 32 of 32)
-
(1 citation)
PDBe - 5DOYView structure
-
(1 citation)
PDBe - 5W4KView structure
-
(1 citation)
PDBe - 2HHHView structure
-
(1 citation)
PDBe - 4V9QView structure
-
(1 citation)
PDBe - 1I95View structure
-
(1 citation)
PDBe - 5O2RView structure
-
(1 citation)
PDBe - 4WFAView structure
-
(1 citation)
PDBe - 5HL7View structure
-
(1 citation)
PDBe - 3CF5View structure
-
(1 citation)
PDBe - 4V9BView structure
-
(1 citation)
PDBe - 1K8AView structure
-
(1 citation)
PDBe - 4V7CView structure
-
(1 citation)
PDBe - 4V53View structure
-
(1 citation)
PDBe - 1VQ9View structure
-
(1 citation)
PDBe - 4WQUView structure
-
(1 citation)
PDBe - 4WT8View structure
-
(1 citation)
PDBe - 4W2HView structure
-
(1 citation)
PDBe - 4W2IView structure
-
(1 citation)
PDBe - 1FJGView structure
-
(1 citation)
PDBe - 4V51View structure
-
(1 citation)
PDBe - 5IWAView structure
-
(1 citation)
PDBe - 4Z8CView structure
-
(1 citation)
PDBe - 4DR5View structure
-
(1 citation)
PDBe - 4W2FView structure
-
(1 citation)
PDBe - 4V8AView structure
-
(1 citation)
PDBe - 4V64View structure
-
(1 citation)
PDBe - 4V7VView structure
-
(1 citation)
PDBe - 4V7WView structure
-
(1 citation)
PDBe - 4V7XView structure
-
(1 citation)
PDBe - 4V7ZView structure
-
(1 citation)
PDBe - 4Z3SView structure
-
(1 citation)
PDBe - 5KCSView structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Blast from the Past: Reassessing Forgotten Translation Inhibitors, Antibiotic Selectivity, and Resistance Mechanisms to Aid Drug Development.
Mol Cell, 61(1):3-14, 12 Nov 2015
Cited by: 34 articles | PMID: 26585390
Review
Antibiotics and the ribosome.
Mol Microbiol, 59(6):1664-1677, 01 Mar 2006
Cited by: 90 articles | PMID: 16553874
Review
Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res, 44(5):2439-2450, 24 Jan 2016
Cited by: 94 articles | PMID: 26809677 | PMCID: PMC4797290
Ribosome-targeting antibiotics and mechanisms of bacterial resistance.
Nat Rev Microbiol, 12(1):35-48, 01 Jan 2014
Cited by: 452 articles | PMID: 24336183
Review





