Abstract
Free full text

Molecular Evolution of the Pathogenicity Island of Enterotoxigenic Bacteroides fragilis Strains
Abstract
Enterotoxigenic Bacteroides fragilis (ETBF) strains, which produce a 20-kDa zinc metalloprotease toxin (BFT), have been associated with diarrheal disease in animals and young children. Studying a collection of ETBF and nontoxigenic B. fragilis (NTBF) strains, we found that bft and a second metalloprotease gene (mpII) are contained in an ~6-kb pathogenicity island (termed B. fragilis pathogenicity island or BfPAI) which is present exclusively in all 113 ETBF strains tested (pattern I). Of 191 NTBF strains, 100 (52%) lack both the BfPAI and at least a 12-kb region flanking BfPAI (pattern II), and 82 of 191 NTBF strains (43%) lack the BfPAI but contain the flanking region (pattern III). The nucleotide sequence flanking the left end of the BfPAI revealed a region with the same organization as the mobilization region of the 5-nitroimidazole resistance plasmid pIP417 and the clindamycin resistance plasmid pBFTM10, that is, two mobilization genes (bfmA and bfmB) organized in one operon and a putative origin of transfer (oriT) located in a small, compact region. The region flanking the right end of the BfPAI contains a gene (bfmC) whose predicted protein shares significant identity to the TraD mobilization proteins encoded by plasmids F and R100 from Escherichia coli. Nucleotide sequence analysis of one NTBF pattern III strain (strain I-1345) revealed that bfmB and bfmC are adjacent to each other and separated by a 16-bp GC-rich sequence. Comparison of this sequence with the appropriate sequence of ETBF strain 86-5443-2-2 showed that in this ETBF strain the 16-bp sequence is replaced by the BfPAI. This result defined the BfPAI as being 6,036 bp in length and its precise integration site as being between the bfmB and bfmC stop codons. The G+C content of the BfPAI (35%) and the flanking DNA (47 to 50%) differ greatly from that reported for the B. fragilis chromosome (42%), suggesting that the BfPAI and its flanking region are two distinct genetic elements originating from very different organisms. ETBF strains may have evolved by horizontal transfer of these two genetic elements into a pattern II NTBF strain.
Enterotoxigenic Bacteroides fragilis (ETBF) strains have been associated with watery diarrheal disease in livestock and young children (27, 28, 34, 35, 38, 40). The primary virulence factor proposed for these strains is a zinc-dependent metalloprotease toxin (B. fragilis toxin [BFT]) (23). This toxin stimulates fluid accumulation in lamb ligated ileal loops (LLIL) and alters the morphology of human intestinal epithelial cells in vitro, especially HT29/C1 cells (3, 17, 27, 30, 40, 49).
We recently reported cloning and sequencing the B. fragilis toxin gene (bft) from a cosmid library of ETBF strain 86-5443-2-2 (piglet isolate) (9) and, at the same time, Kling et al. reported sequencing the bft gene from ETBF strain VPI 13784 (lamb isolate) (16). The bft gene from both strains encodes a preprotoxin of 44 kDa that is processed to yield a biologically active toxin of ca. 20 kDa, which is secreted into the culture supernatant. By comparing the reported bft sequences from ETBF strains 86-5443-2-2 and VPI 13784, we developed specific oligonucleotide probes derived from an area of reduced homology in the sequence of these bft genes. Hybridization of these oligonucleotide probes to ETBF strains 86-5443-2-2 and VPI 13784 revealed that each gene could be independently identified (9). Furthermore, examination of a collection of ETBF strains revealed that ETBF strains contain one of these bft genes but not both (9). We designated these alleles bft-1 and bft-2 representing the genes derived from strains VPI 13784 and 86-5443-2-2, respectively. In additional studies (50), we found that (i) BFT purified from culture supernatant of ETBF strain 86-5443-2-2 (BFT-2) had modest but consistently greater biological activity when tested on HT29/C1 cells than BFT purified from ETBF VPI 13784 (BFT-1) and that (ii) BFT-1 and BFT-2 were eluted with different concentrations of NaCl from a high-resolution anion-exchange column (MonoQ) and exhibit different electrophoretic mobilities on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. These results indicate that ETBF VPI 13784 and 86-5443-2-2 produce two distinct isotypes of BFT.
In studying a collection of ETBF and nontoxigenic B. fragilis (NTBF) strains, we also found that bft is located in an ~6-kb region present only in ETBF strains (7). Later, sequence analysis of this 6-kb region revealed that, in addition to bft, this region contains another metalloprotease distinct from bft. Together, these data indicate that bft is contained in a pathogenicity island (8). Moncrief et al. recently reported the sequence of this 6-kb region (termed a pathogenicity islet by the authors) from strain VPI 13784 (24). In the present study we analyzed the DNA sequence of this specific 6-kb region and its flanking region from ETBF 86-5443-2-2 and a large collection of ETBF and NTBF strains. Our results suggest that the specific 6-kb region and its flanking region form different mobile genetic elements and that ETBF strains may have evolved by acquisition of these two genetic elements.
MATERIALS AND METHODS
Bacterial strains, vectors, and growth conditions.
ETBF 86-5443-2-2 (piglet isolate) was used for the identification and characterization of the B. fragilis pathogenicity island (BfPAI). B. fragilis strains used for sequencing, determination of integration site, and the mobilization of BfPAI are listed in Table Table1.1. For colony blot hybridizations, a total of 113 ETBF and 191 NTBF strains were tested. Of 75 intestinal ETBF strains, 39 were from the United States (human), 22 were from Bangladesh (human), 4 were from Thailand (human), 7 were from the United States (calf), and 3 were from the United States (lamb). Of 38 extraintestinal ETBF strains, 4 were isolated in the United States (1 from blood, 1 from pleural fluid, and 2 from wound), and 34 were isolated in Korea (12 from blood, 6 from wound, 4 from pus, 3 from peritoneal fluid, 2 from bile, and 7 from other body fluids). Of 75 intestinal NTBF strains, 6 were from the United States (human), 25 were from Bangladesh (human), 38 were from Thailand (human), 5 were from the United States (calf), and 1 was from the United States (lamb). Of the 116 extraintestinal NTBF strains, 60 were from the United States (27 from blood, 6 from pleural fluid, 6 from abscess, 6 from bile, 11 from wound, and 4 from other body fluids), and 56 were isolated from Korea (11 from blood, 18 from wound, 11 from pus, 4 from peritoneal fluid, and 12 from other body fluids). Isolates were identified as ETBF or NTBF on the basis of the activity on HT29/C1 cells and/or the LLIL assay.
TABLE 1
B. fragilis strains used in this study
study
| Strain | Origin | Characteristica | Source or reference |
|---|---|---|---|
| 86-5443-2-2 | Pig | ETBF (intestinal) | L. L. Myers |
| VPI 13784 | Lamb | ETBF (intestinal) | T. D. Wilkins |
| J38-1 | Human | ETBF (intestinal) | 26, 49 |
| 20793-3 | Human | ETBF (intestinal) | 26, 49 |
| DS-64 | Human | ETBF (intestinal) | 26, 49 |
| DS-233 | Human | ETBF (intestinal) | 26, 49 |
| DS-49 | Human | ETBF (intestinal) | 26, 49 |
| I-1356-1 | Calf | ETBF (intestinal) | L. L. Myers |
| TM4000 | Human | NTBF (abscess) | M. H. Malamy |
| 077225-2 | Human | NTBF (intestinal) | L. L. Myers |
| I-1345 | Calf | NTBF (intestinal) | L. L. Myers |
| LM-20 | Human | NTBF (blood) | 26 |
| LM-12 | Human | NTBF (blood) | 26 |
| K518 | Human | NTBF (abscess) | G.-T. Chung |
Human intestinal and extraintestinal B. fragilis strains from the United States were gifts from R. B. Sack, L. L. Myers, or the microbiology laboratory of the Johns Hopkins Hospital, as previously reported (26, 49). Strains from Korea, Bangladesh, and Thailand and the animal (calf and lamb) B. fragilis strains were gifts from G.-T. Chung, R. B. Sack, P. Escheverria, and L. L. Myers, respectively.
Bacteroides thetaiotaomicron BT4107 and BT400, used as bacterial mating controls, were gifts from N. B. Shoemaker and A. A. Salyers. Escherichia coli HB101 and DH5α were used as hosts for maintaining cosmid and plasmid clones, respectively. The cosmid vector pHC79 was used to construct the B. fragilis 86-5443-2-2 genomic library, whereas pBluescript II SK(+) (Stratagene, Inc., La Jolla, Calif.) was used for subcloning cosmid DNA. B. fragilis strains were propagated anaerobically on BHC medium (3.7 g of brain heart infusion base [Difco Laboratories] per liter and 0.1 mg of vitamin K per liter, 0.5 mg of hemin per liter, 50 mg of l-cysteine per liter [all from Sigma, St. Louis, Mo.]), and E. coli strains were grown on Luria agar or in Luria broth. The following antibiotic concentrations were used unless otherwise noted: ampicillin, 200 μg/ml; tetracycline, 3 μg/ml; rifampin, 20 μg/ml; and trimethoprim, 100 μg/ml.
DNA isolation analysis.
Plasmid DNA was extracted by the alkali lysis method (37) or by using Qiagen columns (Qiagen, Inc.). The CsCl-ethidium bromide density gradient centrifugation technique was used to identify plasmids in ETBF strain 86-5443-2-2. Cosmids 5G11 and 11F11 containing bft-2 were restriction mapped by Southern blot walking according to the conditions described previously (9). Purification of DNA fragments and extraction from gel slices were performed with Qiaex II gel extraction kit (Qiagen, Inc.).
Colony blot hybridizations.
Colony blots of B. fragilis strains were prepared by the technique described previously (9). Briefly, B. fragilis organisms grown overnight on BHC agar were transferred to Whatman 541 filters. The filters were microwave processed in alkali solution (0.5 M NaOH, 1.5 M NaCl) followed by neutralization in 2 M ammonium acetate. The filters were hybridized with either fragment probes or specific oligoprobes. The fragment probes were labeled with [α-32P]dATP by random priming (Random-Prime DNA-Labeling Kit; Boehringer GmbH, Mannheim, Germany), hybridized in 50% formamide–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS–1 mM EDTA–1× Denhardt’s solution at 37°C, and washed with 5× SSC–0.1% SDS at 65°C. Synthetic oligonucleotide probes were end labeled with [γ-32P]dATP by using T4 polynucleotide kinase (37), hybridized in 6× SSC–5× Denhardt’s solution–1 mM EDTA (pH 8) for 16 h at 56°C and washed twice in 1× SSC–1% SDS at 56°C for 10 min.
Nucleotide sequence analysis.
Recombinant plasmids and PCR products were sequenced by the fluorescent dideoxy terminator method of cycle sequencing (20) on a PE/ABd 373a automated DNA sequencer at the DNA Analysis Facility of Johns Hopkins University. Sequences were compiled with Sequencher software (Gene Codes Corp., Ann Arbor, Mich.). DNA and amino acid sequences were analyzed by using programs developed by the Genetic Computer Group at the University of Wisconsin (4), and a sequence homology search was performed by using the National Center for Biotechnology Information (NCBI) BLAST server (1).
Expression of MPII in E. coli.
The T7 promoter polymerase expression system (Novagen pET-26b[+]) was used to express metalloprotease MPII in E. coli. Primers PrimA and PrimB.1 (sequences are described in Table Table2)2) generated a PCR product of the DNA region encoding the proprotein and mature protein of MPII and containing a BamHI and XhoI at the 5′ and 3′ ends of the product, respectively. Vector pET-26b(+) carries an N-terminal pelB signal sequence for potential periplasmic location. Cloning of the PCR product in the BamHI and XhoI sites of the pET-26b(+) vector generated a fusion protein containing pelB signal sequence-MPII proprotein-mature protein, whose expression was under the control of the T7 promoter. As recommended by the manufacturer, the mpII construction was first cloned into a nonexpression host (E. coli HB101) lacking the T7 RNA polymerase. Once established in a nonexpression host, plasmids were transferred into the expression host (E. coli BL21) containing a copy of the T7 RNA polymerase gene, which permitted expression of MPII. Expression of MPII in E. coli was determined by Western blot with anti-ETBF 86-5443-2-2 antibodies.
TABLE 2
Primers and oligonucleotide probes used in this study
study
| Primer or oligoprobe | Sequence (5′ to 3′) |
|---|---|
| Primers | |
 P1T7 P1T7 | GCTGGTAGACTACCTGAGTAAGGAGTC |
 P1T7-1 P1T7-1 | GCTTCCGTACCCAGGTATCTCTCCATA |
 P1T3 P1T3 | TTCAACCTGATCGATCCGGAAGATCCG |
 P1T3-1 P1T3-1 | GGTAGTGCTTATGTCCCTGCAACCCTA |
 PrimA PrimA | GCATGGATCCATGTGCTGACGATCTTCTT |
 PrimB.1 PrimB.1 | CGCGCTCGAGTTTTTGTATGCATTCCAAC |
 Confir1 Confir1 | CAGCTTCATTAGAACATGCAGCTAATAA |
 Confir2 Confir2 | CTGTCCATAACTGTATTCCATTTGATAC |
 Confir3 Confir3 | CACCGACAGAACATATCCACTCAACTG |
 Confir4 Confir4 | GGTGTTGCAAGAGAGGCTCAAACACAT |
 OlirteB1 OlirteB1 | GACTACGCCGAAGTTAATACCG |
 OlirteB2 OlirteB2 | TCGTGCAGGCGGTACAGAAGAT |
| Oligoprobes | |
 MT086 MT086 | GGTGCTAGGCATGCGGATGATC |
 MTVPI MTVPI | GGCGCTGAGCATACGGATAATT |
 K469 K469 | CAGATGCAGGATGCGGCGAACT |
 Olibft3 Olibft3 | CGAGGTGTCTGCTCTTGAATC |
Activity of MPII on HT29/C1 cells and E-cadherin.
The cell-free supernatant and the supernatants of whole-cell lysate of strain F2 (E. coli expressing MPII) were tested for biological activity on HT29/C1 cells and E-cadherin. Activity on HT29/C1 cells was determined at 3 or 24 h as described previously (26, 49). Activity of MPII on E-cadherin was determined as described previously (51). Briefly, HT29/C1 cells were treated with sterile MPII-containing filtrates for 16 h at 37°C. Cleavage of E-cadherin was determined by Western blot by using antibodies to the cytoplasmic domain of E-cadherin (a gift of James Nelson, Stanford University, Palo Alto, Calif.) as the primary antibody (1:1,000 dilution) and anti-rabbit immunoglobulin G-horseradish peroxidase as the secondary antibody (1:10,000 dilution). Purified BFT from ETBF strain 86-5443-2-2 was used as a positive control in both assays. Negative controls included medium alone and crude supernatant filtrates of the NTBF strain 077225-2 and E. coli BL21 containing plasmid pET26.
Bacterial matings.
Thymidine-dependent mutant strains 086Thy−3, I-1356-1Thy−1, I-1345Thy−2, and K518Thy−2 derived from ETBF 86-5443-2-2 (contains bft-2), ETBF I-1356-1 (contains bft-1), NTBF I-1345 (pattern III; see Results), and NTBF K518 (pattern II; see Results), respectively, were used as donor strains. All donor strains were resistant to tetracycline (Tcr) and sensitive to rifampin (Rifs). The presence of the tetracycline-inducible transposon (Tcr element) in each Tcr strain was confirmed by PCR by using a set of primers derived from the regulatory gene rteB (OlirteB1 and OlirteB2 [sequences in Table Table2])2]) (46). NTBF TM4000 (Tcs Rifr) was used as the recipient strain. B. thetaiotaomicron BT4107 (a thymidine-dependent donor containing the TcrEmr-DOT element) and B. thetaiotaomicron BT4001 (a rifampin-resistant recipient) strains were used as bacterial mating controls. Conjugation matings were done by using the filter mating procedure as described by Shoemaker et al. (42). Briefly, exponential cultures of donor and recipient bacteria were centrifuged, washed, and resuspended in fresh medium. Donor and recipient cells were mixed in a final ratio of 2:1, and 20 μl of this mix was applied to nitrocellulose filters placed on BHC medium containing thymidine (100 μg/ml). The filter matings were incubated overnight at 35°C, and transconjugants were selected on BHC medium containing rifampin (20 μg/ml) and tetracycline (3 μg/ml) and lacking thymidine to avoid the growth of donor Rifr spontaneous mutants. Transconjugants were confirmed to contain the Tcr element by PCR by using primers OlirteB1 and OlirteB2 (Table (Table2).2). Donor strains were grown in medium containing 1 μg of tetracycline per ml before mating to induce factors in the Tcr element. The frequency of transfer of the Tcr element was determined by dividing the number of transconjugants by the number of recipients.
PCR conditions.
PCR primers used in this study are listed in Table Table2.2. PCRs were performed with Taq polymerase (1.5 U) in 50 μl containing either chromosomal DNA (20 to 50 ng) or plasmid (5 to 10 ng), primers (25 pmol), deoxynucleoside triphosphates (200 μM), and MgCl2 (1.5 M). Amplification used a hot start (94°C for 5 min), followed by denaturation at 94°C for 1 min, annealing at 66°C for 2 min, and extension at 72°C for 1 min. The amplification cycle was repeated either 29 times for determination of the integration site of BfPAI or 19 times for generating mpII for the cloning experiments and was followed by a final extension at 72°C for 5 min. The amplified products were directly sequenced or cloned into the TA cloning vector (Invitrogen, San Diego, Calif.).
Data analysis.
Data were analyzed by the chi-square formula; a P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
We originally submitted the mpII gene sequence from ETBF strain 86-5443-2-2 to GenBank as bft3 (GenBank accession number AF056297). However, we adopt here the nomenclature mpII in accordance with that used by Moncrief et al. (24). The bfmA, bfmB, bfmC, and open reading frame (ORF) ORF-A genes were assigned accession numbers AF118241, AF118242, AF118243, and AF118244, respectively.
RESULTS
The region flanking the bft gene is present only in ETBF strains.
To determine whether the DNA region flanking bft-1/bft-2 was specific for ETBF, two overlapping cosmids (11F11 and 10G11) containing bft-2 (from ETBF strain 86-5443-2-2) were mapped with several restriction enzymes. Eight adjacent restriction fragments were used to probe colony blots of 113 ETBF and 191 NTBF strains. Five patterns of hybridization were identified (Fig. (Fig.1).1). All of the probes spanning an 18-kb sequence hybridized to the 113 ETBF strains (pattern I). Of 191 NTBF strains, 100 (52%) lacked this 18-kb chromosomal region (pattern II), and 82 (43%) lacked only an ~6-kb region (containing probes G, Tox, H, and B) flanking bft-2 (pattern III). Of the 191 NTBF strains, 7 (3.6%) lacked the region spanning probe E in addition to the 6-kb region flanking bft (pattern IV). Two NTBF strains containing the bft gene (i.e., hybridized to the Tox probe) did not hybridize to probes J, I, D, B, and H in the region spanning ca. 11.5 kb upstream of bft-2 (pattern V). These two strains hybridized with an oligoprobe specific for the DNA region downstream of the metalloprotease motif of the bft-2 allele (oligoprobe MT086), but neither of them hybridized to an oligoprobe (oligoprobe K469) derived from a common region of bft-1 and bft-2 that encodes the N-terminal region of the mature protein, suggesting that these two NTBF strains contain a deletion in the DNA encoding the amino terminus of the mature BFT protein, as well as the region upstream of the gene.
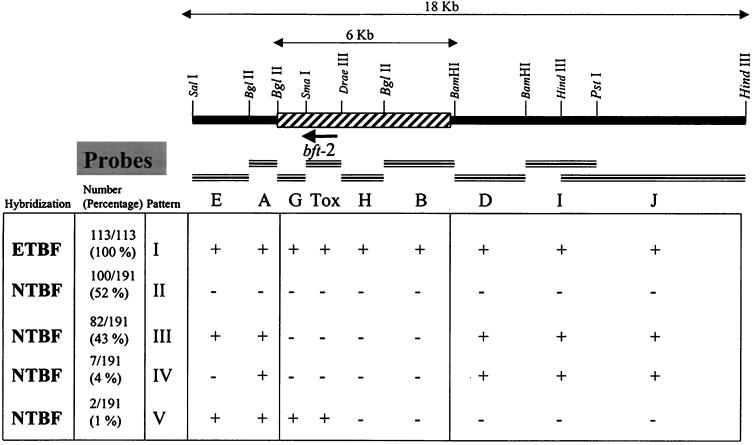
Partial restriction map of the region flanking bft in ETBF 86-5443-2-2. Arrow shows bft and the direction of its transcription. Probes used in the colony blot hybridizations are shown in the black bars. The results of hybridization of these probes to B. fragilis strains (Table (Table2)2) are indicated beneath each probe. The 6-kb region spanning probes G, Tox, H, and B (the striped bar) is the specific locus present only in ETBF strains (BfPAI). Only the restriction sites used for the construction of the probes are shown in the figure.
Overall, pattern I ETBF strains were isolated at a significantly higher frequency from intestinal (75 of 150 [50%]) than extraintestinal (38 of 154 [25%]) samples (P < 0.001) (Table (Table3).3). All B. fragilis strains with hybridization patterns IV and V were isolated from extraintestinal sources (Table (Table3).3). In the United States and Thailand, only 6% (4 of 64) of extraintestinal B. fragilis strains or 10% (4 of 42) of intestinal B. fragilis strains, respectively, are ETBF strains (pattern I). Interestingly, in these locations the NTBF strains with pattern II were identified with higher frequency than the NTBF strains with pattern III (Table (Table4).4). In contrast, in intestinal B. fragilis strains from the United States and Bangladesh, as well as extraintestinal B. fragilis strains from Korea, places where ETBF strains have been detected with higher frequency (3a, 34, 35), either NTBF strains with pattern II were not detected (intestinal, United States) or a similar proportion of NTBF strains with patterns II and III were identified (Table (Table4).4). Because no significant differences in the proportion of pattern II versus pattern III NTBF strains were found when areas with lower (extraintestinal, United States; intestinal, Thailand) and higher (extraintestinal, Korea; intestinal, Bangladesh) numbers of ETBF strains were compared, the groups were combined and analyzed. This comparison revealed that pattern II NTBF strains were identified in a significantly higher proportion from areas with lower recovery of ETBF isolates (P < 0.02).
TABLE 3
Frequency of patterns of hybridization in B. fragilis strains isolated from intestinal and extraintestinal sources
sources
| Pattern | No. of strains/total (%)
| |
|---|---|---|
| Intestinal | Extraintestinal | |
| I | 75/113 (66) (66) | 38/113 (33.6) (33.6) |
| II | 35/100 (35) (35) | 65/100 (65) (65) |
| III | 40/82 (48.7) (48.7) | 42/82 (51) (51) |
| IV | 0/7 | 7/7 (100) (100) |
| V | 0/2 | 2/2 (100) (100) |
TABLE 4
Frequency of patterns of hybridization in intestinal B. fragilis strains isolated from The United States, Bangladesh, and Thailand and in extraintestinal B. fragilis strains isolated from The United States and Korea
Korea
| Source and pattern | No. of strains/total (%), type |
|---|---|
| Intestinal (United States) | |
 I I | 49/49 (100), (100), ETBF ETBF |
 III III | 12/12 (100), (100), NTBF NTBF |
| Extraintestinal (United States) | |
 I I | 4/4 (100), (100), ETBF ETBF |
 II II | 36/60 (60), (60), NTBF NTBF |
 III III | 15/60 (25), (25), NTBF NTBF |
 IV IV | 7/60 (11), (11), NTBF NTBF |
 V V | 2/60 (3), (3), NTBF NTBF |
| Intestinal (Bangladesh) | |
 I I | 22/22 (100), (100), ETBF ETBF |
 II II | 11/25 (44), (44), NTBF NTBF |
 III III | 14/25 (56), (56), NTBF NTBF |
| Intestinal (Thailand) | |
 I I | 4/4 (100), (100), ETBF ETBF |
 II II | 24/38 (63), (63), NTBF NTBF |
 III III | 14/38 (37), (37), NTBF NTBF |
| Extraintestinal (Korea) | |
 I I | 34/34 (100), (100), ETBF ETBF |
 II II | 29/56 (52), (52), NTBF NTBF |
 III III | 27/56 (48), (48), NTBF NTBF |
Nucleotide sequence of the specific 6-kb region.
The colony blot hybridization results indicate that an ~6-kb region flanking bft is present exclusively in all ETBF strains (n = 113). To examine this region for the presence of additional potential virulence genes, in addition to the bft-2 sequence already reported (9), we determined the sequence of 3,045 bp downstream of bft-2 and 5,799 bp upstream of bft-2 (total sequenced region, 10,038 bp) (Fig. (Fig.2a).2a). Analysis of the specific 6-kb region revealed that, in addition to bft-2, this region contains two other ORFs (ORF-A and ORF-B), which are 375 and 1,180 bp in length, respectively. ORF-A is transcribed in the opposite direction from bft-2, and its stop codon is located 270 bp downstream of stop codon of bft-2 (Fig. (Fig.2a).2a). The predicted amino acid sequence of the protein encoded by ORF-A showed no significant similarity to any sequence in the database. ORF-B also transcribes in the opposite direction of bft-2, with the start codon located 1.6 kb upstream of the start codon of the bft-2 gene (Fig. (Fig.2a).2a). The predicted protein encoded by ORF-B is 44.36 kDa with a predicted lipoprotein signal peptide (18 amino acids) and a zinc-binding metalloprotease motif similar to BFT-1 and BFT-2. However, this predicted protein is only 29 and 28% identical to BFT-1 and BFT-2, respectively, and contains an ATP/GTP-binding site motif A (48) not found in BFT-1 or BFT-2. The predicted protein encoded by ORF-B was termed metalloprotease II (MPII) in accordance with the results of Moncrief et al. (24).
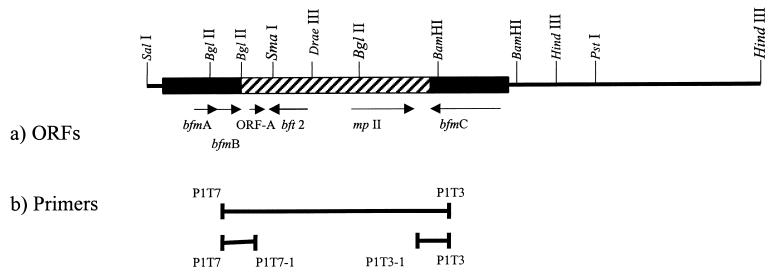
Partial restriction map of the region flanking bft in ETBF 86-5443-2-2. The thicker bar (both solid and striped) shows the region sequenced, and the striped bar shows the BfPAI. (a) ORFs detected in the BfPAI and flanking regions. Arrows show the locations of the ORFs and the direction of their transcription. (b) Relative positions of the primers and the expected PCR products to determine the integration site of the BfPAI. Only the restriction sites important for the construction of the probes (Fig. (Fig.1)1) are shown in the figure.
To correlate the presence of bft-1/bft-2 and mpII, a specific oligoprobe derived from mpII sequence (Olibft3) was designed. In colony blot hybridization, Olibft3 hybridized with all 113 ETBF strains containing either bft-1 or bft-2, but none of 191 NTBF strains. These results strongly suggest that bft-1/bft-2 and mpII colocalize in ETBF strains.
The G+C content of the specific 6-kb region (35%) differs substantially from that reported for the B. fragilis chromosome (42%) (45). This result and the presence of bft-2 and mpII in the 6-kb locus, which is unique to ETBF strains, suggest that this element is a pathogenicity island or islet (herein designated BfPAI for B. fragilis pathogenicity island).
Nucleotide sequence of the DNA region flanking the BfPAI.
Analysis of the sequence of the left end flanking the BfPAI revealed two ORFs of 540 and 948 bp (Fig. (Fig.2a),2a), which encode predicted proteins of 316 and 180 amino acids residues with calculated molecular masses of 21.37 and 37.24 kDa, respectively. A search in the GenBank and EMBL databases for related sequences revealed that these two ORFs, as well as the region immediately upstream of the ORFs, have the same organization as the mobilization region of the 5-nitroimidazole and clindamycin resistance plasmids of B. vulgatus (pIP417) and B. fragilis (pBFTM10), respectively (12, 47). The mobilization regions of both antibiotic resistance plasmids consist of two genes, named btgA and btgB in the clindamycin resistance plasmid, and mobA and mobB in the 5-nitroimidazole resistance plasmid. BtgA/MobA and BtgB/MobB proteins share 49 and 70% identity, respectively. Both genes are organized in an operon, and immediately upstream of the operon there is a putative origin of transfer (oriT). By using the NCBI BLAST server (1), the highest homology score for the predicted 316-amino-acid protein was found with the mobilization protein MobB [P(N)4.5e-15 by BLAST algorithm], with 41% of identity between amino acids 42 and 84, 46% identity between amino acids 176 and 203, 33% identity between amino acids 214 and 234, 46% identity between amino acids 17 and 31, and 38% identity between amino acids 139 and 151. The predicted 180-amino-acid protein shared 25 and 40% identity and similarity, respectively, with a limited region (102 amino acids) of MobA (with both the Gapped BLAST and PSI-BLAST searches). Due to the sequence similarity of the predicted proteins encoded by the ORF of 948 bp (316 amino acids) to MobB and the ORF of 540 bp (180 amino acids) to MobA and the analogous gene arrangement, we designated these ORFs bfmB (B. fragilis mobilization gene B) and bfmA (B. fragilis mobilization gene A), respectively.
Figure Figure3A3A shows the comparison of this region upstream of the BfPAI and the mobilization region of plasmid pBFTM10. Similar to btgB/mobB and btgA/mobA, the ATG initiation codon of bfmB overlaps the final codon of bfmA, suggesting that both genes are transcribed together, and the region immediately upstream to these genes revealed several features consistent with the origin of transfer (oriT) of conjugative or mobilizable plasmids (Fig. (Fig.3B).3B). This region is ca. 200 bp long and has a higher A+T content than flanking regions (72 versus 56%). The region contains three inverted repeat (IR) sequences (IR1, IR2, and IR3) with lengths of 7, 13, and 9 bp, respectively (Fig. (Fig.3B).3B). None of these three IR sequences have similarity with those found in the putative oriT from plasmid pIP417 or pBFTM10. The IR1 and IR3 elements have perfect right (R)- and left (L)-arm inverted nucleotide sequences. Only 11 of the 13 bp of IR2R and IR2L are identical. IR1(R)-IR1(L) and IR3(R)-IR3(L) are separated by 3 and 8 bp, respectively. Approximately 165 bp upstream of the start codon of bfmA a putative nick site was identified according to the consensus sequence established by Pansegrau and Lanka (31) from the IncP plasmid (RK2/RP4) (Fig. (Fig.3C).3C). There was no sequence similarity with the proposed nick region of plasmids pIP417 and pBFTM10.
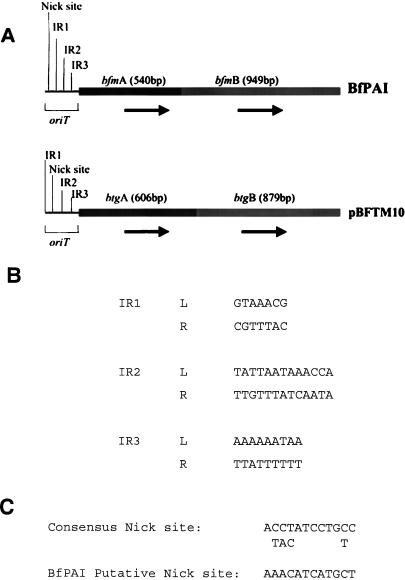
Putative mobilization region of the BfPAI. (a) Comparison between the putative mobilization region of BfPAI and the mobilization region of the clindamycin resistance plasmid pBFTM10. Arrows show the direction of gene transcription. (b) Nucleotide sequence (5′ to 3′) of the three IRs. (c) Nick site present in the putative oriT gene of the BfPAI.
The right end flanking the BfPAI contained an ORF (designated bfmC) of 1,602 bp which transcribes in the same direction as bft-2 but in the opposite direction of bfmA and bfmB (Fig. (Fig.2a).2a). The bfmC gene yields a predicted protein product (BfmC) of 534 residues with a molecular mass of 61.0 kDa. By using the BLAST search program, BfmC shares the highest homology score with a conjugal transfer protein TrsK of conjugative bacteriocin-producing plasmid pMRC01 from Lactococcus lactis [P(N)5e-09] (5) sharing 22% identity and 41% similarity over 395 amino acids. BfmC also shares significant similarity with mobilization proteins TraG of Helicobacter pylori [P(N)1.5e-07], TrsK of plasmid pGO1 from Staphylococcus aureus [P(N)3e-04], TraD of plasmids F and R100 from E. coli [P(N)0.003], and TrbC of Salmonella typhimurium [P(N)0.092] (14, 25, 29, 52). BfmC contains a perfect ATP/GTP-binding motif A (48) at the amino-terminal region; the same motif appears to be present in TrsK encoded by plasmids pMRCO1, as well as TraD encoded by plasmids F and R100. Alignment of these proteins in the region surrounding this motif is shown in Fig. Fig.4.4. The organization of the left and right ends flanking the BfPAI suggests that the BfPAI may have integrated into a plasmid. However, several different types of plasmid preparation procedures performed on ETBF strain 86-5443-2-2 failed to reproducibly reveal detectable plasmid DNA (data not shown).

Alignment of the region surrounding the ATP/GTP-binding site motif A in BfmC, TrsK (pMRCO1), TrsK (pGO1), TraD (R100), and TraD (F). Amino acids similar to those of the BfmC sequence are in capital letters and boldface. The ATP/GTP-binding site motif A in BfmC is underlined.
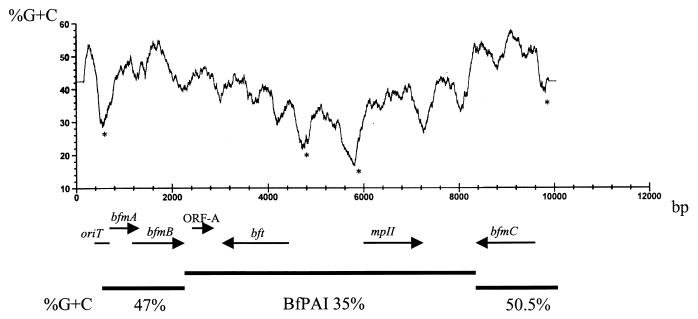
Analysis of the G+C content of the left-end (47%) and right-end (50%) sequences flanking the BfPAI revealed that the G+C content of these regions is significantly different from that of the BfPAI (Fig. (Fig.5).5). Interestingly, the G+C content of the regions immediately upstream of the bfmA/bfmB operon, bft, mpII, and bfmC all decrease sharply (asterisks in Fig. Fig.5).5). Similar results were found by Smith in the DNA sequences of transposon Tn4555 and plasmid pBI143 (45a). This may be due to the fact that promoter regions often have higher A+T content to facilitate separation of DNA for the initiation of transcription. The differing G+C contents of the BfPAI, the flanking region, and the B. fragilis chromosome suggest that the BfPAI is contained in yet another foreign genetic element.
Determination of the integration site of the BfPAI.
Based on the results of colony blot hybridization (Fig. (Fig.1)1) and the above sequence analysis, we hypothesized that ETBF strains could be generated by the integration of the BfPAI in certain NTBF strains (hybridization pattern III). Since the exact left and right borders of the BfPAI were contained in the DNA region spanning probes A and D, respectively (Fig. (Fig.1),1), two primers (P1T7 and P1T3 sequences in Table Table2)2) for these regions were designed (Fig. (Fig.2b)2b) and used in PCR experiments to amplify DNA from one strain of each pattern of hybridization. As expected, PCR yielded a product (ca. 1.6 kb) only in the NTBF strain with patterns III (strain I-1345) and IV (strain LM20) but not in any strain with the other patterns of hybridization (Fig. (Fig.6A).6A). NTBF strains of patterns II and V did not hybridize with probes A and/or D, which contain primers P1T7 and/or P1T3, respectively (Fig. (Fig.1),1), and in ETBF strains (pattern I) primers P1T7 and P1T3 flank a ca. 7-kb region (Fig. (Fig.2b),2b), a fragment too large to be amplified by Taq DNA polymerase under the conditions tested. To determine whether the integration of ETBF is site specific, we examined five additional NTBF strains with hybridization pattern III. All strains yielded products of 1.6 kb similar to strain I-1345 (data not shown). In addition, we designed two new primers (P1T7-1 and P1T3-1) containing sequences for the region in the BfPAI spanning probes G and B, respectively (Fig. (Fig.2b).2b). When P1T7 and P1T7-1 were used as primers, PCR experiments with six ETBF strains yielded the same predicted product (1.3 kb) (Fig. (Fig.6B).6B). Similarly, the six ETBF strains yielded the predicted 1.1-kb product when P1T3 and P1T3-1 were used as primers (Fig. (Fig.6C).6C). All of these results indicate that the integration of the BfPAI is in the same small region and in the same orientation in the cases examined and suggest that the integration of the BfPAI is site specific and unidirectional.
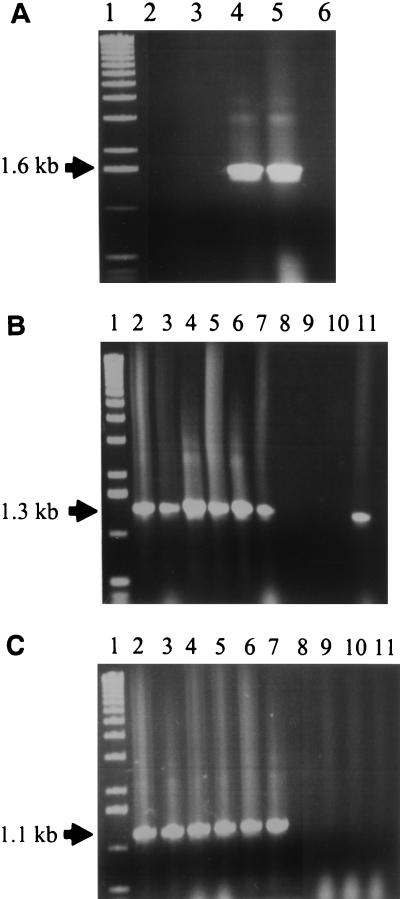
PCR analysis of the left and right junctions of BfPAI. (A) Screening for the integration site by using primers P1T7 and P1T3 (see Fig. Fig.2b).2b). PCR yielded a product (1.6 kb) only in NTBF strains with pattern III (strain I-1345) and IV (strain LM20). Lanes: 1, 1-kb ladder molecular weight marker; 2, ETBF 86-5443-2-2 (pattern I, positive control); 3, NTBF 077225-2 (pattern II); 4, NTBF I-1345 (pattern III); 5, NTBF LM-20 (pattern IV); 6, NTBF LM-12 (pattern V). (B) Screening for the left junction by using primers P1T7 and P1T7-1 (see Fig. Fig.2b).2b). PCR yielded a similar-sized fragment (1.3 kb) in six ETBF strains (pattern I) and NTBF LM12 (pattern V). Lanes: 1, 1-kb ladder molecular weight marker; 2, ETBF 86-5443-2-2 (pattern I); 3, ETBF VPI 13784 (pattern I); 4, ETBF J-38-1 (pattern I); 5, ETBF DS-64 (pattern I); 6, ETBF DS-233 (pattern I); 7, ETBF DS-49 (pattern I); 8, NTBF 077225-2 (pattern II); 9, NTBF I-1345 (pattern III); 10, NTBF LM-20 (pattern IV); 11, NTBF LM-12 (pattern V). (C) Screening for the right junction by using primers P1T3-1 and P1T3 (see Fig. Fig.2b).2b). PCR yielded a similar-sized fragment (1.1 kb) in all six ETBF strains (pattern I). Lanes are as described for panel B.
To determine the actual integration site and the exact borders of the BfPAI, we sequenced the 1.6-kb PCR product obtained from NTBF strain I-1345 by using primers P1T7 and P1T3. Comparison of this sequence and the appropriate sequence of ETBF strain 86-5443-2-2 defined the integration site and the putative left and right borders of the BfPAI (Fig. (Fig.7).7). Integration of BfPAI in ETBF strains is predicted to lie between the stop codons of bfmB and bfmC. ETBF strains lack a span of 16 bp at the integration site which is present in pattern III NTBF strain I-1345. According to this alignment, the BfPAI is calculated to be 6,036 bp in length. Analysis of the borders of the BfPAI revealed nearly perfect 12-bp direct repeats present 45 and 40 bp from the left and right ends of the BfPAI, respectively (data not shown).
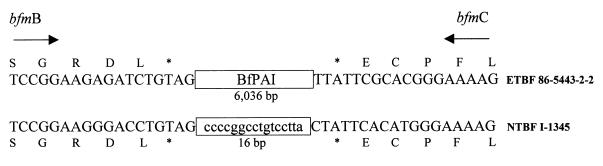
Comparison of the nucleotide-amino acid sequences of the region flanking the BfPAI of ETBF strain 86-5443-2-2 with the equivalent region of NTBF strain I-1345. Nucleotide sequences belonging to bfmB and bfmC are given in capital letters. Arrows show direction of gene transcription, and stop codons are indicated by asterisks. The 16-bp sequence lost in ETBF is given in lowercase letters.
Sequence comparison of 86-5443-2-2 and VPI 13784 BfPAIs.
Moncrief et al. (24) recently reported the sequence of the pathogenicity island (islet) from an ETBF strain containing bft subtype 1 (strain VPI 13784). Comparison of the BfPAI sequence from strain 86-5443-2-2 and the reported sequence from ETBF strain VPI 13784 reveals that the two sequences are not identical. A summary of this comparison is shown in Table Table5.5. Overall, both sequences share 97% identity. However, the BfPAI region containing the mpII gene is more conserved than the region containing bft (99 versus 95% identity, respectively). Similarly, the predicted proteins encoded by mpII from ETBF 86-5443-2-2 and VPI 13784 are 99% identical; whereas BFT-1 from VPI 13784 and BFT-2 from 86-5443-2-2 are 92% identical (9).
TABLE 5
Sequence comparison of the BfPAI from strains 86-5443-2-2 and VPI 13784
13784
| Nucleotide sequence or coding region | Value |
|---|---|
| Nucleotide sequence | |
 Lengtha Lengtha | 6,036 bp (86-5443-2-2), 6,032 bp (VPI 13784) |
 Overall % identity Overall % identity | 97 |
 Overall number of gaps Overall number of gaps | 33 |
 % Identity of region surrounding bft (bp 1 to 3,232) % Identity of region surrounding bft (bp 1 to 3,232) | 95 |
 No. of gaps in region surrounding bft (bp 1 to 3,232) No. of gaps in region surrounding bft (bp 1 to 3,232) | 33 |
 % Identity of region surrounding mpII (bp 3,233 to 6,036) % Identity of region surrounding mpII (bp 3,233 to 6,036) | 99 |
 No. of gaps in region surrounding mpII (bp 3,233 to 6,036) No. of gaps in region surrounding mpII (bp 3,233 to 6,036) | 0 |
 % Identity of partial direct repeats at the ends % Identity of partial direct repeats at the ends | 100 |
| Coding region | |
 % Identity of BFT % Identity of BFT | 92 |
 % Identity of MPII % Identity of MPII | 99 |
 ORF-A ORF-A | Not present in VPI 13784 |
 ORF1 ORF1 | Not present in 86-5443-2-2 |
Near the left end of the bft gene from strain VPI 13784, Kling et al. identified an ORF (ORF1) of 261 nucleotides that encodes a predicted protein with homology to a snake cytotoxin (16). This ORF1 was not identified in the BfPAI from strain 86-5443-2-2. Deletion of nucleotide 16 and the addition of one nucleotide between bases 47 and 48 and bases 142 and 143 of ORF1 changes the frame three times in strain 86-5443-2-2. To determine whether ORF1 was specific for ETBF strains containing bft-1, we amplified by PCR and sequenced the region spanning ORF1 from a strain containing bft-1 (strain J38-1) and a strain containing bft-2 (strain 20793-3). We found that the nucleotide sequence of strains J38-1 and 20793-3 were identical to the appropriate sequences of strains VPI 13784 and 86-5443-2-2, respectively. Conversely, we also found that ORF-A detected in BfPAI from strain 86-5443-2-2 (Fig. (Fig.2a)2a) was not present in strain VPI 13784. The deletion of nucleotide 60 and the addition of one nucleotide between bases 172 and 173 of ORF-A change the frame twice in strain VPI 13784. Sequence analysis revealed that ORF-A was not present in either strain J38-1 or 20793-3. However, the region spanning this ORF in strain 20793-3 shared 99% similarity with that from 86-5443-2-2 but only 95% similarity with that of VPI 13784, whereas the same region from strain J38-1 shares 99% similarity with that of strain VPI 13784 and 95% similarity with that of strain 86-5443-2-2. The role of the proteins encoded by ORF1 and ORF-A in the virulence of strains VPI 13784 and 86-5443-2-2, respectively, if any, remains to be determined.
The near-direct repeat sequences present at the borders of both BfPAIs are identical. We found that the insertion site of the BfPAI in strain 86-5443-2-2 is similar to that of strain VPI 13784 and that the precise location of the BfPAI is between the stop codons of bfmB and bfmC (Fig. (Fig.7).7). Our results indicate that the first nucleotide (G) of the 17 bp found in the NTBF strains (but not in the ETBF strains) reported by Moncrief et al. (24) is part of the bfmC stop codon, suggesting that 16 bp may be the target sequence for the integration site of the BfPAI and that the size of the VPI 13784 BfPAI is 6,032 bp. However, we cannot rule out the possibility that the last base of the bfmC codon is part of the target sequence for the integration of the BfPAI.
Mobilization of the BfPAI.
Analysis of the nucleotide sequence of the DNA region flanking the BfPAI suggested that bft-1/bft-2 and mpII are in a genetic element that may be mobilized from ETBF to NTBF strains. Transfer of Bacteroides plasmids pIP417 and pBFTM10 is linked to the presence of the Bacteroides tetracycline resistance transposons (termed Tcr elements) (12, 36, 47). Tcr elements are large chromosomal self-transmissible elements (ca. 65 to 150 kb) that also facilitate the mobilization of other elements, such as coresident plasmids, other transposons, and discrete unlinked 10- to 12-kb segments of chromosomal DNA nonreplicating Bacteroides units (NBUs) (36). Interestingly, the transfer efficiency of Tcr elements is enhanced by pretreatment of donor cells with concentrations up to 1 μg of tetracycline per ml. Under this condition, Tcr elements may be transferred in a frequency of 10−3 to 10−7, and ca. 10% of transconjugants will contain Tcr elements which cotransferred with plasmids, other transposons, or NBU elements (36). To determine whether Tcr elements can mobilize the transfer of BfPAI, we performed mating experiments between ETBF strain 086Thy−3 (a thymidine-dependent mutant strain derived from ETBF 86-5443-2-2 which contains an inducible Tcr transposon [see Materials and Methods]) and NTBF strain TM4000 (tetracycline sensitive). However, under different conditions of induction, we failed to detect tetracycline-resistant transconjugants, indicating that the Tcr element of ETBF 086Ty−3 was not mobilized. When we used the mating positive controls, B. thetaiotaomicron strains BT4107 (donor) and BT4001 (recipient), transfer of Tcr element was observed at a frequency of 1.3 × 10−5 to 2.4 × 10−6. These results suggested that the Tcr element is not conjugative in strain 086Thy−3, or, alternatively, that BFT and/or MPII may inhibit the mobilization of the Tcr element. To test these possibilities, we selected ETBF strain I-1356-1Thy-1 (containing bft subtype 1) as donor strains, as well as NTBF strains K518Thy−2 (pattern II) and I-1345Thy−2 (pattern III). No transfer of Tcr element was observed under any of the conditions of induction used when ETBF I-1356-1Thy-1 or NTBF K518Thy−2 were used as the donor strains. However, the Tcr element was transferred at a frequency of 1.5 × 10−5 to 1.0 × 10−6 when NTBF I-1345Thy−2 was used as a donor strain. To determine whether the region flanking BfPAI was cotransferred with the Tcr element in strain I-1345Thy−2, we performed colony blot of 600 Tcr transconjugants by using the region spanning bfmB as a probe. No transfer of the region flanking BfPAI was detected in any of the 600 tetracycline-resistant transconjugants probed.
Evaluation for the biological activity of MPII.
BFT (BFT-1 or BFT-2) alters the morphology and physiology of human intestinal epithelial cells (HT29/C1, T84) (49); recently, we demonstrated that BFT cleaves the zonula adherens protein, E-cadherin (51). To determine whether MPII had a biological activity similar to that of BFT-1 and BFT-2, MPII was overexpressed in E. coli by using the T7 polymerase system (see Materials and Methods), and its biological activity was tested on HT29/C1 cells. By Western blot analysis, proteins with a molecular mass consistent with the proprotein-mature protein (44 kDa) and mature protein (20 kDa) were detected in the supernatants of whole-cell lysates, but no protein was detected in the cell-free supernatants (data not shown), suggesting that MPII is synthesized as a precursor protein similar to BFT (9, 16) and then processed to a mature 20-kDa protein. However, only a small amount of the putative pre-pro-mature protein was processed to a 20-kDa protein, indicating that MPII is not fully processed or secreted in E. coli. The supernatant of whole-cell lysates of MPII clones neither had biological activity on HT29/C1 cells nor cleaved E-cadherin (data not shown).
DISCUSSION
The data contained in this report strongly suggest that ETBF strains arose via the chromosomal acquisition of a 6-kb region containing the bft gene from a foreign source. By studying a collection of B. fragilis strains isolated from different countries and from different sources (intestinal and extraintestinal), we showed that this 6-kb region of DNA is found exclusively in ETBF strains. Of note, at least an additional 12 kb of flanking DNA were also found in about half of the NTBF strains, suggesting a second recombination event potentially modulating B. fragilis virulence. We have further found that the DNA flanking the ETBF-specific 6-kb region contains mobilization genes and have confirmed that the 6-kb region contains both bft and a second metalloprotease gene. We propose that, consistent with the report of Moncrief et al. (24), the specific 6-kb locus be called a BfPAI.
Recently, it has been found that genes involved in virulence, the presence of which distinguish pathogenic and nonpathogenic strains of a species, are often clustered in pathogenicity islands (10, 19). For example, enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli, and uropathogenic E. coli all contain pathogenicity islands inserted at the identical site in the nonpathogenic E. coli chromosome (2, 21). In addition, pathogenicity islands have been identified in Yersinia pestis (6), Salmonella typhimurium (22, 41), Clostridium difficile (11), Vibrio cholerae (15), and other bacterial pathogens. The BfPAI is similar to other pathogenicity islands in that (i) it contains virulence genes (bft and possibly mpII), (ii) it has direct repeat sequences at the ends of the locus and is flanked by putative mobilization genes, and (iii) it has a G+C content different from the rest of the chromosome.
Our previous study (9) showed that there are two isotypes of bft (bft-1 [VPI-bft] and bft-2 [086-bft]) whose predicted encoded proteins share 92% identity (95% identity in nucleotide sequence). In studying a collection of ETBF strains we found that all ETBF strains contain bft but the two alleles are found in different amounts (9). The results found in this study suggest that the divergence found between bft-1 and bft-2 is perceived in the BfPAI sequence. B. fragilis strains are composed of different genetic groups (33), and no predominant clones have been found between the ETBF and NTBF strains (43). The identification of two groups of ETBF strains based on the sequence of the BfPAI supports two paths of evolution for these organisms. Perna et al. (32), suggested that the variability found between the locus of enterocyte effacement from an enterohemorrhagic E. coli O157:H7 strain and that of an EPEC strain might originate by natural selection after specific host adaptation or evasion of the host immune response. We have not found any correlation between bft subtype and source of the strains (intestinal or extraintestinal and human or animal) (9, 9a). Interestingly, we found that the region of the BfPAI surrounding mpII is more conserved than the region surrounding bft. The conservation of the MPII sequence in strains 86-5443-2-2 and VPI 13784 suggests that this protein is intolerant of even conservative amino acid changes and that MPII may have an important role in ETBF strains and/or in ETBF-induced disease. However, no biological activity has yet been identified for this protein.
The BfPAI is integrated specifically between potential mobilization genes (Fig. (Fig.2a)2a) having significant homology with proteins encoded by conjugal plasmids. It has been reported that Bacteroides mobilizable plasmids and transposons utilize a common mobilization strategy. That is, the genes for DNA-processing enzymes and the cis-acting oriT are located on an adjacent DNA sequence in a small, compact mobilization region (45). The region flanking the left end of the BfPAI has an organization similar to these mobilization regions: two genes organized in an operon (bfmA/bfmB) and an oriT located adjacent to these genes (Fig. (Fig.3A).3A). Phylogenetic analysis of the protein coding sequence of the mobilization region of Bacteroides mobilizable plasmids and transposons has shown that they can be clustered together in three major groups based on protein sequence homology (44). We found that the putative mobilization region of BfPAI has the same organization as the mobilization regions of Bacteroides antibiotic resistance plasmids pBFTM10 and pIP417. These two plasmids are clustered in the second major homology group of Bacteroides mobilization proteins, a plasmid system that requires two gene products for transfer (btgA and btgB in pBFTM10 and mobA and mobB in pI417) (12, 47). Parsimony analysis of the alignment of BfmB with the mobilization proteins of plasmids pIP417 and pBFTM10, as well as other Bacteroides mobilizable elements, showed that the mobilization proteins of BfPAI are clustered in the same group as plasmids pIP417 and pBFTM10 (data not shown). It has been proposed that BtgA/MobA and BtgB/MobB bind to the oriT region and catalyze the nicking reaction that initiates the transfer process (45, 47). BfmB shares significant identity with BtgB/MobB; however, BfmA shares weak identity with MobA, and the sequence of the putative oriT of BfPAI does not share sequence similarity with those of plasmids pIP417, pBFTM10, or other Bacteroides mobilizable elements, suggesting that the mobilization region of the BfPAI is unique.
Despite the similarity of the organization of the putative BfPAI mobilization region to that of the mobilization regions of Bacteroides mobilizable plasmids and transposons, it was not mobilized by Tcr elements under the conditions studied. This result indicates that the BfPAI mobilization region is not activated by the Tcr element regulatory proteins RteA and RteB as occurs with other Bacteroides mobilizable elements (46). The right end flanking the BfPAI contains an ORF, whose encoded protein (BfmC) shares significant amino acid similarity with TraD-related proteins. It has been demonstrated that TraD is essential for DNA transfer (13), and it has been proposed that TraD has some role in coupling transfer of the unwound T strand to the transfer pore (18). The similarity of BfmC to TraD proteins suggests that BfPAI might be contained in a self-transmissible element which contains its own transfer regulatory proteins. Alternatively, it has been suggested that certain pathogenicity islands were originally plasmids that were integrated into chromosomal sites and subsequently lost their replication functions (2, 11).
The presence of the BfPAI in a region that appears to be of plasmid origin suggests that ETBF strains might have originated by horizontal transfer of the BfPAI and its flanking element. The different G+C content of the BfPAI (35%) and the flanking elements (47 to 50%) versus that for the B. fragilis chromosome (42%) indicates that these two genetic elements come from very different sources. We found that 52% of NTBF strains, in addition to lacking the BfPAI, do not contain the mobilization genes flanking the BfPAI (pattern II), whereas 43% of the NTBF strains lack the BfPAI but present these mobilization genes (pattern III). One hypothesis to explain the origin of these NTBF strains as well as ETBF strains (pattern I) is that the 6-kb region (BfPAI) and the flanking region may be different mobile elements that can be spread independently, namely, pattern II NTBF strains may acquire by horizontal transfer the element containing the mobilization genes, yielding pattern III NTBF strains. Subsequently, these pattern III strains may acquire the BfPAI, yielding ETBF. The 16-bp GC-rich sequence present in NTBF strains with pattern III (Fig. (Fig.7)7) may be the target sequence for integration of the BfPAI into the flanking element. Consistent with this hypothesis, higher numbers of NTBF strains containing the mobilization genes (pattern III) have been isolated in regions where ETBF strains are more common than in places where ETBF were isolated at a low frequency (Table (Table4).4). Alternatively, the flanking element may have previously acquired the BfPAI, forming a single mobile element. In this case, ETBF strains might originate by horizontal transfer of this element into NTBF with pattern II, and pattern III NTBF strains might originate by spontaneous deletion of the 6-kb BfPAI by recombination within the 12-bp direct repeats at the ends of the BfPAI.
We found that a small percentage of strains lack portions of the BfPAI as well as its flanking regions (patterns IV and V). All of these NTBF strains were isolated from extraintestinal sources, suggesting that these strains might result from spontaneous deletions after adaptation to the extraintestinal environment. Schubert et al. (39) found that some E. coli strains can present the “high-pathogenicity island” of Yersinia species but that in 3% of these E. coli strains the pathogenicity island manifests deletions of different sizes. These authors suggested that E. coli strains acquired the Yersinia pathogenicity island by horizontal transfer and then suffered spontaneous deletions after adaptation to a new environment.
In summary, the present study indicates that a BfPAI is a constant feature of the recently recognized ETBF strains. However, divergence within this BfPAI suggests two paths of evolution for ETBF organisms. The BfPAI and its flanking region (with homology to mobilizable plasmids) appear to be two distinct foreign genetic elements, suggesting that ETBF evolved by horizontal transfer of these two elements into NTBF strains. Our future studies will focus on determining the size and chromosomal location of the locus flanking the BfPAI, as well as on determining whether these loci are self-transmissible elements.
ACKNOWLEDGMENTS
We thank Dwight Derr for tissue culture assistance; Jeffrey C. Smith, James Nataro, and Davis Karaolis for review of the manuscript; Nadja B. Shoemaker for the gifts of B. thetaiotaomicron strains and suggestions about the mobilization experiments; Jeffrey C. Smith for the parsimony analysis of bfmB; and Lyle L. Myers, R. Bradley Sack, Jay Reuben, Tracy D. Wilkins, and Peter Escheverria for the gifts of B. fragilis strains.
This work was supported by National Research Service Award AI09863-01 (A.A.F.) and National Institutes of Health Award DK45496 (C.L.S.).
REFERENCES
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.181.21.6623-6633.1999
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/jb/181/21/6623.full.pdf
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/content/full/181/21/6623
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/181/21/6623
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/181/21/6623
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jb.181.21.6623-6633.1999
Article citations
A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.
Nat Commun, 15(1):8794, 10 Oct 2024
Cited by: 0 articles | PMID: 39389974 | PMCID: PMC11467351
Intestinal Bacteroides modulates inflammation, systemic cytokines, and microbial ecology via propionate in a mouse model of cystic fibrosis.
mBio, 15(2):e0314423, 05 Jan 2024
Cited by: 4 articles | PMID: 38179971 | PMCID: PMC10865972
Bile acid fitness determinants of a Bacteroides fragilis isolate from a human pouchitis patient.
mBio, 15(1):e0283023, 08 Dec 2023
Cited by: 2 articles | PMID: 38063424 | PMCID: PMC10790697
Microdiversity sustains the distribution of rhizosphere-associated bacterial species from the root surface to the bulk soil region in maize crop fields.
Front Plant Sci, 14:1266218, 12 Oct 2023
Cited by: 0 articles | PMID: 37905168 | PMCID: PMC10613529
Toxin-linked mobile genetic elements in major enteric bacterial pathogens.
Gut Microbiome (Camb), 4:e5, 17 Mar 2023
Cited by: 0 articles | PMID: 39295911 | PMCID: PMC11406385
Review Free full text in Europe PMC
Go to all (45) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of the Bacteroides fragilis pathogenicity island in human blood culture isolates.
Anaerobe, 12(1):17-22, 15 Aug 2005
Cited by: 10 articles | PMID: 16701608
Enterotoxigenic and non-enterotoxigenic Bacteroides fragilis from fecal microbiota of children.
Braz J Microbiol, 46(4):1141-1145, 27 Oct 2015
Cited by: 5 articles | PMID: 26691473 | PMCID: PMC4704618
Modulation of bft expression by the Bacteroides fragilis pathogenicity island and its flanking region.
Mol Microbiol, 45(4):1067-1077, 01 Aug 2002
Cited by: 18 articles | PMID: 12180925
The toxins of Bacteroides fragilis.
Toxicon, 39(11):1737-1746, 01 Nov 2001
Cited by: 70 articles | PMID: 11595636
Review
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: AI09863-01
Grant ID: F32 AI009863
NIDDK NIH HHS (2)
Grant ID: DK45496
Grant ID: R01 DK045496