4V8Y
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
4V9I
 
 | | Crystal structure of thermus thermophilus 70S in complex with tRNAs and mRNA containing a pseudouridine in a stop codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S Ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4V91
 
 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 25S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4V8Z
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
4V92
 
 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 18S RRNA, ES1, ES10, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
8RM0
 
 | |
8RM1
 
 | |
3FIM
 
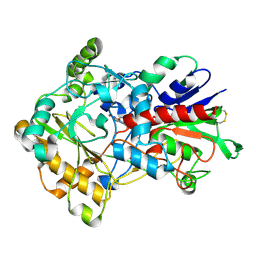 | | Crystal structure of aryl-alcohol-oxidase from Pleurotus eryingii | | Descriptor: | Aryl-alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fernandez, I.S. | | Deposit date: | 2008-12-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel structural features in the GMC family of oxidoreductases revealed by the crystal structure of fungal aryl-alcohol oxidase
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4JYA
 
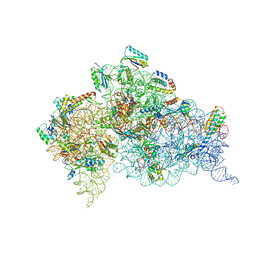 | | Crystal structures of pseudouridinilated stop codons with ASLs | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-26 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4JV5
 
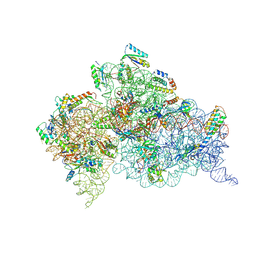 | | Crystal structures of pseudouridinilated stop codons with ASLs | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein 20, 30S ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-03-25 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4K0K
 
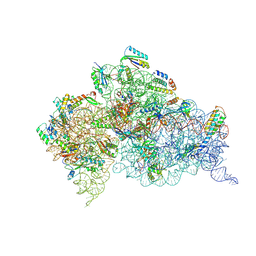 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit complexed with a serine-ASL and mRNA containing a stop codon | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
7UFI
 
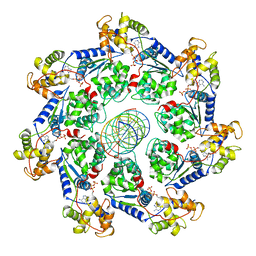 | | VchTnsC AAA+ ATPase with DNA, single heptamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*TP*CP*CP*AP*GP*TP*AP*CP*AP*GP*CP*GP*CP*GP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*AP*GP*CP*CP*GP*CP*GP*CP*TP*GP*TP*AP*CP*TP*GP*GP*AP*G)-3'), ... | | Authors: | Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
7UFM
 
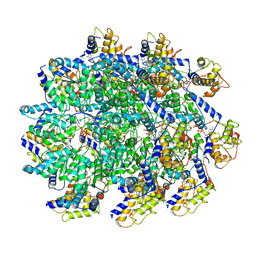 | | VchTnsC AAA+ with DNA (double heptamer) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (36-MER), VchTnsC | | Authors: | Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
1BR0
 
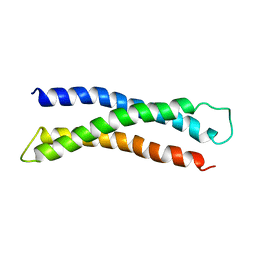 | | THREE DIMENSIONAL STRUCTURE OF THE N-TERMINAL DOMAIN OF SYNTAXIN 1A | | Descriptor: | PROTEIN (SYNTAXIN 1-A) | | Authors: | Fernandez, I, Ubach, J, Dubulova, I, Zhang, X, Sudhof, T.C, Rizo, J. | | Deposit date: | 1998-08-25 | | Release date: | 1998-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A.
Cell(Cambridge,Mass.), 94, 1998
|
|
1K5W
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE SYNAPTOTAGMIN 1 C2B-DOMAIN: SYNAPTOTAGMIN 1 AS A PHOSPHOLIPID BINDING MACHINE | | Descriptor: | CALCIUM ION, Synaptotagmin I | | Authors: | Fernandez, I, Arac, D, Ubach, J, Gerber, S.H, Shin, O, Gao, Y, Anderson, R.G.W, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-10-12 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine.
Neuron, 32, 2001
|
|
1KTX
 
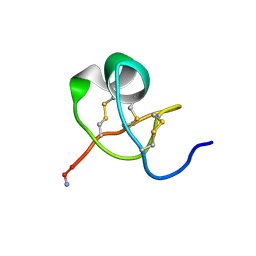 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
2YMZ
 
 | | Crystal structure of chicken Galectin 2 | | Descriptor: | GALECTIN 2, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Fernandez, I.S, Ruiz, F.M, Solis, D, Gabius, H.-J, Romero, A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fine-Tuning of Prototype Chicken Galectins: Structure of Cg-2 and Structure-Activity Correlations
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3BSQ
 
 | | Crystal structure of human kallikrein 7 produced as a secretion protein in E.coli | | Descriptor: | Kallikrein-7, SULFATE ION | | Authors: | Fernandez, I.S, Standker, L, Magert, H.J, Forssmann, W.G, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human epidermal kallikrein 7 (hK7) synthesized directly in its native state in E. coli: insights into the atomic basis of its inhibition by LEKTI domain 6 (LD6)
J.Mol.Biol., 377, 2008
|
|
7Q3R
 
 | |
7PC2
 
 | | HIV-1 Env (BG505 SOSIP.664) in complex with the IgA bNAb 7-269 and the antibody 3BNC117. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC IgG Fab heavy chain, ... | | Authors: | Fernandez, I, Bontems, F, Pehau-Arnaudet, G, Rey, F. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller.
J.Exp.Med., 219, 2022
|
|
7Q3Q
 
 | |
6UZ7
 
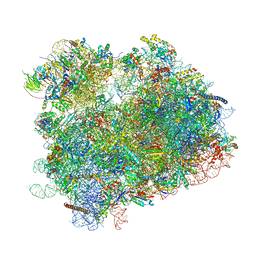 | | K.lactis 80S ribosome with p/PE tRNA and eIF5B | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Fernandez, I.S, Huang, B.Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Long-range interdomain communications in eIF5B regulate GTP hydrolysis and translation initiation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7QF1
 
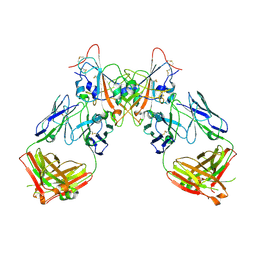 | | Crystal structure of the SARS-CoV-2 RBD in complex with the human antibody CV2.6264 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV2.6264 heavy chain, CV2.6264 light chain, ... | | Authors: | Fernandez, I, Pederzoli, R, Rey, F.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2.
J.Exp.Med., 219, 2022
|
|
7QF0
 
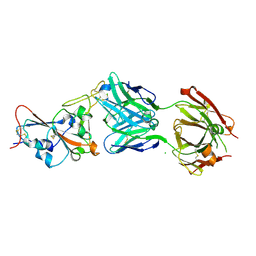 | | Crystal structure of the SARS-CoV-2 RBD in complex with the human antibody CV2.2325 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CV2.2325 heavy chain, ... | | Authors: | Fernandez, I, Pederzoli, R, Rey, F.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2.
J.Exp.Med., 219, 2022
|
|
7QEZ
 
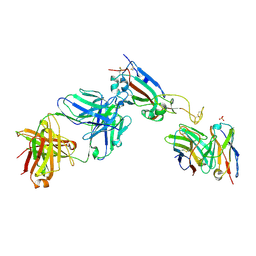 | |