Abstract
Basal cell carcinoma is the most prevalent cancer in the western world, showing a rapid increase in incidence. Activation of the Sonic hedgehog/Patched (PTCH) signaling pathway because of PTCH1 inactivation is a key event in sporadic and familial basal cell carcinoma development in humans and is associated with transcriptional activation of specific target genes, including PTCH1 itself. These changes are analogous to the situation in Drosophila where hedgehog activates the zinc-finger transcription factor Cubitus interruptus, leading to increased transcription of target genes. In the present study, we show that mice ectopically expressing the human Cubitus interruptus homolog GLI-1 in the skin develop tumors closely resembling human BCCs as well as other hair follicle-derived neoplasias, such as trichoepitheliomas, cylindromas, and trichoblastomas. Furthermore, examination of the tumors revealed wild-type p53 and Ha ras genes. These findings firmly establish that increased GLI-1 expression is central and probably sufficient for tumor development and suggest that GLI-1-induced tumor development does not depend on additional p53 or Ha ras mutations.
Genetic studies on patients with nevoid basal cell carcinoma syndrome (NBCCS), which predisposes affected individuals to the development of multiple basal cell carcinomas (BCCs), have led to the identification of inactivating mutations in the human homolog of the Drosophila gene patched (PTCH1) as the defect underlying this syndrome (1, 2). In addition, a number of surveys have provided evidence that mutations in the PTCH1 locus is a significant cause of sporadic BCCs as well as other hair follicle-derived neoplasias such as trichoepitheliomas (TEs) (3–5). The other common genetic alteration in BCCs is a mutation in the tumor-suppressor gene p53 (6).
In biochemical assays, Ptch1 has been shown to bind the ligand Sonic hedgehog (Shh) (7, 8). Ptch1 is a transmembrane protein that together with Smoothened (Smoh), a seven-transmembrane protein, forms a receptor complex for Shh (9, 10). Ligand binding results in derepression of signaling from Smoh and subsequently to activation of the transcription factor Gli, the mammalian homolog to Drosophila Cubitus interruptus (Ci). Three Ci homologs have been identified in mammals, Gli-1, Gli-2, and Gli-3, which share a highly conserved zinc-finger domain with Ci and are believed to function as the most downstream components in the vertebrate Sonic hedgehog-Patched signaling pathway (11, 12). The precise roles played by the three Gli genes have not yet been fully defined. GLI-1 was originally isolated as an amplified gene in a glioma and can transform primary rat cells in cooperation with adenovirus E1A (13). Several lines of evidence suggest a role for Gli-1 in mediating the Shh signal: first, Gli-1 is expressed in cells that are responsive to Shh (14, 15). Second, elevated Gli-1 expression is observed in Shh-treated cells, and third, ectopic expression of Gli-1 in the dorsal midbrain and hindbrain mimics the effects of ectopically expressed Shh (15, 16). Moreover, increased expression of GLI-1 is observed in human BCCs (17, 18).
BCCs cannot be grown in culture and are not induced in classical mouse skin tumorigenesis experiments. Ptch1+/− mice develop medulloblastomas and rhabdomyosarcomas, and recently BCC formation has been reported after UV radiation (19–21). Moreover, transgenic mice overexpressing Shh or a mutated variant of SMOH show epidermal proliferations in late embryonic skin that partially resembles BCC (22, 23). To test the hypothesis that GLI-1 is the downstream effector that drives tumorigenesis, transgenic mice were generated specifically overexpressing GLI-1 in the basal layer of epidermis and outer root sheet of hair follicles. These mice develop several types of spontaneous skin tumors, including BCC, TEs, cylindromas, and trichoblastomas. Furthermore, mutation analysis of the p53 and Ha ras genes, respectively, did not reveal mutations in their hot spot regions, exons 4–8 for p53 and codons 12, 13, and 61 for Ha ras, in any of the tumors examined. This suggests a p53- and Ha ras-independent mechanism by which GLI-1 induce these tumors in mouse skin.
Materials and Methods
Transgenic Mice.
A bluescript KS (−) plasmid (p5′BK5II) containing a 5.2-kb fragment from the bovine keratin 5 (K5) promoter, a 0.6-kb fragment containing the rabbit β-globin intron, and a 1.1-kb fragment containing two 3′ poly(A) signal sequences was obtained from Jose Jorcano (CIEMAT, Madrid). A 3.5-kb fragment containing the full-length human GLI-1 cDNA was amplified from pGLI-1 K12 (24) by using GLI-1 specific primers (fwd: 5′-GCGCTAGCATGTTCAACTCGATGACCC-3′, rev: 5′-GCTACGTATTAGGCACTAGAGTTGAGGAA-3′) containing restriction half sites for SnaBI and NheI, respectively. The amplified product was ligated into the SnaBI and NheI sites of the p5′BK5II vector. The construct was verified by sequencing. The transgene was isolated by cutting with NotI/SalI. Founder transgenic mice were made by microinjecting the transgene into the pronuclei of fertilized [C57BL/6J × CBA] F2 oocytes and were genotyped by PCR amplification of sequences specific for the β-globin intron and the human GLI-1 cDNA respectively from tail genomic DNA. All experiments were performed with mice hemizygous for the transgene, and transgenic mice were always directly compared with nontransgenic siblings. All transgenic mice were generated within the transgenic core facility at Novum/Huddinge hospital.
Immunohistochemical Staining.
Tissues used for immunohistochemistry were formalin fixed and paraffin embedded. Before being incubated in 1% H2O2 in methanol for 30 min, the paraffin sections were dewaxed, rehydrated, and submerged in 10 mM citrate buffer while heated in a 97°C water bath, for 20 min. Sections were incubated overnight at +4°C with 1:500 diluted rabbit polyclonal antibodies directed against either mouse K1, K5, K6, or loricrin (all from Berkeley Antibody Company, Berkeley, CA) or 1:200 diluted goat polyclonal antibodies against Gli-1 or Ptch 1 (both from Santa Cruz Biotechnology) or 1:500 diluted rabbit polyclonal antibodies against phosphohistone H3 (Upstate Biotechnology, Lake Placid, NY) or 1:200 diluted mouse monoclonal antibodies against Bcl-2 (Transduction Laboratories, Lexington, KY) or 1:700 diluted rabbit polyclonal antibodies against wild-type p53 (NovoCastra, Newcastle, U.K.). All antibodies were diluted in PBS containing 0.1% BSA. Detection was carried out with the Vectastain Elite Kit (Vector Laboratories) by using rabbit or goat IgG. Sections were washed with PBS before biotinylated secondary antibodies were applied for 1 h at room temperature, followed by extensive rinses and incubation with avidin–biotin immunoperoxidase. Immunohistochemical staining was visualized with 3-amino-9-ethylcarbazole (AEC, Vector Laboratories), and counterstaining was performed with Mayer's hematoxylin.
RNA Isolation and Reverse Transcription (RT)–PCR.
Total cellular RNA was isolated from the back skin, containing dermis as well as epidermis, of transgenic and wild-type sibling control mice by using guanidium thiocyanate-phenol-chloroform extraction as described previously (25). RT-PCR was carried out by using the Access RT-PCR system kit (Promega). The RT reaction was performed at 48°C for 45 min by using 1 μg of total RNA. The following primer pairs were used: 5′-TGCTCTTTCTCTCCAGCACCTCGGATC-3′ (sense: corresponding to the vector sequence) and 5′-CAGTGCCCGCTTCTTGGTCAA-3′ (antisense: corresponding to the human GLI-1 cDNA sequence from 301–327). These transgene-specific GLI-1 primers yielded a 369-bp PCR product. In the same reaction, β-actin-specific primers giving a 553-bp PCR product were used as an internal control.
Analysis of p53 and Ha ras Mutations.
Microdissection of 10-μm-thick paraffin sections was performed, and normal cells were separated from tumor cells by using a scalpel. Microdissected tissue was incubated in digestion buffer (10 mM KCl/1.5 mM MgCl2/10 mM Tris, pH 9.0/0.5% Tween 20) containing 200 μg/ml proteinase K at 55°C for 6 h. All microdissected tissue contained >80% tumor cells. Amplification of exons 4–8 of the mouse p53 gene or exons 1 and 2 of the mouse Ha ras gene was performed as follows: 10 μl aliquots of tumor lysate were used for PCR amplification in a 20-μl reaction volume containing 2 μl 10 × reaction buffer without MgCl2 (Promega); 250 μM of each dNTP; 3.2 pmol of each primer; and 0.5 units Taq polymerase (Promega). The primers used for amplification of the murine p53 or Ha ras exons and the individual MgCl2 concentrations have previously been described (26, 27). Thirty-five cycles of amplification were carried out, with denaturation at 94°C for 1 min, annealing at 54 or 58°C (p53, depending on the exon), or 60°C (Ha ras) for 2 min, and extension at 72°C for 2 min. PCR products were purified by using MicroSpin columns (Amersham Pharmacia Biotech) and subjected to sequencing.
Results
Spontaneous Tumor Formation in K5 GLI-1 Transgenic Mice.
To examine the effect of deregulated GLI-1 expression in mammalian skin, we generated transgenic mice expressing the human GLI-1 gene under the control of the bovine K5 promoter (Fig. 1a). This K5 promoter fragment has previously been shown to direct transgene expression to the basal cell compartment of stratified squamous epithelia as well as the outer root sheet of hair follicles (28). Three independent K5 GLI-1 transgenic founders were generated, all of which developed tumors within 1–13 wk. In addition to tumor formation, the GLI-1 transgenic mice exhibited failure to thrive and died prematurely at 1 to 6 mo of age. Founder 1 developed a hyperkeratotic tumor on the back within the first week and died at 4 wk of age. Founder 2 showed the first signs of tumor development at 4 wk of age and developed a 1-cm tumor-ulcer at 10 wk of age, and subsequently tumors developed on the back (Fig. 1b). The third founder showed the weakest phenotype, because 13 wk passed before tumors developed on the back. RT-PCR analysis of total RNA from dorsal back skin showed K5 GLI-1 mRNA in all three founders, demonstrating expression of the transgene in these mice (Fig. 1c). A summary of the macroscopically detected tumors on each founder is described in Table 1.
Figure 1.
Generation of transgenic mice. (a) Schematic drawing of the K5 GLI-1 transgene. The transgene contains the bovine K5 promoter, the rabbit β-globin intron, the human GLI-1 cDNA, and the SV40 poly(A) signal. (b) GLI-1 transgenic founder (founder 2) showing a tumor ulceration and the nodule (arrow) on the back. (c) RT-PCR analysis of K5 GLI-1 and β-actin mRNA in dorsal skin of the transgenic founders 1 to 3 (1–3). Sizes of the specific PCR products are 553 bp for β-actin and 370 bp for K5 GLI-1. Lane M pBR322-MspI ladder (New England Biolabs).
Table 1.
Spontaneous skin tumors in K5 GLI-1 mice
| Founder | Macroscopically observed tumors | Tumor site | Age, wk | p53 status | Ha-ras status |
|---|---|---|---|---|---|
| 1 | Trichoepithelioma (1) | Back | 1 | wt | wt |
| 2 | Trichoepithelioma (1) | Back | 4 | wt | wt |
| Trichoepithelioma (1) | Nose | 8 | − | − | |
| BCC (1) | Back | 10 | wt | wt | |
| 3 | Trichoblastoma (6) | Back | 13 | wt | wt |
Age is in weeks at time of tumor detection. Numbers in parentheses for different tumors indicate the number of each tumor type found. wt, wild type.
Four Different Types of Adnexal Skin Tumors in GLI-1 Overexpressing Mice.
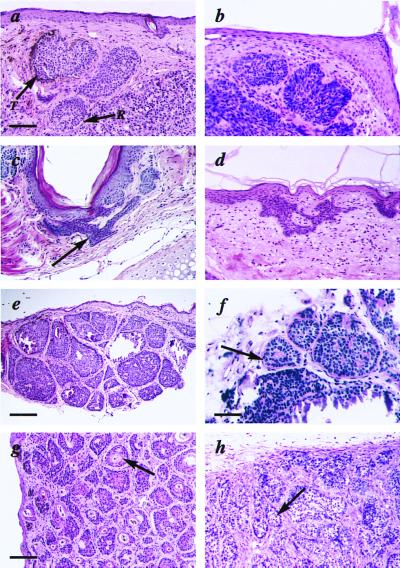
Histologic examination of the observed nodules on founder 2 identified two different skin tumors separated by nontumorigenic skin. The first nodule showed a large primitive tumor (Fig. 2a). The peripheral layer of these tumor formations showed palisading similar to human BCCs, and the cells had a uniform nonanaplastic appearance (Fig. 2 a and b). The dermis was completely filled with tumor masses also lined with palisading basaloid cells but with large necrotic areas in the center of the tumor nests. Inside the same tumor islands, abundant amyloid was identified, indicating the presence of phagocytosis and apoptosis of tumor cells. Colloid bodies and scattered calcinosis were also observed in this tumor. The stroma was arranged in parallel bundles around the tumor masses with a retraction zone resulting in peritumoral lacuna also seen in human BCCs (Fig. 2b). Such retraction zones were observed in some but not all tumor nests. In addition to the nodular BCC observed on the back, several BCCs were found on the nose, including a superficial BCC showing strong resemblance to its human counterpart (Fig. 2 c and d).
Figure 2.
Histological features of the K5 GLI-1 transgenic skin. (a) Nodular BCC from founder 2 with palisading basaloid cells forming tumor nests (T) and retraction space of stroma from tumor islands (R). (b) Human nodular BCC. (c) Superficial BCC from founder 2 with buds and irregular proliferations of tumor tissue attached to the undersurface of the epidermis (arrow). (d) Human superficial BCC. (e) Cylindroma from founder 2 with tumor islands forming a jigsaw puzzle. (f) The periodic acid/Schiff reagent-positive hyalin membrane surrounding the cylindroma tumor islands (arrow). (g) TE from founder 1 with keratinized centers (arrow) surrounded by basophilic tumor cells. (h) Trichoblastoma from founder 3 illustrating tumor islands consisting mostly of germinative cells (arrow). Bar = 135 μm (e), 54 μm (a–d and g, h), and 33 μm (f).
A second type of tumor observed was macroscopically similar to the BCC-like tumor described above but showed different features under the microscope. This tumor consisted of numerous tumor islands of epithelial cells appearing like pieces in a jigsaw puzzle (Fig. 2e). The thick hyaline sheath that surrounded the tumor islands like a cylinder was periodic acid/Schiff reagent positive (Fig. 2f). All the above characteristics are typical for a cylindroma, and this tumor was consequently classified as a cutaneous cylindroma (29).
A third type of tumor developed on both founder 1 and 2, consisting of large well-circumscribed tumors located in the dermis reaching down to the muscle layer without direct connection with the epidermis. The tumors consisted of multiple tumor nests of epithelial origin all showing horn cysts in the center (Fig. 2g). The centers were fully keratinized and surrounded by basophilic tumor cells, similar to the hair matrix cells. These tumors are very similar to human TE, a benign human skin tumor believed to develop from pluripotential epithelial cells in the hair follicle (29).
Six different tumors were examined from founder 3. Histologically, all six tumors were shown to be benign well-circumscribed noduli with a silhouette placed deep in the dermis (Fig. 2h). No connection with epidermis was observed, even though the overlying epidermis seemed thickened and hyperplastic. The numerous aggregations consisted mostly of follicular germinative cells arranged in a palisade pattern in the periphery. No keratinizing cysts were observed. These tumors were classified as counterparts to the benign human skin tumor, trichoblastoma (29).
In summary, four different types of skin tumors were found on the K5 GLI-1 mice: BCCs, TE, cylindromas, and trichoblastomas. The tumors described above were macroscopically detected; however, more tumors of similar types were detected when examining additional (n = 5) skin samples from these mice (data not shown).
Tumors Show Differentiation Marker Expression Similar to Human BCCs.
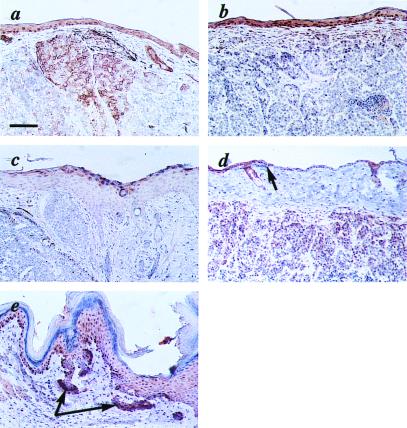
Terminal differentiation of keratinocytes involves the sequential expression of specific keratins in the basal and spinous layers and loricrin, a component of the cornified envelope, in the granular layer. To determine whether the histologic features seen in the K5 GLI-1 mice are accompanied by an altered pattern of differentiation marker expression, immunohistological analysis was performed. Staining of K5 GLI-1 BCC with antibodies against the basal K5 revealed high expression in the tumor, consistent with the pattern seen in human BCC (Fig. 3a) (30). K1, which is expressed only in cells committed to terminal differentiation, was found to be abundantly expressed in the suprabasal layer of the transgenic skin but not in tumor cells (Fig. 3b). In addition, loricrin, a major component of the epidermal cornified envelope, showed normal expression in the transgenic stratified epithelia (Fig. 3c). Taken together, these findings suggest that the interfollicular epidermal keratinocytes present in the K5 GLI-1 transgenic mice differentiate normally. Analysis of K6 and histone 3, both associated with hyperproliferation, showed abundant expression in tumor cells and in overlaying hyperproliferative epidermis but not in histologically normal epidermis (Fig. 3 d and e). This observation indicates that, whereas tumor cells and reactive epidermal keratinocytes are actively proliferating, uninvolved epidermal cells proliferate at normal rate.
Figure 3.
Expression of marker proteins in the K5 GLI-1 mouse skin. (a) Positive immunoreactivity for K5 both in the epidermis and in the tumor cells. (b) K1 is expressed in the suprabasal layers of the epidermis but not in the tumor cells. (c) The granular layer of the epidermis shows positive immunoreactivity with the loricrin antibody, but the closely located tumor cells are negative. (d) Strong expression of K6 in the tumor cells and the overlying hyperproliferative epidermis, whereas the histologically normal epidermis (arrow) is negative. (e) A strong immunoreactivity with phosphohistone 3 (H3) antibodies is seen in the tumor cells (arrows) with a weaker signal in the hyperplastic epidermis. Bar = 54 μm (a–e).
Increased Expression of GLI-1 Target Genes in Tumor Cells.
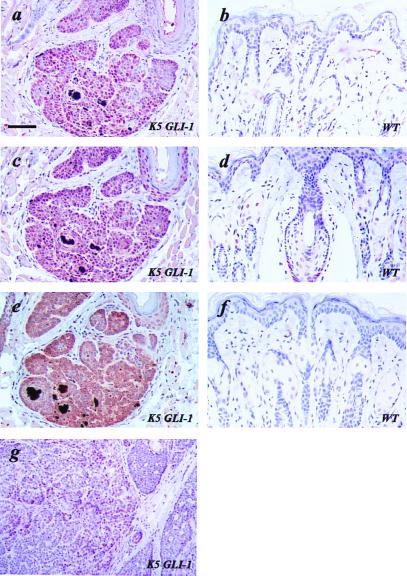
Human BCCs and TEs consistently overexpress PTCH1 and GLI-1 (5, 17, 18, 31). Furthermore, earlier studies in frogs and mice have shown that ectopic expression of GLI-1 causes increased expression of endogenous Gli-1 and Ptch1 (15, 16, 18). We used immunohistochemical analysis to examine the expression of these genes in transgenic tumors as well as in wild-type skin. Gli-1 was strongly expressed in tumor cells, and expression was also observed in adjacent reactive epidermis, although at a lower level (Fig. 4a). Even though the antibody used does not distinguish between the human and murine Gli-1, we interpret the observed difference in signal as reflecting increased expression of the endogenous murine gene because no positive signal was observed in histologically normal epidermis. No Gli-1 expression could be detected in wild-type skin (Fig. 4b). Consistent with the Gli-1 expression, increased expression of murine Ptch1 was observed in transgenic skin compared with wild type (Fig. 4 c and d).
Figure 4.
Expression of GLI-1 target genes in K5 GLI-1 transgenic skin and tumors (K5 GLI-1) (a, c, e, and g) and in wild-type skin (WT) (b, d, and f). High expression of Gli-1 is seen in a transgenic BCC and adjacent epidermis (a), but no signal was detected in wild-type skin (b). Ptch1 (c and d) and Bcl-2 (e and f) are both overexpressed in transgenic epidermis and tumor cells compared with wild-type skin. However, single cells in the lower parts of the hair follicle in wild-type skin show positive immunostaining for Ptch1 (d). (g) Sections of a BCC showing scattered tumor cells with immunoreactivity for p53 (brown precipitate). Bar = 92 μm (a–f), 54 μm (g).
Human BCCs have demonstrated a marked increase in expression of Bcl-2, in contrast to the other common epidermal malignancy, squamous cell carcinoma (30). In the BCC from a K5 GLI-1 transgenic mouse Bcl-2 expression was markedly elevated with no detectable expression in wild-type skin (Fig. 4 e and f), suggesting a potential role in GLI-1-induced BCC formation.
Wild-Type p53 and Ha ras in Transgenic Skin Tumors.
The tumor-suppressor gene p53 is commonly mutated in most types of human epithelial malignancies, including BCCs (6, 32). The level of p53 protein is low in normal cells; however, increased expression because of stabilization is observed in cells harboring a mutated p53. When analyzing a tumor sample from a K5 GLI-1 mouse for p53 protein expression, a moderately intense dispersed pattern was observed primarily in peripheral cells, suggestive of a possible involvement of p53 in the formation of this tumor (Fig. 4g). Similar results were obtained when analyzing additional tumors (n = 3; data not shown). However, direct sequencing of p53 exons 4–8 after microdissection of this particular BCC did not reveal any mutations (Table 1). Moreover, when the p53 mutation analysis was extended to involve all macroscopically observed tumors (n = 10), no p53 mutation was found in any tumor. This finding suggests that GLI-1 can drive tumor formation independently of a mutated p53 protein.
Other genes commonly mutated in nonmelanoma skin cancer are members of the ras oncogene family, comprised of the N, Ki, and Ha ras genes. Mutations in all three ras genes have been found in BCCs, with Ha ras showing the highest mutation frequency (33). In addition, in patients with nonmelanoma skin cancer, there is a high incidence of Ha ras loss of heterozygosity (36% for patients with BCC) in normal-appearing skin (34). These findings suggest a potential involvement of the Ras signaling pathway in BCC development. However, examination of the transgenic mouse tumors (n = 10) for Ha ras mutations revealed no mutations in exons 1 and 2, including the hot spot regions codons 12, 13, and 61, suggesting a mechanism whereby GLI-1 induce tumor growth independently of a mutated Ha ras gene (Table 1). However, we cannot exclude presence of mutations in other ras genes, and further analysis is ongoing to address this issue.
Discussion
In the present study, we have shown that inappropriate expression of the transcription factor GLI-1 is the probable mechanism by which mutations in the SHH-PTCH signaling pathway elicit development of skin tumors. Consistent with this, GLI-1 expression is up-regulated in most human BCCs as well as in medulloblastomas and rhabdomyosaromas appearing in Ptch+/− heterozygous mice (17–20). In addition, ectopic expression of GLI-1 in murine brain or frog epidermis is associated with enhanced proliferation (16, 18). The GLI-1 cDNA used in this study corresponds to the tumor-derived GLI-1 gene originally isolated by Kinzler et al. and is hence possibly mutant (16, 24). However, recent functional studies revealed no differences in terms of transactivating ability or potential to interact with SUFUH (suppressor of fused), a known inhibitor of GLI-1, in comparison to GLI-1 cloned from normal tissue (35). Furthermore, the sequence alteration occurs in a region of GLI-1 that is highly variable between different species, indicating that the two forms represent polymorphic variants.
A surprising and very interesting outcome of our study was the development of four different types of skin tumors in K5-GLI-1 mice, in each instance closely mimicking their human counterparts. In the case of BCCs, presence of palisading basaloid cells in the periphery of tumor nests, appearance of retraction spaces, lack of keratinizing centers, as well as expression of Bcl-2, endogenous Gli-1, and Ptch1, are all characteristics typical of human BCCs. Moreover, tumors resembling different histological subtypes like nodular and superficial BCCs were identified. In TEs, advanced follicular differentiation including central horn cysts and rudimentary hair structures combined with lack of retraction spaces were key features. The critical features supporting classification as cylindroma were the jigsaw puzzle arrangement, presence of basaloid cells in the periphery, light-staining cells in the tumor centers, and a periodic acid/Schiff reagent-positive hyalin sheath surrounding the tumor islands. Similarly, the trichoblastomas located in the dermis and penetrance into the subcutaneous fat layer with absence of keratinizing cysts were highly similar to the corresponding human tumor. Consistent with the human situation, no metastasis was observed. Thus, K5-GLI-1 transgenic mice represent a highly relevant experimental model of spontaneous BCC and related skin cancers having the same pathogenetic background as in humans.
In human BCCs, TEs, trichoblastomas, and cylindromas are all classified as adnexal skin tumors with the cell of origin most likely residing in the apocrine/follicular unit, a view supported by a very similar cytokeratin expression pattern in BCC, TE, trichoblastoma, and fetal hair follicles (36). Furthermore, TEs and cylindromas often coexist in patients with the tumor predisposition syndrome hereditary multiple epithelioma (MIM 132700) (37, 38). TE and BCC occur in the multiple familial TE syndrome (MIM 60/606), and trichoblastoma and BCC often develop within a congenital malformation called sebaceous nevus, in which deletions in the PTCH1 gene region were recently reported (39–41). A link to constitutive activation of the SHH-PTCH1 signaling pathway is clear for BCCs and TEs displaying both PTCH1 mutations and consistent up-regulation of PTCH1 mRNA in familial as well as sporadic tumors (5, 31). Our finding that all the above tumor types appear in the K5-GLI-1 transgenic mice now suggests that the different predisposing genetic defects converge on activation of GLI-1 expression.
An important question is how overexpression of GLI-1 can result in various tumor phenotypes. We favor the hypothesis that GLI-1 serves as a dose-dependent cell fate/differentiation determinant during hair follicle development and cycling. According to this view, the level of GLI-1 expression underlies the cellular phenotype with a low-level driving proliferation, resulting in trichoblastomas and with higher levels in addition, partly inducing the hair follicle differentiation program yielding BCCs, cylindromas, and TEs. This interpretation is supported by a stronger up-regulation of PTCH1 expression and by inference, GLI-1 expression in human TEs compared with BCCs (5).
As a consequence, the tumors we observe may be regarded as failed attempts to make a hair follicle, a failure because proliferation and hair follicle cell fate is induced in an adnexal progenitor cell within the epithelial compartment without a concomitant recruitment of mensenchymal cells to form a dermal papilla, which can signal back and promote further maturation. Consistent with this interpretation, secretion of Shh by the epithelial placode cells is required for proper papilla formation (42–44), and epithelial outgrowths stimulated by Shh overexpression eventually differentiate into hair follicles (22).
Tumor development is believed to require a number of genetic alterations and is often associated with genetic instability. BCCs are unusual in that they are apparently genetically stable and no precursor lesion has been identified. In humans, p53 mutations occur very frequently in BCCs as well as in patches of morphologically normal skin, especially in UV-exposed areas (32, 45). On the basis of these observations, it has been suggested that p53 mutations are an early and required event in BCC development. Immunohistochemical analysis of p53 expression in K5-GLI-1-induced tumors showed scattered p53-positive cells (Fig. 4g) in a pattern previously observed in some human BCCs, suggesting that p53 mutations may be present also in the murine tumors. Surprisingly, direct sequencing of exons 4–8 of p53 in 10 tumors failed to reveal any sequence alterations, supporting the conclusion that development of BCCs or other hair follicle-associated tumors does not absolutely depend on p53 mutations. Consistent with this, there is no reported predisposition for development of BCCs in Li-Fraumeni patients harboring germline p53 mutations or in p53−/ mice. However, we can at present not exclude the presence of mutations in other genes involved in the same signaling pathway as p53, such as p21waf and Mdm2.
A similar analysis of the Ha ras oncogene also failed to reveal any mutations in the transgenic tumors, indicating that GLI-1-induced tumor formation does not depend on secondary Ha ras mutations. Again, this observation is consistent with earlier reports describing development of squamous cell carcinomas but not BCCs in mice overexpressing Ha ras in epidermal cells (46).
If a single oncogenic event, i.e., GLI-1 expression above a certain threshold, is sufficient to initiate BCC and TE development, then all precursor cells receiving this signal should develop into tumors. In fact, in support of such a view, regions of macroscopically normal dorsal skin from founder 1 having the highest level of transgenic expression when analyzed histologically were found to be filled with tumors in various stages of development, leaving only a few normally appearing hair follicles (data not shown). Fewer tumors in mice with lower expression of the K5-GLI-1 transgene could be explained by not all precursor cells achieving a GLI-1 expression level above the threshold in combination with a slower tumor growth rate. An alternative explanation for a reduced number of tumors could be mosaic transgene expression, sparing potential tumor precursor cells from the expression of GLI-1. In any event, ectopic GLI-1 expression is able to initiate frequent tumor formation originating from hair follicle structures, and these tumors may or may not be associated with secondary genetic hits.
Finally, the observation that GLI-1 is not or is only to a very low level expressed in normal human skin (19) and that there is no apparent phenotype in Gli-1−/ mice make GLI-1 a promising therapeutic target (47, 48). The availability of a transgenic mouse model for the most common human cancer will make possible detailed mechanistic studies of BCC pathogenesis and development of new therapeutic strategies.
Acknowledgments
We thank K. W. Kinzler (Program in Human Genetics, The Johns Hopkins University School of Medicine) for the human GLI-1 cDNA, J. Jorcano for the K5 expression vector, Mari-Anne Hedblad for help with analyzing the tumors, Tobias Skarin and Åsa Bergström for technical assistance, and P. Söderkvist for advice regarding p53 mutation analysis. This study was supported by grants from the Swedish Cancer Fund, the Swedish Children Cancer Fund, the Swedish Medical Society, the Welander–Finsen Foundation, and Svenska Läkarsällskapet.
Abbreviations
- BCC
basal cell carcinoma
- RT
reverse transcription
- K
keratin
- TE
trichoepithelioma
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050467397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050467397
References
- 1.Johnson R L, Rothman A L, Xie J, Goodrich L V, Bare J W, Bonifas J M, Quinn A G, Myers R M, Cox D R, Epstein E H, Jr, Scott M P. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 2.Hahn H, Wicking C, Zaphiropoulous P G, Gailani M R, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Undèn A B, Gillies S, et al. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 3.Aszterbaum M, Rothman A, Johnson R L, Fisher M, Xie J, Bonifas J M, Zhang X, Scott M P, Epstein E H., Jr J Inv Dermatol. 1998;110:885–888. doi: 10.1046/j.1523-1747.1998.00222.x. [DOI] [PubMed] [Google Scholar]
- 4.Gailani M R, Stahle-Backdahl M, Leffell D J, Glynn M, Zaphiropoulos P G, Pressman C, Undèn A B, Dean M, Brash D E, Bale A E, et al. Nat Genet. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- 5.Vorechovsky I, Undèn A B, Sandstedt B, Toftgård R, Stahle-Backdahl M. Cancer Res. 1997;57:4677–4681. [PubMed] [Google Scholar]
- 6.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 7.Marigo V, Davey R A, Zuo Y, Cunningham J M, Tabin C J. Nature (London) 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 8.Stone D M, Hynes M, Armanini M, Swanson T A, Gu Q, Johnson R L, Scott M P, Pennica D, Goddard A, Phillips H, et al. Nature (London) 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 9.Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper J E. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 10.Murone M, Rosenthal A, de Sauvage F J. Curr Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz i Altaba A. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 12.Ruppert J M, Kinzler K W, Wong A J, Bigner S H, Kao F T, Law M L, Seuanez H N, O'Brien S J, Vogelstein B. Mol Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruppert J M, Vogelstein B, Kinzler K W. Mol Cell Biol. 1991;11:1724–1728. doi: 10.1128/mcb.11.3.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marigo V, Johnson R L, Vortkamp A, Tabin C J. Dev Biol. 1996;180:273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki H, Hui C, Nakafuku M, Kondoh H. Development (Cambridge, UK) 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 16.Hynes M, Stone D, Dowd M, Pitts-Meek S, Goddard A, Gurney A, Rosenthal A. Neuron. 1997;19:15–26. doi: 10.1016/s0896-6273(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 17.Green J, Leigh I M, Poulsom R, Quinn A G. Br J Dermatol. 1998;139:911–915. doi: 10.1046/j.1365-2133.1998.02598.x. [DOI] [PubMed] [Google Scholar]
- 18.Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Nature (London) 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 19.Goodrich L V, Milenkovic L, Higgins K M, Scott M P. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 20.Hahn H, Wojnowski L, Zimmer A M, Hall J, Miller G, Zimmer A. Nat Med. 1998;4:619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 21.Aszterbaum M, Epstein J, Oro A, Douglas V, LeBoit P E, Scott M P, Epstein E H., Jr Nat Med. 1999;5:1285–1291. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- 22.Oro A E, Higgins K M, Hu Z, Bonifas J M, Epstein E H, Jr, Scott M P. Science. 1997;276:817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 23.Xie J, Murone M, Luoh S M, Ryan A, Gu Q, Zhang C, Bonifas J M, Lam C W, Hynes M, Goddard A, et al. Nature (London) 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 24.Kinzler K W, Ruppert J M, Bigner S H, Vogelstein B. Nature (London) 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Hegi M E, Söderkvist P, Foley J F, Schoonhoven R, Swenberg J A, Kari F, Maronpot R, Anderson M W, Wiseman R W. Carcinogenesis. 1993;14:803–810. doi: 10.1093/carcin/14.5.803. [DOI] [PubMed] [Google Scholar]
- 27.Devereaux T R, Foley J F, Maronpot R R, Kari F, Anderson M W. Carcinogenesis. 1993;14:795–801. doi: 10.1093/carcin/14.5.795. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez A, Bravo A, Jorcano J L, Vidal M. Differentiation (Berlin) 1994;58:53–64. doi: 10.1046/j.1432-0436.1994.5810053.x. [DOI] [PubMed] [Google Scholar]
- 29.Ackerman A B, de Viragh P A, Chongchitnant N. Neoplasms with Follicular Differentiation. Philadelphia: Lea & Febiger; 1993. [Google Scholar]
- 30.Markey A C, Lane E B, Macdonald D M, Leigh I M. Br J Dermatol. 1992;126:154–160. doi: 10.1111/j.1365-2133.1992.tb07813.x. [DOI] [PubMed] [Google Scholar]
- 31.Undèn A B, Zaphiropoulos P G, Bruce K, Toftgård R, Stahle-Backdahl M. Cancer Res. 1997;57:2336–2340. [PubMed] [Google Scholar]
- 32.Pontén F, Berg C, Ahmadian A, Ren Z P, Nistér M, Lundeberg J, Uhlén M, Pontén J. Oncogene. 1997;15:1059–1068. doi: 10.1038/sj.onc.1201435. [DOI] [PubMed] [Google Scholar]
- 33.Pierceall W E, Goldberg L H, Tainsky M A, Mukhopadhyay T, Ananthaswamy N. Mol Carcinog. 1991;4:196–202. doi: 10.1002/mc.2940040306. [DOI] [PubMed] [Google Scholar]
- 34.Ananthaswamy H N, Applegate L A, Goldberg L H, Bales E S. Mol Carcinog. 1989;2:298–301. doi: 10.1002/mc.2940020510. [DOI] [PubMed] [Google Scholar]
- 35.Kogerman P, Grimm T, Kogerman L, Krause D, Undèn A B, Sandstedt B, Toftgård R, Zaphiropoulos P G. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto O, Asahi M. Br J Dermatol. 1999;140:8–16. doi: 10.1046/j.1365-2133.1999.02601.x. [DOI] [PubMed] [Google Scholar]
- 37.Puig L, Nadal C, Fernandez-Figueras M T, Alegre M, de Moragas J M. Am J Dermatopathol. 1998;1:56–60. doi: 10.1097/00000372-199802000-00011. [DOI] [PubMed] [Google Scholar]
- 38.Verhoef S, Schrander-Stumpel C T, Vuzevski V D, Temelaars A, Jansen L A, Malfeyt G A, Ceelen T L, Lindhout D, Halley D J, van den Ouweland A M. J Med Genet. 1998;10:841–845. doi: 10.1136/jmg.35.10.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson S C, Bennett R G. J Am Acad Dermatol. 1993;2:322–326. doi: 10.1016/0190-9622(93)70046-v. [DOI] [PubMed] [Google Scholar]
- 40.Mehregan A H, Pinkus H. Arch Dermatol. 1965;91:574–588. doi: 10.1001/archderm.1965.01600120006002. [DOI] [PubMed] [Google Scholar]
- 41.Xin H, Matt D, Qin J Z, Burg G, Boni R. Cancer Res. 1999;8:1834–1836. [PubMed] [Google Scholar]
- 42.St-Jacques B, Dassule H R, Karavanova I, Botchkarev V A, Li J, Danielian P S, McMahon J A, Lewis P M, Paus R, MacMahon A P. Curr Biol. 1998;19:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 43.Karlsson L, Bondjers C, Betsholtz C. Development (Cambridge, UK) 1999;126:2611–2621. doi: 10.1242/dev.126.12.2611. [DOI] [PubMed] [Google Scholar]
- 44.Chiang C, Swan R Z, Grachtchouk M, Bolinger M, Litingtung Y, Robertson E K, Cooper M K, Gaffield W, Westphal H, Beachy P A, et al. Dev Biol. 1999;1:1–9. doi: 10.1006/dbio.1998.9103. [DOI] [PubMed] [Google Scholar]
- 45.Jonason A S, Kunala S, Price G J, Restifo R J, Spinelli H M, Persing J A, Leffell D J, Tarone R E, Brash D E. Proc Natl Acad Sci USA. 1996;24:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailleul G, Surani M A, White S C, Barton S C, Brown K, Blessing M, Jorcano J, Balmain A. Cell. 1990;62:697–708. doi: 10.1016/0092-8674(90)90115-u. [DOI] [PubMed] [Google Scholar]
- 47.Matise M P, Epstein D J, Park H L, Platt K A, Joyner A L. Development (Cambridge, UK) 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 48.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui C C. Development (Cambridge, UK) 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]