Abstract
The forkhead box m1 (Foxm1) transcription factor is essential for initiation of carcinogen-induced liver tumors; however, whether FoxM1 constitutes a therapeutic target for liver cancer treatment remains unknown. In this study, we used diethylnitrosamine/phenobarbital treatment to induce hepatocellular carcinomas (HCCs) in either WT mice or Arf–/– Rosa26-FoxM1b Tg mice, in which forkhead box M1b (FoxM1b) is overexpressed and alternative reading frame (ARF) inhibition of FoxM1 transcriptional activity is eliminated. To pharmacologically reduce FoxM1 activity in HCCs, we subjected these HCC-bearing mice to daily injections of a cell-penetrating ARF26–44 peptide inhibitor of FoxM1 function. After 4 weeks of this treatment, HCC regions displayed reduced tumor cell proliferation and angiogenesis and a significant increase in apoptosis within the HCC region but not in the adjacent normal liver tissue. ARF peptide treatment also induced apoptosis of several distinct human hepatoma cell lines, which correlated with reduced protein levels of the mitotic regulatory genes encoding polo-like kinase 1, aurora B kinase, and survivin, all of which are transcriptional targets of FoxM1 that are highly expressed in cancer cells and function to prevent apoptosis. These studies indicate that ARF peptide treatment is an effective therapeutic approach to limit proliferation and induce apoptosis of liver cancer cells in vivo.
Introduction
Human hepatocellular carcinoma (HCC) is the fifth most common cancer, yet it is among the most lethal cancers worldwide because late detection and high frequency of tumor recurrence render current HCC therapy ineffective (1). The primary etiology of human HCC involves HBV and HCV infections, which are primarily responsible for the high incidence of HCC in Africa and Asia and increasing occurrence of HCC in Europe and America (1). Persistent hepatic infection by either human HBV or HCV results in chronic hepatic inflammatory injury and activation of hepatic stellate cells, which oversecrete collagen, leading to hepatic fibrosis, cirrhosis, and subsequent development of HCC (1). Other causes of human HCC involve hepatic damage and fibrosis resulting from iron or copper deposition, alcohol, or nonalcoholic steatohepatitis (NASH, or fatty liver disease) as well as exposure to the potent hepatic carcinogen aflatoxin B1 produced by specific strains of mold (1).
Activation of the Ras/MAPK signaling pathway drives cell-cycle progression by temporal expression of cyclin regulatory subunits, which activate their corresponding cyclin-dependent kinases (CDKs) through complex formation and phosphorylate substrates critical for cell-cycle progression (2). Development of cancer is a multistep process involving gain-of-function mutations that activate the Ras/MAPK and PI3K/Akt signaling pathways that stimulate cell-cycle progression and enhance cell survival (2, 3). Cancer progression also requires inactivation of tumor suppressor genes that function to arrest cell proliferation in response to oncogenic stimuli (4). In mouse models of liver cancer, loss-of-function mutations in the p53 tumor suppressor gene or gain-of-function mutations in either the Ras/MAPK, PI3K/Akt, or TGF-α signaling pathways are known to stimulate formation of HCC tumors (5–7). A well-established mouse liver tumor induction and promotion protocol is available and consists of a single postnatal injection of the DNA-damaging diethylnitrosamine (DEN; tumor initiator) and continuous administration of the tumor promoter phenobarbital (PB) (8). Gene expression profiling studies demonstrated that mouse HCCs induced by DEN treatment express genes similar to those found in the poorer survival group of human HCCs (9), supporting the relevance of using DEN-induced mouse liver tumors as a model for the study of human liver tumors.
Expression of the alternative reading frame (ARF) tumor suppressor protein is induced in response to oncogenic stimuli and prevents abnormal cell proliferation through a p53-dependent G1 cell-cycle arrest by increasing stability of the p53 tumor suppressor through nucleolar targeting of the p53 ubiquitin ligase protein Mdm2 (10). The ARF protein also mediates p53-independent cell-cycle arrest, as the mouse ARF protein targets both the E2F1 and c-Myc transcription factors to the nucleolus, thus preventing their transcriptional activation of S-phase–promoting target genes (11–14). Loss of ARF function is a critical event for tumor promotion, as evidenced by extinguished expression of the ARF protein in a variety of tumors through DNA methylation and silencing of the ARF promoter region (4).
The mammalian forkhead box (Fox) family of transcription factors consists of more than 50 mammalian proteins (15, 16) that share homology in the winged helix DNA-binding domain (17, 18). Expression of FoxM1 (or FoxM1b) is ubiquitous in all proliferating mammalian cells, and its expression is induced during the G1 phase of the cell cycle and continues during S-phase and mitosis (19–23). FoxM1b transcriptional activity requires activation of the RAS/MAPK pathway and binding of activated CDK-cyclin complexes to the activation domain, which mediate phosphorylation-dependent recruitment of the CREB-binding protein (CBP) transcriptional coactivator (24). Liver regeneration studies that used the albumin promoter/enhancer cre recombinase (Alb-Cre) transgene to mediate hepatocyte-specific deletion of the mouse Foxm1 LoxP/LoxP targeted allele (Foxm1fl/fl) demonstrated that Foxm1 is required for hepatocyte DNA replication and mitosis (25). Foxm1-deficient hepatocytes accumulate nuclear levels of the CDK inhibitor (CDKI) proteins p21Cip1 and p27Kip1 (8, 25) because FoxM1 regulates expression of S-phase kinase-associated protein 2 (Skp2) and CDK subunit 1 (Cks1) proteins (26), which are involved in targeting these CDKI proteins for degradation during the G1/S transition (27). For G2/M progression, FoxM1 regulates transcription of cyclin B1 and the Cdk1-activating Cdc25B phosphatase (25, 28), and FoxM1 is essential for transcription of the mitotic regulatory genes polo-like kinase 1 (PLK1), aurora B kinase, survivin, centromere protein A (CENPA), and CENPB (26, 29, 30).
We previously used the Alb-Cre transgene to conditionally delete the mouse Foxm1fl/fl allele in hepatocytes prior to DEN/PB liver tumor induction. FoxM1-deficient hepatocytes failed to proliferate and were resistant to the development of hepatic tumors (8, 31). This tumor resistance was associated with increased nuclear levels of the CDKI protein p27Kip1 and undetectable levels of the CDK1 activator Cdc25B phosphatase (8, 31). The FoxM1 transcription factor was identified as a novel inhibitory target of the mouse ARF tumor suppressor, and structure-function studies demonstrated that amino acid residues 26–46 of the mouse ARF protein were sufficient to inhibit FoxM1 (8). Furthermore, treatment of osteosarcoma U2OS cells with a cell-penetrating ARF26–44 peptide containing 9 N-terminal d-arginine (D-Arg) residues (WT ARF26–44) (32, 33) significantly reduced FoxM1 function in this cancer cell line (8). Moreover, we previously developed Tg mice in which the Rosa26 promoter was used to drive ubiquitous expression of the human FoxM1b cDNA, and increased FoxM1b levels stimulated proliferation of pulmonary cells in response to lung injury (34) and stimulated development and progression of prostate cancers in both TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double-Tg mice (35).
In this study, we used the Mx promoter–driven Cre recombinase (Mx-Cre) transgene (36) to conditionally delete the Foxm1fl/fl allele in preexisting mouse liver tumors induced by DEN/PB treatment and demonstrated that FoxM1 is required for tumor progression of hepatic cancer cells. We show that administering the WT ARF26–44 peptide to mice following DEN/PB exposure is an effective treatment to diminish Foxm1 function in vivo, causing selective HCC apoptosis and reduced proliferation and angiogenesis in HCC regions. We also show that Arf–/– Rosa26-FoxM1b Tg mice, in which FoxM1b is overexpressed and ARF inhibition of FoxM1 transcriptional activity is eliminated, developed highly proliferative liver tumors following DEN/PB exposure. We showed that WT ARF26–44 peptide treatment of these Arf–/– Rosa26-FoxM1b Tg mice efficiently diminished HCC proliferation and selectively induced apoptosis of the HCC region.
Results
The mouse Foxm1 transcription factor is required for hepatic tumor progression.
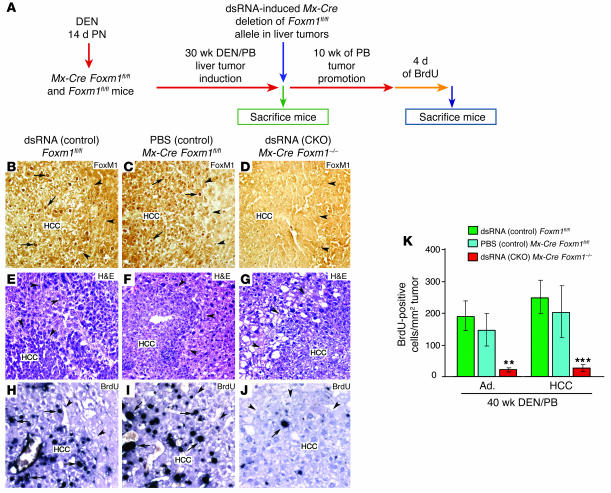
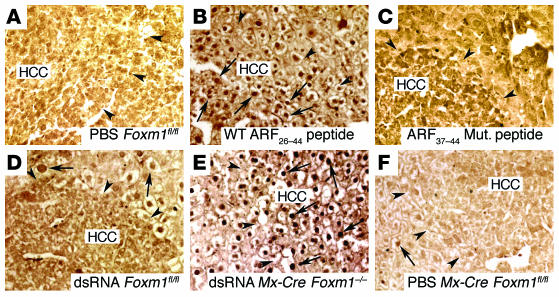
We previously showed that conditional deletion of Foxm1 in hepatocytes prior to DEN/PB liver tumor induction is sufficient to inhibit hepatic tumor initiation (8, 31). Here, we determined that Foxm1 is required for hepatic tumor progression. In order to do so, we used the IFN-α/β–regulated Mx-Cre transgene (36) to conditionally knock out (CKO) or delete the Foxm1fl/fl targeted allele in preexisting liver tumors induced by the DEN/PB exposure (8). We induced HCC in mice with 30 weeks of DEN/PB exposure, and then induced Mx-Cre expression with synthetic double-stranded RNA (dsRNA) to CKO the Foxm1fl/fl targeted allele. Mice were then subjected to an additional 10 weeks of PB tumor promotion protocol (Figure 1A). To achieve long-term BrdU labeling of the liver tumors, the mice were then given drinking water containing 1 mg/ml of BrdU for 4 days (8, 37). The Mx-Cre transgene efficiently deleted the Foxm1fl/fl targeted allele, as evidenced by the absence of detectable nuclear staining of FoxM1 protein in liver tumors of dsRNA CKO Mx-Cre Foxm1–/– mice compared with control liver tumors (Figure 1, B–D).
Figure 1. The mouse Foxm1 transcription factor is required for hepatic tumor progression.
(A) Diagram depicting experimental design of conditional deletion of Foxm1fl/fl in preexisting liver tumors. See Methods for details of DEN/PB treatment of mice to induce HCCs as described previously (8). To create the Foxm1 CKO, mice were injected with synthetic dsRNA to induce expression of the Mx-Cre recombinase transgene (Mx-Cre) to delete the mouse Foxm1fl/fl targeted allele at 30 weeks after DEN/PB treatment, and the mice were then subjected to 10 additional weeks of PB tumor promotion and were labeled with BrdU as described in Methods. Controls included dsRNA-treated Foxm1fl/fl mice and PBS-treated Mx-Cre Foxm1fl/fl mice. PN, postnatally. (B–D) dsRNA CKO Mx-Cre Foxm1–/– liver tumors display no detectable nuclear staining of Foxm1 protein as determined by immunostaining with Foxm1 antibody. (E–G) H&E staining of the indicated HCC liver sections after 40 weeks of DEN/PB exposure (tumor margins indicated by arrowheads). (H–J) BrdU incorporation was detected by immunostaining of liver tumor sections with monoclonal BrdU antibody from the indicated mice at 40 weeks following DEN/PB exposure. Arrows indicate nuclear staining for either Foxm1 protein or BrdU. (K) Graph of mean number of BrdU-positive cells per square millimeter liver tumor (±SD) as described in Methods. The asterisks indicate statistically significant changes: **P ≤ 0.01 and ***P ≤ 0.001. Ad., hepatic adenomas. Magnification: ×200 (B–G); ×400 (H–J).
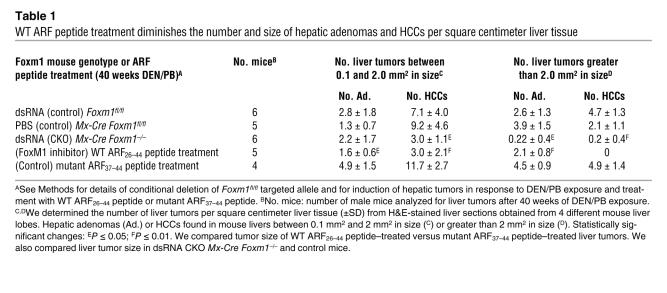
We used liver sections stained with H&E to determine the number of tumors per square centimeter of liver tissue (Figure 1, E–G). To calculate the area or size of liver tumors, we used micrographs of H&E-stained liver tumor sections taken with an Axioplan 2 microscope (Zeiss) and the AxioVision program (version 4.3; Zeiss). After 40 weeks of DEN/PB exposure, control mice displayed hepatic adenomas and HCCs that were larger than 2 mm2 in size (Table 1). Deletion of Foxm1 in preexisting hepatic tumors in dsRNA CKO Mx-Cre Foxm1–/– mice caused a significant reduction in the number of liver tumors larger than 2 mm2 in size compared with control liver tumors after 40 weeks of DEN/PB exposure (Table 1). We next measured tumor cell proliferation by determining the number of hepatic tumor cells that immunostained positive for BrdU incorporation (Figure 1, H–J). Compared with controls, dsRNA CKO Mx-Cre Foxm1–/– mice displayed an 80% reduction in the number of liver tumor cells that stained positive for BrdU after 40 weeks of DEN/PB treatment (Figure 1K). Taken together, these results indicate that deletion of Foxm1 in preexisting liver tumors significantly diminished proliferation and growth of hepatic cancer cells.
Table 1 .
WT ARF peptide treatment diminishes the number and size of hepatic adenomas and HCCs per square centimeter liver tissue
The cell-penetrating WT ARF26–44 peptide targets the endogenous mouse FoxM1 protein to the nucleolus of hepatic tumor cells.
We previously synthesized a cell-penetrating ARF26–44 peptide fused to 9 N-terminal D-Arg residues (32, 33), which efficiently transduced into osteosarcoma U2OS cells and inhibited FoxM1b transcriptional activity (8). Treatment of U2OS cells with 12 μM of (D-Arg)9-ARF26–44 (WT ARF26–44) peptide fluorescently tagged with tetramethylrhodamine (TMR) targeted nuclear GFP-FoxM1b green fluorescence colocalized with red WT ARF26–44 peptide fluorescence in the nucleolus (Figure 2, C and D). In contrast, GFP-FoxM1b protein remained nuclear in U2OS cells when treated with a TMR fluorescently tagged mutant (D-Arg)9-ARF37–44 (mutant ARF37–44) peptide (Figure 2E), which lacked the amino acids 26–37 required to interact with the FoxM1b protein (8). Because Arg-rich sequences are sufficient for nucleolar targeting (38), the mutant ARF37–44 peptide fluorescence also localized to the nucleolus of U2OS cells (Figure 2F). We did not observe any signal in the absence of the ARF peptide (data not shown).
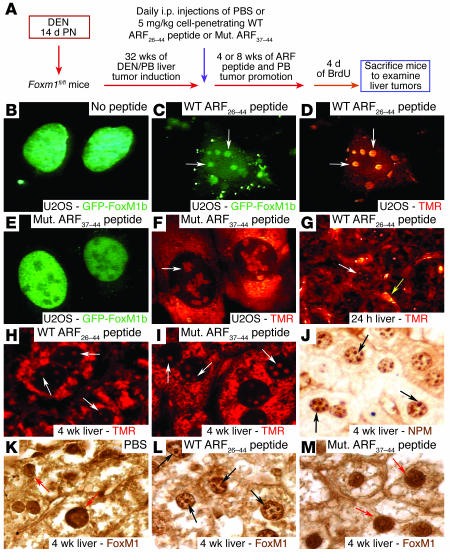
Figure 2. The WT ARF26–44 peptide targets the liver tumor FoxM1 protein to the nucleolus.
(A) Experimental design diagram of ARF peptide treatment of liver tumor–bearing mice. Liver tumors were induced in mice with DEN/PB exposure, and then they were subjected to daily i.p. injections of the WT ARF26–44 peptide or mutant ARF37–44 peptide (Mut. ARF37–44 peptide) as described in Methods. (B–F) GFP-FoxM1b protein is targeted to the nucleolus by the WT ARF26–44 peptide. U2OS cells were transfected with GFP-FoxM1b expression vector and were either left untreated or incubated for 48 hours with TMR fluorescently tagged WT ARF26–44 peptide (C and D) or mutant ARF37–44 peptide (E and F) and then analyzed for GFP (green) or peptide (red) fluorescence. (G) TMR fluorescence labeling revealed that the WT ARF26–44 peptide was localized to the hepatocyte cytoplasm and nucleolus (white arrow) and in the hepatic mesenchymal cells (yellow arrow). (H and I) Both mutant ARF37–44 peptide and WT ARF26–44 peptide are targeted to the hepatocyte cytoplasm and nucleolus (white arrows) as determined by laser confocal microscopy. Immunostaining of tumor sections with antibody specific to either NPM protein (black arrows) (J) or FoxM1 protein (K–M). WT ARF26–44 peptide targets tumor FoxM1 to the nucleolus (black arrows), whereas FoxM1 remains nuclear after treatment with mutant ARF37–44 peptide or PBS (red arrows). Magnification: ×400 (B–F and J–M); ×200 (G); and ×600 (H and I).
Dose-response curve determined that only i.p. injection of 5 mg/kg body weight or greater of the TMR fluorescently tagged WT ARF26–44 peptide was detectable in cytoplasm and nucleolus of hepatocytes and in hepatic mesenchymal cells at 24 hours after administration (Figure 2G; see Methods). After 32 weeks of DEN/PB liver tumor induction, we subjected Foxm1fl/fl mice to daily i.p. injections of 5 mg/kg body weight of the WT ARF26–44 peptide or mutant ARF37–44 peptide for 4 weeks and of WT ARF26–44 peptide for 8 weeks (Figure 2A). After 4 weeks of treatment with TMR fluorescently tagged ARF peptides, laser confocal microscopy of paraffin-embedded mouse liver tumor sections revealed that ARF peptide fluorescence localized to the hepatocyte cytoplasm and nucleolus (Figure 2, H and I) and was uniformly distributed throughout the liver parenchyma (data not shown). The FoxM1 protein staining in WT ARF26–44 peptide–treated liver tumor sections was partially localized to the nucleolus in hepatic tumor cells (Figure 2L, black arrows), which was similar to the immunostaining pattern of the nucleolar protein nucleophosmin (NPM; Figure 2J, black arrows). In contrast, mutant ARF37–44 peptide– or PBS-treated liver tumor cells displayed only nuclear FoxM1 staining (compare Figure 2K and Figure 2M, red arrows). These studies demonstrate that the WT ARF26–44 peptide reduces in vivo function of FoxM1 by partially targeting the endogenous FoxM1 protein to the nucleolus of hepatic tumor cells.
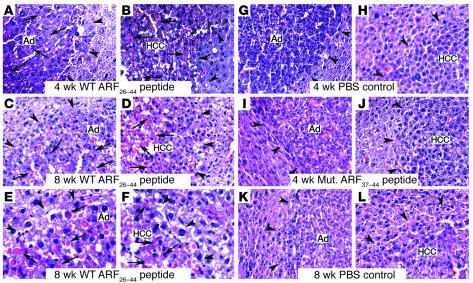
WT ARF26–44 peptide diminishes proliferation and size of liver tumors.
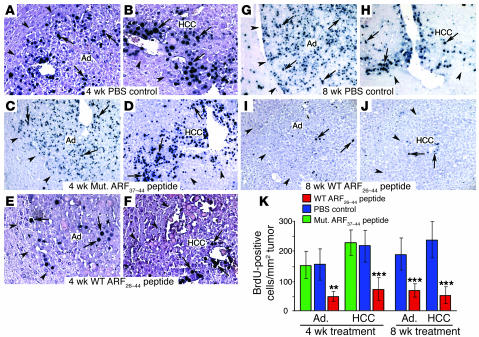
We next determined the number of hepatic tumor cells that incorporated BrdU in mice treated with WT ARF26–44 peptide, mutant ARF37–44 peptide, or PBS. Significant reduction in BrdU incorporation was found in liver tumors that had been treated with the WT ARF26–44 peptide for 4 or 8 weeks compared with mouse liver tumors treated with mutant ARF37–44 peptide or PBS (Figure 3, A–K). Compared with control treatment, treatment with the WT ARF26–44 peptide for 8 weeks significantly reduced tumor growth and prevented development of HCCs larger than 2 mm2 in size (Table 1). These results indicate that treatment with the WT ARF26–44 peptide is an effective method with which to reduce proliferation and growth of HCCs.
Figure 3. Treatment of mice with the WT ARF26–44 peptide diminishes proliferation of mouse hepatic tumors.
Hepatic tumors were induced in Foxm1fl/fl mice with DEN/PB treatment, and then they were treated with daily i.p. injections of 5 mg/kg body weight of WT ARF26–44 peptide or mutant ARF37–44 peptide or PBS for 4 or 8 weeks as described in the Figure 2A legend. These ARF peptide–treated mice were given drinking water with 1 mg/ml of BrdU for 4 days before sacrifice in order to obtain long-term labeling of the liver tumors (37). Arrows indicate nuclear staining for BrdU incorporation, and arrowheads show liver tumor margins. (A–J) BrdU incorporation was detected by immunohistochemical staining of liver tumor sections with monoclonal BrdU antibody from mice treated with the indicated ARF peptides. (K) Graph of mean number of BrdU-positive cells per square millimeter liver tumor (±SD) following treatment with WT ARF26–44 peptide or mutant ARF37–44 peptide or PBS. We calculated the mean number (±SD) of BrdU-positive hepatocyte nuclei per square millimeter liver tumor from 3 distinct mice treated with ARF peptide as described in Methods. The asterisks indicate statistically significant changes: **P ≤ 0.01 and ***P ≤ 0.001. Magnification: ×200 (A–J).
We previously demonstrated that tumor resistance in Foxm1–/– hepatic tumors was associated with persistent nuclear accumulation of the CDKI protein p27Kip1 (8). WT ARF26–44 peptide–treated HCC cells displayed increased nuclear levels of the p27Kip1 protein, which were similar to those found with dsRNA CKO Mx-Cre Foxm1–/– liver tumors (Figure 4, B and E). In contrast, p27Kip1 immunostaining was predominantly cytoplasmic in mutant ARF37–44 peptide– or PBS-treated mouse HCCs (Figure 4, A, C, D, and F). These studies indicate that the WT ARF26–44 peptide is effective in reducing FoxM1 function in vivo and that nuclear accumulation of p27Kip1 protein was associated with reduced hepatic tumor proliferation.
Figure 4. WT ARF26–44 peptide treatment causes nuclear accumulation of p27Kip1 protein in mouse HCC tumors.
(A–F) Nuclear accumulation of p27Kip1 protein in HCC tumors from WT ARF26–44 peptide–treated mice and dsRNA Mx-Cre Foxm1–/– mice. Hepatic tumors were induced in Foxm1fl/fl mice with DEN/PB treatment, and then they were treated with daily i.p. injections of 5 mg/kg body weight of WT ARF26–44 peptide (B) or mutant ARF37–44 peptide (C) or PBS (A) for 4 weeks as described in the Figure 3A legend. (D–F) The Foxm1 gene was genetically deleted in preexisting liver tumors of dsRNA Mx-Cre Foxm1–/– mice versus control dsRNA Foxm1fl/fl and PBS Mx-Cre Foxm1fl/fl as described in the Figure 1 legend. Liver tumor sections from the indicated mice were immunohistochemically stained with the p27Kip1 antibody. Arrows indicate nuclear staining for p27Kip1 protein, and arrowheads show liver tumor margins. Magnification, ×200.
WT ARF26–44 peptide causes selective apoptosis of hepatic tumor cells.
Analysis of H&E-stained liver tumor sections from mice treated with the WT ARF26–44 peptide revealed that many of the hepatic adenomas and HCC tumor cells stained red and exhibited disruption of nuclear membrane, which is indicative of apoptosis (Figure 5, A–F). These red-staining cells were found neither in the surrounding normal liver tissue (Figure 5, A–F) nor in hepatic tumors from mice treated with either the mutant ARF37–44 peptide or PBS (Figure 5, G–L). Furthermore, these apoptotic tumor cells were not apparent in deficient livers in dsRNA CKO Mx-Cre Foxm1–/– mice (Figure 1, E–G), a finding consistent with our previous tumor studies with Alb-Cre Foxm1–/– livers (8).
Figure 5. H&E-stained liver tumors from mice treated with WT ARF26–44 peptide.
Hepatic tumors were induced in Foxm1fl/fl mice with DEN/PB treatment, and then they were treated with daily i.p. injections of 5 mg/kg body weight of WT ARF26–44 peptide or mutant ARF37–44 peptide for 4 or 8 weeks (see Figure 2A legend). Arrows indicate red-staining cells undergoing apoptosis, and arrowheads show liver tumor margins. (A–F) H&E-stained liver tumor sections from WT ARF26–44 peptide–treated mice revealed that many of the hepatic adenomas and HCC tumor cells stained red and were rounded up, which is indicative of apoptosis. E and F are higher-magnification images of C and D. No red-staining apoptotic cells were found in either the surrounding, normal liver tissue or in liver tumors from dsRNA CKO Mx-Cre Foxm1–/– mice (see Figure 1, E–G). (G–L) No red-staining tumor cells were found in H&E-stained liver tumor sections from mice treated with either PBS or mutant ARF37–44 peptide. Magnification, ×200 (A–D and G–L); ×400 (E and F).
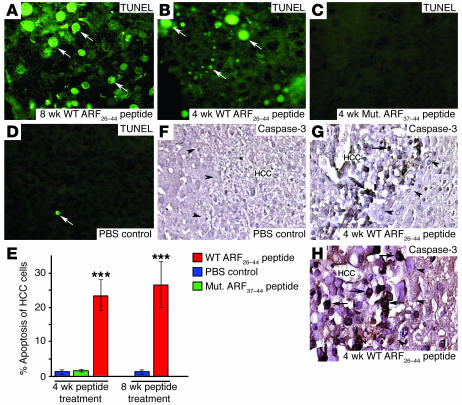
We used a fluorescence-based TUNEL assay to determine that mouse HCC cells treated with WT ARF26–44 peptide exhibited a significant, 22% increase in apoptosis (Figure 6, A, B, and E). In contrast, very few apoptotic HCC cells were found after treatment with mutant ARF37–44 peptide or PBS (Figure 6, C–E). Immunostaining of liver tumor sections with proteolytically cleaved activated caspase-3 protein confirmed this selective apoptosis of mouse HCC cells treated with WT ARF26–44 peptide with no proapoptotic staining in the adjacent normal liver tissue (Figure 6, F–H). These studies show that the WT ARF peptide selectively induces apoptosis of HCC cells without damaging adjacent normal hepatocytes.
Figure 6. WT ARF26–44 peptide treatment induces selective apoptosis of mouse HCC.
(A–D) Liver tumor sections were stained for apoptotic cells using the TUNEL assay. Mice treated with WT ARF26–44 peptide for either 4 or 8 weeks exhibited a significant increase in tumor cell apoptosis as evidenced by TUNEL-positive staining (green fluorescence; indicated by white arrows), whereas mutant ARF37–44 peptide– or PBS-treated mice displayed very few apoptotic cells in HCC tumors. (E) Quantitation of TUNEL-positive staining cells. Triple asterisks indicate statistically significant change at ***P ≤ 0.001. (F–H) Selective apoptosis is detected in HCC tumor cells in mice treated with WT ARF26–44 peptide by immunostaining with antibody specific to proteolytically cleaved activated caspase-3 protein. Arrows indicate nuclear staining for activated caspase-3 protein, and arrowheads show liver tumor margins. Magnification, ×400 (A–D and H); ×200 (F and G).
DEN/PB treatment induced highly proliferative HCCs in Arf–/– Rosa26-FoxM1b Tg mice that were responsive to WT ARF26–44 peptide treatment.
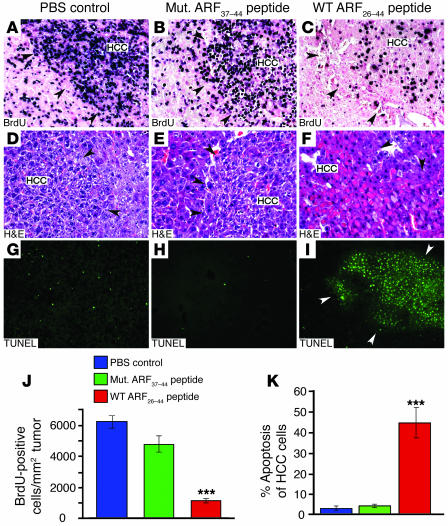
We previously showed that the ARF tumor suppressor targets the FoxM1b protein to the nucleolus and inhibits its transcriptional function (8). Furthermore, we showed that increased expression of the human FoxM1b cDNA in Rosa26-FoxM1b Tg mice (34) stimulated development and progression of prostate cancers in both TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double-Tg mice (35). In order to develop a new genetic model of HCC that is highly dependent on the FoxM1b transcription factor, we crossed the Rosa26-FoxM1b Tg mice into the Arf–/– mouse background. After 33 weeks of DEN/PB treatment, Arf–/– Rosa26-FoxM1b Tg mice developed highly proliferative HCCs, and their HCC cells displayed a proliferation rate of 6,000 BrdU-positive cells per square millimeter tumor (Figure 7J), which is approximately 30-fold greater than that observed in DEN/PB-induced HCCs in WT mice (Figure 3K; 200 BrdU-positive cells per mm2 tumor). The DEN/PB-treated Arf–/– Rosa26-FoxM1b Tg livers also exhibited development of necrosis and fibrosis/cirrhosis (data not shown). These HCC tumor–bearing Arf–/– Rosa26-FoxM1b Tg mice were subjected to daily treatment with either the WT ARF26–44 peptide or mutant ARF37–44 peptide for 4 weeks. In Arf–/– Rosa26-FoxM1b Tg mice, WT ARF26–44 peptide treatment resulted in a significant, 84% reduction in BrdU labeling of HCC cells compared with treatment of these mice with either mutant ARF37–44 peptide or PBS (Figure 7, A–C and J). Red-staining HCC cells with disruption of nuclear membrane indicative of apoptosis were found in H&E-stained liver tumor sections from Arf–/– Rosa26-FoxM1b Tg mice treated with the WT ARF26–44 peptide but not from those treated with mutant ARF37–44 peptide or PBS (Figure 7, D–F). We used a fluorescence-based TUNEL assay to determine that Arf–/– Rosa26-FoxM1b Tg HCC cells treated with WT ARF26–44 peptide exhibited a 42% increase in apoptosis (Figure 7K), which was twice as high as in liver tumors from WT mice (Figure 6E). Furthermore, TUNEL-positive cells were restricted to the HCC region (white arrowheads) and were not detected in adjacent normal liver tissue (Figure 7I). In contrast, very few apoptotic HCC cells were found after treatment of Arf–/– Rosa26-FoxM1b Tg mice with mutant ARF37–44 peptide or PBS (Figure 7, G, H, and K). These Arf–/– Rosa26-FoxM1b Tg liver tumor studies show that the WT ARF26–44 peptide is effective in diminishing BrdU labeling of highly proliferative HCC cells and selectively induces apoptosis of HCC cells in these mice without damaging adjacent normal liver tissue.
Figure 7. WT ARF26–44 peptide treatment reduces proliferation and increases apoptosis of HCCs, which were induced in Arf–/– Rosa26-FoxM1b Tg mice by DEN/PB.
Highly proliferative HCC tumors were induced in Arf–/– Rosa26-FoxM1b Tg mice following 33 weeks DEN/PB treatment. The Arf–/– Rosa26-FoxM1b Tg mice received daily i.p. injections of the WT ARF26–44 peptide (inhibitor of FoxM1 function) or mutant ARF37–44 peptide or PBS for 4 weeks. (A–C) Liver tumor sections were subjected to immunohistochemical staining with BrdU monoclonal antibody to determine HCC proliferation. Liver tumor sections were histologically stained with H&E (D and E) to identify red apoptotic cells or stained for apoptosis using the TUNEL assay (G–I). Black arrowheads indicate the boundaries of the HCC tumor, and white arrowheads (I) indicate the boundary of the HCC region. (J) We counted the BrdU-positive cells in HCCs and used this information to calculate the number of BrdU-positive cells per square millimeter liver tumor tissue (±SD). (K) We counted the TUNEL-positive cells in HCCs and used this information to calculate the percent HCC apoptosis (±SD). P values calculated by Student’s t test: ***P ≤ 0.001. Magnification: ×200 (A–F); ×100 (G–I).
WT ARF26–44 peptide inhibits angiogenesis of the HCC region.
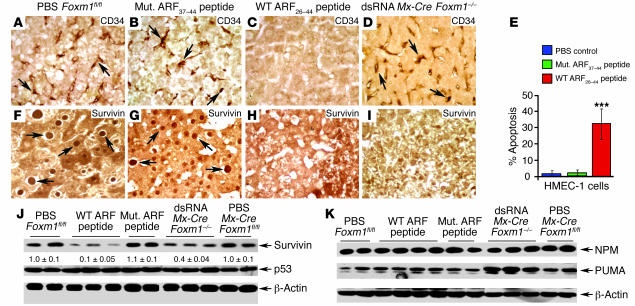
Angiogenesis is critical to mediating HCC growth, and the endothelial cells of new HCC capillaries exhibit expression of the CD34 protein (39–41). Abundant CD34 staining was found in endothelial cells of HCC regions in PBS or mutant ARF37–44 peptide–treated mice (Figure 8, A and B) and from dsRNA CKO Mx-Cre Foxm1–/– mice (Figure 8D). In contrast, expression of the CD34 protein was not detected in the HCC region from WT ARF26–44 peptide–treated mice (Figure 8C). These results suggest that WT ARF26–44 peptide treatment was preventing HCC angiogenesis, which was likely caused by apoptosis of new HCC endothelial cells (Figure 6B; small apoptotic cells). In order to determine whether WT ARF26–44 peptide induces apoptosis of endothelial cells, we treated human microvascular endothelial cells (HMEC-1 cells) for 48 hours with 100 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide or with PBS and then assayed for apoptosis as described in Methods. This analysis revealed that WT ARF26–44 peptide treatment induced a significant increase in apoptosis of HMEC-1 cells compared with treatment with mutant ARF37–44 peptide or PBS (Figure 8E). These studies suggest that WT ARF26–44 peptide is able to induce apoptosis of endothelial cells, which contributes to WT ARF peptide–mediated reduction in HCC angiogenesis.
Figure 8. WT ARF26–44 peptide reduces angiogenesis and survivin expression in mouse HCC.
The CD34 protein is a marker for newly formed sinusoid-like capillaries in HCC regions (39–41), whereas survivin is critical in preventing apoptosis of tumor cells (45–48). Antibodies specific to either CD34 or survivin were used to immunostain HCC tumor sections from mice treated with mutant ARF37–44 peptide, WT ARF26–44 peptide, or PBS or from dsRNA CKO Mx-Cre Foxm1–/– mice. (A–D) Mice treated with WT ARF26–44 peptide display no CD34-positive endothelial cells in HCC capillaries, whereas CD34 staining (indicated by arrows) was abundant in endothelial cells of control mouse HCCs. (E) WT ARF26–44 peptide induces apoptosis of HMEC-1 cells. HMEC-1 cells were treated for 48 hours with 100 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide or with PBS and then assayed for apoptosis as described in Methods. Shown graphically is the percent apoptosis of HMEC-1 cells in response to ARF peptide treatment. (F–I) Reduced survivin expression in WT ARF26–44 peptide–treated and Foxm1–/– liver tumors. Arrows indicate nuclear staining for survivin protein. (J) Western blot analysis reveals significant decrease in survivin protein expression in WT ARF26–44 peptide–treated mouse tumors. (K) No decrease in expression of NPM protein or p53-regulated proapoptotic PUMA protein is found in WT ARF26–44 peptide–treated mouse tumors. A slight increase in hepatic tumor levels of PUMA was found in mice treated with dsRNA. Magnification, ×400.
Reduced levels of the antiapoptotic survivin protein contribute to WT ARF26–44 peptide–induced HCC apoptosis.
We have shown that FoxM1 regulates transcription of survivin (26), which complexes with aurora B kinase to mediate its proper localization during mitosis (42–44). Survivin is also a member of the inhibitor of apoptosis (IAP) protein family and is selectively overexpressed in tumor cells to prevent their apoptosis (45–48). Mutant ARF37–44 peptide– or PBS-treated liver tumors displayed abundant nuclear and cytoplasmic staining of survivin protein (Figure 8, E and F), and survivin expression was restricted to the mouse HCC region (data not shown). Nuclear levels of survivin were diminished in HCC regions from WT ARF26–44 peptide–treated and dsRNA CKO Mx-Cre Foxm1–/– mice (Figure 8, G and H). Western blot analysis revealed that Foxm1–/– liver tumors displayed a 60% decrease in expression of survivin protein (Figure 8, I and J), and no apoptosis was detected in these FoxM1-deficient liver tumors (Figure 1, E–G). A more drastic, 90% decrease in survivin protein levels was found in hepatic tumors from WT ARF26–44 peptide–treated mice (Figure 8, H–J). Our hepatoma data presented below (Figure 9) support the hypothesis that WT ARF26–44 peptide treatment results in hypomorphic levels of FoxM1 activity, which reduces expression of mitotic regulators to levels that are insufficient to properly execute mitosis leading to apoptosis, whereas depleting FoxM1 levels leads to mitotic arrest.
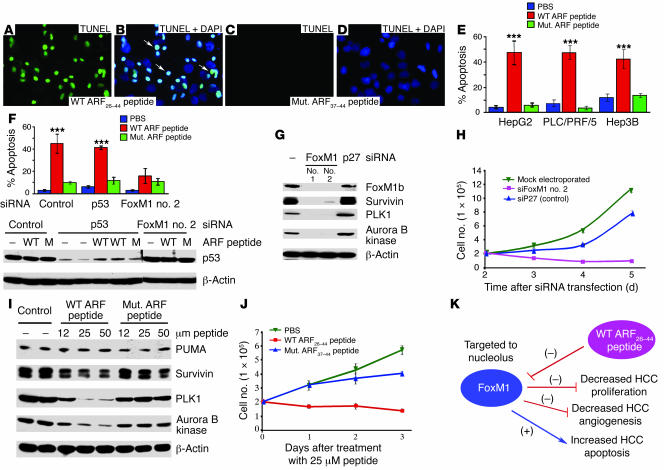
Figure 9. WT ARF26–44 peptide–induced apoptosis of human hepatoma cell lines.
Human hepatoma HepG2 (A–E), PLC/PRF/5 (expressing p53 mutant protein), or Hep3B (p53-deficient) cells were treated for 24 hours with 25 μM of WT ARF26–44 or mutant ARF37–44 peptide; they were then analyzed for apoptosis by TUNEL assay, and percent apoptosis (±SD) was calculated (E; ***P ≤ 0.001). Merged DAPI and TUNEL staining (A and C) images show TUNEL-positive nuclei of HepG2 cells (white arrows; B and D). (F) Graphic representation of WT ARF26–44 peptide–treated HepG2 cells showing that apoptosis is induced in p53-depleted cells but not in FoxM1-deficient cells. Western blot analysis is presented below the graph, showing effective downregulation of p53 protein levels following p53 siRNA electroporation and that treatment with WT ARF26–44 (WT) or mutant ARF37–44 peptide (M) does not alter p53 protein levels. (G and I) At 48 hours after electroporation with siFoxM1 no. 2 or p27 siRNA duplexes (G), or treatment with WT or mutant ARF peptide (I), HepG2 cells were analyzed for protein expression of survivin, PLK1, and aurora B kinase by Western blot analysis. We also determined a growth curve of HepG2 cells at the indicated days following siRNA transfection (H) or at the indicated days after ARF peptide treatment (J) as described in Methods. (K) Model summarizing findings in this article with WT ARF26–44 peptide. Magnification, ×400.
The nucleolar nucleophosmin/B23 (NPM/B23) protein (49, 50) and the p53 negative regulator, Mdm2 protein (51), associated with the ARF tumor suppressor protein through ARF amino acid sequences 1–15 and 26–37, which partially overlapped with our WT ARF26–44 peptide sequence. Despite this partial overlap, Western blot analysis enabled us to determine that neither WT ARF26–44 peptide treatment nor Foxm1 deficiency altered expression of NPM, p53, or Mdm2 proteins in liver tumor extracts (Figure 8, J and K, and data not shown), suggesting that the ARF26–44 peptide sequence was insufficient to influence their expression levels in vivo. The proapoptotic Bcl-2 family member p53-upregulated modifier of apoptosis (PUMA) is a transcriptional target gene of p53 that is essential for both p53 transcriptional and cytoplasmic-dependent apoptosis (52–55). Western blot analysis showed that neither WT ARF26–44 peptide nor mutant ARF37–44 peptide treatment caused significant change in expression of PUMA in liver tumor extracts (Figure 8K), suggesting that this apoptosis of HCC cells did not involve the p53/PUMA proapoptotic pathway.
WT ARF peptide–induced apoptosis of human hepatoma HepG2 cells correlates with diminished expression of survivin, PLK1, and aurora B kinase.
We used the TUNEL assay to determine that human hepatoma HepG2 cells (Figure 9, A–E), PLC/PRF/5 cells that express mutant p53 protein, and p53-deficient Hep3B cells exhibited 50% apoptosis after 24 hours of treatment with 25 μM of WT ARF26–44 peptide (Figure 9E), whereas only low levels of apoptosis were detected in these cells following treatment with mutant ARF37–44 peptide or PBS (Figure 9E). Diminished levels of p53 protein through p53 siRNA silencing of HepG2 cells did not influence apoptosis in response to WT ARF26–44 peptide treatment (Figure 9F). In addition, p53 protein levels were unaltered in HepG2 cells after 24 hours of treatment with WT ARF26–44 or mutant ARF37–44 peptide (Figure 9F). Furthermore, protein expression of the p53 downstream proapoptotic target PUMA was unchanged in HepG2 cells in response to increasing concentrations of the WT ARF26–44 peptide (Figure 9I). These results suggest that WT ARF26–44 peptide–induced apoptosis was independent of the p53/PUMA proapoptotic pathway (52–55). Moreover, HepG2 cells in which FoxM1 levels were depleted by electroporation of FoxM1 no. 2 siRNA duplexes were resistant to apoptosis in response to WT ARF26–44 peptide treatment (Figure 9F), suggesting that induction of apoptosis by the WT ARF peptide was dependent on FoxM1 levels.
Tumor cells are known to express high levels of the mitotic regulators PLK1, aurora B kinase, and survivin proteins, which function to prevent their apoptosis (45, 48, 56–62). Previous studies demonstrated that U2OS cells transfected with siFoxM1 #2 duplex were blocked in mitotic progression and exhibited undetectable levels of FoxM1 and its downstream target mitotic regulators PLK1, aurora B kinase, and survivin (26). Consistent with these studies, FoxM1-depleted HepG2 cells exhibited undetectable protein levels of survivin, PLK1, and aurora B kinase (Figure 9G). We electroporated HepG2 cells with siFoxM1 #2 or control p27Kip1 siRNA (siP27) and grew the cells in culture for 2 days to allow for siRNA silencing, and then 2 × 105 HepG2 cells were plated in triplicate, and viable HepG2 cells were counted at 2, 3, 4, or 5 days following electroporation. These cell-growth studies showed that FoxM1-deficient HepG2 cells were unable to grow in culture and gradually detached from the plate with time in culture (Figure 9H). In contrast, HepG2 cells treated with WT ARF26–44 peptide exhibited a less severe reduction in levels of survivin (50%), PLK1 (80%), and aurora B kinase (80%) proteins compared with controls (Figure 9I). We also determined a growth curve of HepG2 cells at 1, 2, or 3 days following treatment with WT ARF26–44 peptide, mutant ARF37–44 peptide, or PBS. Although the WT ARF26–44 peptide–treated HepG2 cells displayed 50% apoptosis (Figure 9E), they were able to sustain the number of cells initially plated (2 × 105), suggesting that the WT ARF peptide–treated cells were able to proceed through the cell cycle (Figure 9J). These results are consistent with recent studies in which hypomorphic levels of FoxM1 protein (40% of WT FoxM1 levels) in breast cancer cell lines transfected with a different Foxm1 siRNA duplex reduced expression of mitotic regulators to levels that are insufficient to properly execute mitosis, leading to mitotic catastrophe and apoptosis (63). Based on these findings, we propose the hypothesis that WT ARF26–44 peptide treatment causes hypomorphic levels of FoxM1 activity, leading to apoptosis, whereas depleting FoxM1 levels results in mitotic arrest. However, we have not ruled out other possibilities.
Discussion
Patients with HCCs have poor prognoses because late detection and high frequency of tumor recurrence render current HCC therapy ineffective (1). One of the potentially curative approaches in HCC therapy is based on interfering with HCC progression by blocking cell division and selectively inducing tumor cell apoptosis. In this study, we demonstrated that FoxM1 is required for proliferation of mouse liver cancer cells during tumor progression. To pharmacologically reduce in vivo activity of FoxM1 in HCC, mice were subjected to daily injections with a cell-penetrating ARF26–44 peptide inhibitor of FoxM1 function. After 4 weeks of ARF peptide treatment, FoxM1 protein was partially localized to the nucleolus of HCC cells, and these hepatic tumor cells displayed reduced proliferation and angiogenesis with selective induction of HCC apoptosis (Figure 9K). Reduced tumor cell proliferation in Foxm1-deficient and WT ARF26–44 peptide–treated tumors was associated with nuclear accumulation of the CDKI protein p27Kip, which is known to negatively regulate proliferation of HCC cells in mouse liver cancer models (64). In contrast, mice treated with the cell-penetrating mutant ARF37–44 peptide, which is missing ARF sequences required for interaction with the FoxM1 protein (8), did not change nuclear localization of FoxM1 protein and failed to influence proliferation, apoptosis, or angiogenesis of HCC cells. Our current studies show that treatment with the WT ARF26–44 peptide effectively diminishes proliferation and induces apoptosis of HCC cells by reducing FoxM1 function in vivo.
In order to develop a new genetic model of HCC that is highly dependent on FoxM1b transcription factor, we crossed Rosa26-FoxM1b Tg mice, which ubiquitously expressed the human FoxM1b cDNA transgene (34), into the Arf–/– mouse background. Use of the Arf–/– Rosa26-FoxM1b Tg mice allowed us to overexpress the FoxM1b protein and eliminated ARF tumor suppressor inhibition of the FoxM1b protein. After 33 weeks of DEN/PB treatment, Arf–/– Rosa26-FoxM1b Tg mice exhibited highly proliferative HCC cells that displayed approximately 30-fold more BrdU incorporation than that found in WT mice in which liver tumors were induced by DEN/PB exposure. After 4 weeks of treatment with the WT ARF26–44 peptide, the Arf–/– Rosa26-FoxM1b Tg mice exhibited a significant reduction in BrdU incorporation, and apoptosis was induced in twice the number of HCC cells. The results suggest that these highly proliferative hepatic tumors with increased levels of FoxM1b protein are more susceptible to WT ARF26–44 peptide–induced apoptosis and display diminished proliferation of HCC cells.
Increased expression of antiapoptotic survivin protein in human HCC and stage II colorectal carcinomas correlates with reduced apoptosis, poor patient outcome, and tumor recurrence following treatment (46–48). Proliferating tumor cells are known to constitutively express high levels of the mitotic regulators PLK1, aurora B kinase, and survivin proteins, while normal cells only express these proteins in the G2 phase and mitosis. Experiments that reduced expression of these mitotic regulators with siRNA transfection or reduced their activity with specific kinase inhibitors caused increased apoptosis and diminished growth of cancer cells (45, 48, 56–62). We found that HCC regions and HepG2 cells expressed high levels of PLK1, aurora B kinase, and survivin, all of which are FoxM1 transcriptional target genes (26). Previous studies demonstrated that Foxm1–/– mouse embryonic fibroblasts or FoxM1-depleted U2OS cells were blocked in mitotic progression, failed to undergo apoptosis, and were associated with undetectable levels of PLK1, aurora B kinase, and survivin (26). Interestingly, we showed that WT ARF26–44 peptide treatment induced significant apoptosis in human hepatoma HepG2 cells, PLC/PRF/5 cells that express mutant p53 protein, and p53-deficient Hep3B cells. We also showed that siRNA silencing of FoxM1 levels in HepG2 cells reduced apoptosis in response to WT ARF26–44 peptide treatment, suggesting that ARF-dependent apoptosis of this hepatoma cell line required FoxM1 expression. Furthermore, we showed that FoxM1-depleted HepG2 cells failed to grow in culture and displayed undetectable levels of the mitotic regulators PLK1, aurora B kinase, and survivin proteins (Figure 9). In contrast, treatment of HepG2 cells with the WT ARF26–44 peptide resulted in less drastic reductions in expression of survivin, PLK1, and aurora B kinase protein and allowed growth of HepG2 cells in culture but resulted in a significant induction of apoptosis (Figure 9). Consistent with these findings, published studies demonstrated that hypomorphic levels of FoxM1 protein (40% of WT FoxM1 levels) in breast cancer cell lines transfected with a different Foxm1 siRNA duplex reduced expression of mitotic regulators to levels that are insufficient to properly execute mitosis, leading to mitotic catastrophe and apoptosis (63). Taken together, our results suggest that WT ARF26–44 peptide treatment of cancer cells results in partial inhibition of Foxm1 activity, leading to mitotic catastrophe and apoptosis caused by reducing expression of the mitotic regulators PLK1, aurora B kinase, and survivin proteins to levels that are insufficient for proper mitotic progression.
Angiogenesis of HCC cells is a critical event that serves to increase blood supply to the growing tumor, and CD34 protein is expressed on new sinusoid-like endothelial cells of HCC capillaries (39–41). Mice treated with our WT ARF26–44 peptide failed to develop new CD34-positive endothelial cells in HCC capillaries (Figure 8C), whereas HCC angiogenesis was not affected in dsRNA CKO Mx-Cre Foxm1–/– mice. One explanation for the difference between these mice is that the WT ARF26–44 peptide may also be inducing apoptosis of CD34-positive endothelial cells in HCC capillaries, thus preventing angiogenesis of HCC regions required for tumor growth and expansion (Figure 9K). This is supported by the fact that WT ARF26–44 peptide treatment induces apoptosis of HMEC-1 cells in vitro (Figure 8E). Although the WT ARF26–44 peptide induces apoptosis of HepG2 cells in vitro, we cannot rule out the possibility that this WT ARF peptide is also inducing apoptosis of HCC cells by preventing tumor angiogenesis, thus limiting blood supply required for efficient growth of HCCs.
The WT ARF26–44 peptide sequences required to inhibit Foxm1 function do not completely overlap with ARF coding regions essential to inactivating other cell-cycle regulators. For example, both amino acid regions 1–14 and 26–37 of the ARF protein are required for association and nucleolar targeting of the p53-negative regulator Mdm2 protein (65) and the NPM/B23 protein (49, 50). Consistent with the specificity of our WT ARF peptide treatment of mice, we found no changes in liver tumor expression of Mdm2, p53, or NPM/B23 proteins in vivo, and apoptosis was independent of increased expression of PUMA, a p53 target gene required for mediating apoptosis (52–55). The ARF tumor suppressor protein also mediates nucleolar targeting of both the proliferation-specific E2F1 and c-Myc transcription factors (11–14). However, there is no overlap with our WT ARF26–44 peptide sequence or the ARF sequences necessary for association with the E2F1 (ARF sequences 6–10 and 21–25) protein (66) and c-Myc (ARF sequences 1–14) protein (12). Cotransfection assays in U2OS cells demonstrated that expression constructs containing ARF 26–37 sequences were unable to effectively inhibit FoxM1b transcriptional activity, suggesting that inhibition of FoxM1 function requires the entire ARF26–44 peptide sequence (data not shown). Furthermore, the in vitro experiments with FoxM1-deficient HepG2 cells showed a clear relationship between FoxM1 and WT ARF26–44 peptide–mediated apoptosis. Moreover, a recent study has shown that translation initiation produces an ARF protein product initiating at methionine amino acid residue 45 and that this N-terminal–truncated ARF protein is localized to the mitochondria and induces caspase-independent cell death (67). Because these ARF26–44 peptide sequences are not contained within this N-terminal–truncated ARF polypeptide, the WT ARF26–44 peptide utilizes a mechanism that is distinct from the caspase-independent function of this N-terminal–truncated ARF peptide. Based on these studies, the WT ARF26–44 peptide will specifically reduce Foxm1 function in vivo to limit growth of HCC and induce HCC apoptosis without diminishing the function of other known inhibitory targets of the ARF tumor suppressor protein.
After i.p. injection, the absorbed WT ARF26–44 peptide enters the portal circulation and then flows to the hepatic sinusoid capillaries, which are lined with fenestrated endothelial cells that allow efficient delivery of the ARF peptide to the underlying hepatocytes and hepatic mesenchymal cells (68). However, WT ARF26–44 peptide treatment of mice did not cause side effects because it could not transverse the endothelial cell barrier of blood vessels to intestinal, colonic, pulmonary, pancreatic, or kidney epithelial cells (data not shown). Likewise, ARF peptide fluorescence also remained in the vessel endothelial cells of bone marrow, spleen, and thymus yet was undetectable in the hematopoietic cells of these organs, which correlated with normal distribution of white and red blood cells in mice treated with this WT ARF26–44 peptide (data not shown). Administration of WT ARF26–44 peptide by i.p. injection is therefore a selective method for hepatic delivery of this ARF peptide in vivo to limit liver tumor progression and selectively induce apoptosis of HCC cells.
Methods
Mx-Cre–mediated deletion of the Foxm1fl/fl allele in mouse liver tumors induced by DEN/PB exposure.
Generation of C57BL/6 mice containing Foxm1fl/fl was described previously (25), and they were bred into the C57BL/6 mouse background for 8 generations. The type I IFN–inducible Mx-Cre Tg C57BL/6 mice (C57BL/6-TgN Mx-Cre mice) were purchased from The Jackson Laboratory. The Mx-Cre Tg C57BL/6 mice were bred with Foxm1fl/fl C57BL/6 mice, and the offspring were screened for Mx-Cre Foxm1fl/+ mice. These mice were then backcrossed with Foxm1fl/fl C57BL/6 mice to generate Mx-Cre Foxm1fl/fl C57BL/6 mice. Mx-Cre Foxm1fl/fl C57BL/6 mice were bred with Foxm1fl/fl C57BL/6 mice to generate a sufficient number of mice for the liver tumor experiments. Charles J. Sherr (St. Jude Children’s Research Hospital, Memphis, Tennessee, USA) provided the Arf–/– C57BL/6 mice (69). The Arf–/– C57BL/6 mice were backcrossed with the Rosa26-FoxM1b FVBN Tg mice, which ubiquitously express the human FoxM1b cDNA (34), to generate Arf–/– Rosa26-FoxM1b Tg mice.
At 14 days after birth, each mouse in the litter received a single, i.p. injection of the tumor initiator DEN (5 μg/g body weight; catalog N0756; Sigma-Aldrich) to induce liver tumors (8). Two weeks later, male mice were given water containing 0.025% PB tumor promoter for the duration of the experiment (8). At 30 weeks of DEN/PB exposure (which leads to HCC formation), we subjected Mx-Cre Foxm1fl/fl mice to 3 consecutive i.p. injections (separated by 1 day) of 250 μg of synthetic sRNA polyinosinic-polycytidylic acid [poly(I-C); Sigma-Aldrich] (36) to induce expression of the Mx-Cre transgene and cause deletion of the Foxm1fl/fl allele in preexisting liver tumors. We continued PB administration in the drinking water for an additional 10 weeks to allow tumor growth. Livers from mice sacrificed by CO2 asphyxiation were dissected and paraffin embedded for histological staining or immunostaining and for isolation of protein extracts as described previously (8). All protocols used in this study were approved by the animal protocol committee at the University of Illinois at Chicago.
Treatment of mice with DEN/PB-induced liver tumors with WT ARF26–44 peptide or mutant ARF37–44 peptide.
Genemed Synthesis Inc. synthesized the WT ARF26–44 peptide (rrrrrrrrrKFVRSRRPRTASCALAFVN) or mutant ARF37–44 peptide (rrrrrrrrrSCALAFVN), both of which contain 9 N-terminal D-Arg (r) residues (32, 33) and which are TMR fluorescently tagged (red) at the N-terminus. We chose to i.p. inject mice with this cell-penetrating WT ARF26–44 peptide to efficiently deliver this peptide to the liver in vivo. In order to determine the effective concentration of dose of the ARF peptide for efficient liver delivery, mice were subjected to i.p. injection of 0.1, 1, 5, or 10 mg/kg body weight of TMR fluorescently tagged WT ARF26–44 peptide and were sacrificed 24 hours later, after which their livers were dissected, formalin fixed, and paraffin embedded as described previously (25). Liver sections were treated with xylene to remove paraffin wax and then examined by fluorescence microscopy for red peptide fluorescence. This dose-response curve determined that i.p. injection of either equal to or greater than 5 mg/kg body weight of TMR fluorescently tagged WT ARF26–44 peptide was detectable in cytoplasm and nucleolus of hepatocytes and in hepatic mesenchymal cells at 24 hours after injection. Based on these studies, hepatic tumors were induced in Foxm1fl/fl mice by 32 weeks of DEN/PB exposure, and then they were subjected to daily i.p. injections of 5 mg/kg body weight of the WT ARF26–44 peptide or mutant ARF37–44 peptide for 4 weeks and with WT ARF26–44 peptide for 8 weeks. After 33 weeks of DEN/PB treatment, Arf–/– Rosa26-FoxM1b Tg mice were subjected to daily i.p. injections of 5 mg/kg body weight of the WT ARF26–44 peptide or mutant ARF37–44 peptide for 4 weeks. Liver tumor–bearing mice were also administered sterile PBS as controls.
BrdU labeling, immunohistochemical staining, and TUNEL apoptosis assay.
To monitor hepatic cellular proliferation, PB was removed 4 days prior to the completion of the experiment, and mice were placed on drinking water with 1 mg/ml of BrdU for 4 days before they were sacrificed (8, 37). Hepatic tumor cell DNA replication in liver sections was determined by immunohistochemical detection of BrdU incorporation as described previously (8). We used an affinity-purified rabbit polyclonal antibody specific to FoxM1b protein (1:500 dilution), which was generated against amino acids 365–748 of the human FoxM1b protein as described previously (26). We also used the following antibodies specific to the mouse anti-BrdU (Bu20a, 1:100; Dako), rabbit anti–cleaved caspase-3 (5A1, 1:100; Cell Signaling Technology), rabbit anti-survivin (1:250; Novus Biologicals), mouse anti-Kip1/p27 (1:100; BD Biosciences), and rat anti-CD34 (RAM34, 1:100; BD Biosciences) for immunohistochemical detection of 5-μm liver sections using methods described previously (8, 23, 25, 29). To measure apoptosis in mouse livers, we used the TUNEL assay on liver sections using the ApopTag Fluorescein In Situ Apoptosis Detection Kit from Intergen according to the manufacturer’s recommendations. We calculated the mean number (±SD) of TUNEL- or DAPI-positive hepatocyte nuclei per 1,000 cells or ×200 field by counting the number of positive hepatocyte nuclei using 5 different ×200 fields of liver tumor sections from male mice at the indicated times of DEN/PB exposure. We used 5 liver tumor sections from 3 mice to calculate the mean number of BrdU-positive cells (±SD) per square millimeter liver tumor. To calculate the area or size of liver tumors, we used micrographs of H&E-stained liver tumor sections taken by an Axioplan 2 microscope (Zeiss) and the AxioVision program (version 4.3; Zeiss).
Western blot analysis.
For Western blot analysis, 75 μg of total protein extracts prepared from liver were separated on SDS-PAGE and transferred to a PVDF membrane (Bio-Rad) as described previously (25). The following commercially available antibodies and dilutions were used for Western blotting: mouse anti–Plk-1 (F-8, 1:500; Santa Cruz Biotechnology Inc.), mouse anti-p53 (FL-393, 1:500; Santa Cruz Biotechnology Inc.), mouse anti-MDM2 (SMP14, 1:500; Santa Cruz Biotechnology Inc.), mouse anti–β-actin (AC-15, 1:5,000; Sigma-Aldrich), mouse anti–aurora B kinase/AIM-1 (1:1,000; BD Biosciences), rabbit anti-survivin (1:2,000; Novus Biologicals), mouse anti-PUMA (1:1,000; Cell Signaling Technology), and mouse anti-NPM/B23 (1:15,000; Zymed). The primary antibody signals were amplified by HRP-conjugated secondary antibodies (Bio-Rad) and detected with Enhanced Chemiluminescence Plus (ECL Plus; Amersham Biosciences). Western blot analysis was performed with liver extracts from 2–4 mice per indicated time point following DEN/PB treatment, and signal intensities were normalized to β-actin signals. Quantitation of expression levels was determined with tiff files from scanned films using the BioMax 1D software program (Kodak).
Treatment of doxycycline-inducible U2OS C3 cells with WT ARF26–44 peptide or mutant ARF37–44 peptide.
We previously reported on the generation and growth of an osteosarcoma U2OS clone C3 cell line (U2OS C3 cells) that allowed doxycycline-inducible (Dox-inducible) expression of the GFP-FoxM1b fusion protein (24). For induced expression of the GFP-FoxM1b fusion protein, we added 1 μg/ml of Dox (Sigma-Aldrich). In order to determine the FoxM1b localization in U2OS C3 cells, they were treated with 12 μM of the TMR fluorescently tagged WT ARF26–44 or mutant ARF37–44 peptide for 24 hours. U2OS C3 cells were fixed with 10% buffered formalin (Fisher Scientific) for 20 minutes at room temperature and rinsed with PBS, and cover glasses were mounted with Vectashield Mounting Medium with DAPI (catalog H-1200; Vector Laboratories). Immunofluorescence was detected using an Axioplan 2 microscope.
Treatment of human hepatoma cell lines with WT ARF26–44 or mutant ARF37–44 peptide and siRNA transfection.
HepG2 cells were plated on 100-mm plates in Ham’s F-12 medium supplemented with 10% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 0.5 U Humulin (insulin; Lilly). HepG2 cells were treated with 12 μM, 25 μM, or 50 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide for 24 hours and used to prepare whole-cell protein extracts using the NP40 lysis buffer as described previously (26), and Western blot analysis was performed as described above. HepG2 cells were treated with 25 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide for 24 hours, and HepG2 cell apoptosis was determined by TUNEL assay using the ApopTag Fluorescein In Situ Apoptosis Detection Kit from Intergen according to the manufacturer’s recommendations. We calculated the percent apoptosis of HepG2 cells (±SD) by counting the number of TUNEL-positive cells (green fluorescence) per 1,000 nuclei as visualized by DAPI (blue fluorescence) counterstaining.
Human hepatoma PLC/PRF/5 and Hep3B cell lines (ATCC) were grown as a monolayer on 100-mm plates in DMEM supplemented with 10% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Human hepatoma PLC/PRF/5 and Hep3B cells were treated with 25 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide for 24 hours, and cells undergoing apoptosis were determined by TUNEL assay as described above.
Transfection of human cells with FoxM1 siRNA duplexes (Dharmacon RNA Technologies), named siFoxM1 no. 1 (CAACAGGAGUCUAAUCAAG) and siFoxM1 no. 2 (GGACCACUUUCCCUACUUU), efficiently depleted FoxM1 levels as described previously (26). We also previously used the human p27Kip1 siRNA duplex (GUACGAGUGGCAAGAGGUGUU) as a control siRNA, which did not influence expression of the FoxM1 gene (26). Human p53 siRNA duplex was purchased from Cell Signaling Technology Inc. These siRNA duplexes were transfected into HepG2 cells using the Nucleofector II apparatus and buffers recommended by the manufacturer (Amaxa Biosystems). Forty-eight hours after electroporation to allow siRNA silencing, HepG2 cells were treated with 25 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide for 24 hours and then examined for apoptosis by TUNEL assay. HepG2 cells were harvested at 72 hours after FoxM1 siRNA or p27 siRNA transfection to prepare protein extracts for Western blot analysis to examine expression of PUMA, survivin, PLK1, and aurora B kinase proteins as described above. HepG2 cells were harvested at 48 hours after p53 siRNA transfection or at 72 hours after p53 siRNA transfection and examined for apoptosis as described above.
Treatment of HMEC-1 cells with WT ARF26–44 or mutant ARF37–44 peptide.
HMEC-1 cells (ATCC) were grown as monolayer cultures on 100-mm plates in MCDB 131 medium supplemented with 15% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 10 ng/ml EGF (Sigma-Aldrich), and 1 μg/ml hydrocortisone (Sigma-Aldrich). HMEC-1 cells were treated for 48 hours with 100 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide and then examined for apoptosis as described above.
Growth curve of HepG2 cells electroporated with FoxM1 or p27Kip1 siRNA or treated with WT or ARF peptide.
HepG2 cells were electroporated with 100 nM of FoxM1 (FoxM1 no. 2) or p27Kip1 (siP27) siRNA duplexes (26) using the Nucleofector II apparatus (Amaxa Biosystems) and eletroporation buffers recommended by the manufacturer for HepG2 cells. HepG2 cells were replated for 2 days to allow siRNA silencing of FoxM1 or p27Kip1 levels, and then 2 × 105 HepG2 cells were plated in triplicate, and viable HepG2 cells were counted at 2, 3, 4, or 5 days following electroporation. Mock-electroporated cells were used as controls. We also plated 2 × 105 HepG2 cells in triplicate, and viable HepG2 cells were counted at 1, 2, or 3 days following treatment with 50 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide. After 2 days in culture, media was replaced with 50 μM of WT ARF26–44 peptide or mutant ARF37–44 peptide. PBS-treated cells were used as controls.
Statistics.
We used the Microsoft Excel program to calculate SD and statistically significant differences between samples using the 2-tailed Student’s t test. The asterisks in each graph indicate statistically significant changes with P values calculated by Student’s t test: *P < 0.05, **P ≤ 0.01, and ***P ≤ 0.001. P values less than 0.05 were considered statistically significant.
Acknowledgments
This work was supported by US Public Health Service Grants R01 DK 54687 from the NIDDK, R01 CA124488 from the National Cancer Institute, and R01 AG 21842 from the National Institute on Aging (to R.H. Costa). We thank H. Yoder, W. Wang, and B. Shin for excellent technical assistance and B. Merrill and P. Raychaudhuri for helpful discussions and critical review of this paper.
Footnotes
Nonstandard abbreviations used: ARF, alternative reading frame; CDK, cyclin-dependent kinase; CDKI, CDK inhibitor; CKO, conditional(ly) KO; D-Arg, d-arginine; DEN, diethylnitrosamine; dsRNA, double-stranded RNA; Fox, forkhead box; HCC, hepatocellular carcinoma; HMEC-1 cell, human microvascular endothelial cell; PB, phenobarbital; PLK1, polo-like kinase 1; PUMA, p53-upregulated modifier of apoptosis; TMR, tetramethylrhodamine.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:99–111 (2007). doi:10.1172/JCI27527
While the revised manuscript was in preparation, Robert H. Costa died in his fight against pancreatic cancer. The authors dedicate this work to his memory.
References
- 1.Bruix J., Boix L., Sala M., Llovet J.M. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 3.Franke T.F., Hornik C.P., Segev L., Shostak G.A., Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003;22:8983–8998. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 4.Sherr C.J. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 5.Calvisi D.F., Thorgeirsson S.S. Molecular mechanisms of hepatocarcinogenesis in transgenic mouse models of liver cancer. Toxicol. Pathol. 2005;33:181–184. doi: 10.1080/01926230590522095. [DOI] [PubMed] [Google Scholar]
- 6.Calvisi D.F., et al. Activation of the canonical Wnt/beta-catenin pathway confers growth advantages in c-Myc/E2F1 transgenic mouse model of liver cancer. J. Hepatol. 2005;42:842–849. doi: 10.1016/j.jhep.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 7.Sandgren E.P., et al. Transforming growth factor α dramatically enhances oncogene-induced carcinogenesis in transgenic mouse pancreas and liver. Mol. Cell. Biol. 1993;13:320–330. doi: 10.1128/mcb.13.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalinichenko V.V., et al. Forkhead box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J.S., et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat. Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 10.Lowe S.W., Sherr C.J. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 11.Datta A., Nag A., Raychaudhuri P. Differential regulation of E2F1, DP1, and the E2F1/DP1 complex by ARF. Mol. Cell. Biol. 2002;22:8398–8408. doi: 10.1128/MCB.22.24.8398-8408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta A., et al. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J. Biol. Chem. 2004;279:36698–36707. doi: 10.1074/jbc.M312305200. [DOI] [PubMed] [Google Scholar]
- 13.Martelli F., et al. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4455–4460. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Y., et al. p19(ARF) directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- 15.Carlsson P., Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 16.Kaestner K.H., Knochel W., Martinez D.E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 17.Clark K.L., Halay E.D., Lai E., Burley S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 18.Marsden I., Jin C., Liao X. Structural changes in the region directly adjacent to the DNA-binding helix highlight a possible mechanism to explain the observed changes in the sequence-specific binding of winged helix proteins. J. Mol. Biol. 1998;278:293–299. doi: 10.1006/jmbi.1998.1703. [DOI] [PubMed] [Google Scholar]
- 19.Korver W., Roose J., Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luscher-Firzlaff J.M., et al. Interaction of the fork head domain transcription factor MPP2 with the human papilloma virus 16 E7 protein: enhancement of transformation and transactivation. Oncogene. 1999;18:5620–5630. doi: 10.1038/sj.onc.1202967. [DOI] [PubMed] [Google Scholar]
- 21.Yao K.M., Sha M., Lu Z., Wong G.G. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J. Biol. Chem. 1997;272:19827–19836. doi: 10.1074/jbc.272.32.19827. [DOI] [PubMed] [Google Scholar]
- 22.Ye H., et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye H., Holterman A., Yoo K.W., Franks R.R., Costa R.H. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S-phase. Mol. Cell. Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Major M.L., Lepe R., Costa R.H. Forkhead box M1B (FoxM1B) transcriptional activity requires binding of Cdk/Cyclin complexes for phosphorylation-dependent recruitment of p300/CBP co-activators. Mol. Cell. Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Kiyokawa H., Dennewitz M.B., Costa R.H. The Forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang I.-C., et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagano M. Control of DNA synthesis and mitosis by the Skp2-p27-Cdk1/2 axis. Mol. Cell. 2004;14:414–416. doi: 10.1016/s1097-2765(04)00268-0. [DOI] [PubMed] [Google Scholar]
- 28.Wang X., et al. Increased levels of Forkhead box M1B transcription factor in transgenic mouse hepatocytes prevents age-related proliferation defects in regenerating liver. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11468–11473. doi: 10.1073/pnas.201360898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krupczak-Hollis K., et al. The mouse Forkhead box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev. Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Laoukili J., et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 31.Costa R.H., Kalinichenko V.V., Major M.L., Raychaudhuri P. New and unexpected: forkhead meets ARF. Curr. Opin. Genet. Dev. 2005;15:42–48. doi: 10.1016/j.gde.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs S.M., Raines R.T. Pathway for polyarginine entry into mammalian cells. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wender P.A., et al. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalinichenko V.V., et al. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell-types following lung injury. J. Biol. Chem. 2003;278:37888–37894. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- 35.Kalin T.V., et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn R., Schwenk F., Aguet M., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 37.Ledda-Columbano G.M., et al. Loss of cyclin D1 does not inhibit the proliferative response of mouse liver to mitogenic stimuli. Hepatology. 2002;36:1098–1105. doi: 10.1053/jhep.2002.36159. [DOI] [PubMed] [Google Scholar]
- 38.Rizos H., Darmanian A.P., Mann G.J., Kefford R.F. Two arginine rich domains in the p14ARF tumour suppressor mediate nucleolar localization. Oncogene. 2000;19:2978–2985. doi: 10.1038/sj.onc.1203629. [DOI] [PubMed] [Google Scholar]
- 39.Cui S., et al. Enhanced CD34 expression of sinusoid-like vascular endothelial cells in hepatocellular carcinoma. Pathol. Int. 1996;46:751–756. doi: 10.1111/j.1440-1827.1996.tb03544.x. [DOI] [PubMed] [Google Scholar]
- 40.Ohmori S., et al. High expression of CD34-positive sinusoidal endothelial cells is a risk factor for hepatocellular carcinoma in patients with HCV-associated chronic liver diseases. Hum. Pathol. 2001;32:1363–1370. doi: 10.1053/hupa.2001.29678. [DOI] [PubMed] [Google Scholar]
- 41.Tanigawa N., Lu C., Mitsui T., Miura S. Quantitation of sinusoid-like vessels in hepatocellular carcinoma: its clinical and prognostic significance. Hepatology. 1997;26:1216–1223. doi: 10.1053/jhep.1997.v26.pm0009362365. [DOI] [PubMed] [Google Scholar]
- 42.Sampath S.C., et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 43.Temme A., et al. Localization, dynamics, and function of survivin revealed by expression of functional survivinDsRed fusion proteins in the living cell. Mol. Biol. Cell. 2003;14:78–92. doi: 10.1091/mbc.E02-04-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wheatley S.P., Henzing A.J., Dodson H., Khaled W., Earnshaw W.C. Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J. Biol. Chem. 2004;279:5655–5660. doi: 10.1074/jbc.M311299200. [DOI] [PubMed] [Google Scholar]
- 45.Altieri D.C. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 46.Ikeguchi M., Ueda T., Sakatani T., Hirooka Y., Kaibara N. Expression of survivin messenger RNA correlates with poor prognosis in patients with hepatocellular carcinoma. Diagn. Mol. Pathol. 2002;11:33–40. doi: 10.1097/00019606-200203000-00007. [DOI] [PubMed] [Google Scholar]
- 47.Kawasaki H., et al. Expression of survivin correlates with apoptosis, proliferation, and angiogenesis during human colorectal tumorigenesis. Cancer. 2001;91:2026–2032. doi: 10.1002/1097-0142(20010601)91:11<2026::aid-cncr1228>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 48.Sarela A.I., Macadam R.C., Farmery S.M., Markham A.F., Guillou P.J. Expression of the antiapoptosis gene, survivin, predicts death from recurrent colorectal carcinoma. Gut. 2000;46:645–650. doi: 10.1136/gut.46.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korgaonkar C., et al. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brady S.N., Yu Y., Maggi L.B., Weber J.D. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol. Cell. Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertwistle D., Sugimoto M., Sherr C.J. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chipuk J.E., Bouchier-Hayes L., Kuwana T., Newmeyer D.D., Green D.R. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 53.Jeffers J.R., et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 54.Nakano K., Vousden K.H. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 55.Villunger A., et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 56.Fan Y., Zheng S., Xu Z.F., Ding J.Y. Apoptosis induction with polo-like kinase-1 antisense phosphorothioate oligodeoxynucleotide of colon cancer cell line SW480. World J. Gastroenterol. 2005;11:4596–4599. doi: 10.3748/wjg.v11.i29.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guan R., et al. Small interfering RNA-mediated Polo-like kinase 1 depletion preferentially reduces the survival of p53-defective, oncogenic transformed cells and inhibits tumor growth in animals. Cancer Res. 2005;65:2698–2704. doi: 10.1158/0008-5472.CAN-04-2131. [DOI] [PubMed] [Google Scholar]
- 58.Gumireddy K., et al. ON01910, a non-ATP-competitive small molecule inhibitor of Plk1, is a potent anticancer agent. Cancer Cell. 2005;7:275–286. doi: 10.1016/j.ccr.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 59.Harrington E.A., et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 60.Liu X., Erikson R.L. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5789–5794. doi: 10.1073/pnas.1031523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reagan-Shaw S., Ahmad N. Silencing of polo-like kinase (Plk) 1 via siRNA causes induction of apoptosis and impairment of mitosis machinery in human prostate cancer cells: implications for the treatment of prostate cancer. Faseb J. 2005;19:611–613. doi: 10.1096/fj.04-2910fje. [DOI] [PubMed] [Google Scholar]
- 62.Spankuch-Schmitt B., Bereiter-Hahn J., Kaufmann M., Strebhardt K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J. Natl. Cancer Inst. 2002;94:1863–1877. doi: 10.1093/jnci/94.24.1863. [DOI] [PubMed] [Google Scholar]
- 63.Wonsey D.R., Follettie M.T. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 64.Santoni-Rugiu E., Jensen M.R., Factor V.M., Thorgeirsson S.S. Acceleration of c-myc-induced hepatocarcinogenesis by Co-expression of transforming growth factor (TGF)-alpha in transgenic mice is associated with TGF-beta1 signaling disruption. Am. J. Pathol. 1999;154:1693–1700. doi: 10.1016/s0002-9440(10)65425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber J.D., et al. Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 2000;20:2517–2528. doi: 10.1128/mcb.20.7.2517-2528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Datta A., et al. ARF directly binds DP1: interaction with DP1 coincides with the G1 arrest function of ARF. Mol. Cell. Biol. 2005;25:8024–8036. doi: 10.1128/MCB.25.18.8024-8036.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reef S., et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol. Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 68.Zhao R., Duncan S.A. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- 69.Kamijo T., Bodner S., van de Kamp E., Randle D.H., Sherr C.J. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]