Abstract
Essential for embryonic development, the polycomb group protein enhancer of zeste homolog 2 (EZH2) is overexpressed in breast and prostate cancers and is implicated in the growth and aggression of the tumors. The tumorigenic mechanism underlying EZH2 overexpression is largely unknown. It is believed that EZH2 exerts its biological activity as a transcription repressor. However, we report here that EZH2 functions in gene transcriptional activation in breast cancer cells. We show that EZH2 transactivates genes that are commonly targeted by estrogen and Wnt signaling pathways. We demonstrated that EZH2 physically interacts directly with estrogen receptor α and β-catenin, thus connecting the estrogen and Wnt signaling circuitries, functionally enhances gene transactivation by estrogen and Wnt pathways, and phenotypically promotes cell cycle progression. In addition, we identified the transactivation activity of EZH2 in its two N-terminal domains and demonstrated that these structures serve as platforms to connect transcription factors and the Mediator complex. Our experiments indicated that EZH2 is a dual function transcription regulator with a dynamic activity, and we provide a mechanism for EZH2 in tumorigenesis.
Initially discovered as epigenetic silencers during embryogenesis, polycomb group (PcG) proteins have been implicated in development and differentiation (34). The biological activities of PcG proteins are expanding and now include the regulation of various adult processes, such as lymphopoiesis, X-inactivation, and cell proliferation, and several PcG genes have been implicated in tumorigenesis (3, 5).
Initial suggestions that EZH2 is involved in cell proliferation came from the observation that EZH2 is preferentially expressed in proliferating, but not resting, mantle cell lymphoma cells (53). Subsequently, EZH2 was found to be overexpressed in metastatic prostate cancer, and knockdown of EZH2 expression inhibited cell proliferation (52). It was also observed that the EZH2 level directly correlates with the aggressiveness of breast cancer, and forced EZH2 expression in immortalized human mammary epithelial cell lines promotes anchorage-independent growth and cell invasion (3, 20).
How EZH2 promotes cell proliferation and tumor progression is still largely unknown. It is believed that EZH2 functions by forming polycomb repressive complex (PRC) with other PcG proteins (29, 35). These protein complexes are characterized by an intrinsic histone lysine methyltransferase (HMTase) activity that is mediated by the SET domain of EZH2 (25) and that targets different lysine residues on histones H1 or H3 in vitro (5, 23). Core histone methylation facilitates the establishment of a stable, repressive chromatin structure to prevent transcription initiation by prebound factors (7). In addition, several PcG proteins interact or colocalize with various non-PcG proteins, including the transcription modulators CtBP (41), E2F6 (2, 51), RYBP (12), AF9 (13), SSX (49), and the mitogen-activated protein/kinase-activated protein kinase 3pK (54). All of these proteins may contribute to the activity of PcG complexes and their ability to bind chromatin and, thus, may be involved in oncogenesis.
Despite the characterization of EZH2 as a component of transcriptional repressors, there has been evidence implying EZH2 activities that are not compatible with the transcription repression model. It is believed that PcG proteins act in conjunction with Trithorax group (TrxG) proteins to maintain repressed or active transcription states of developmentally regulated genes. However, gene ablation of E(z) in Drosophila melonagaster produced phenotypes similar to those of trithorax loss-of-function mutants (46), and E(z) loss-of-function mutations led to a complete loss of the accumulation of the homeotic selector gene products SCR, ANTP, and UBX and the segmentation gene product EN (27). More recently, it has been shown in mammalian cells that knockdown EZH2 expression by RNA interference (RNAi) led to a significant decrease, rather than increase, of G1/S-expressed cyclins (3), suggesting that EZH2 may play an activation role in the expression of these genes. Here we show that EZH2 activates rather than represses the transcription of c-Myc and cyclin D1 in breast cancer cells, and this activation activity is independent of the SET domain that is believed to mediate its transcriptional repression. We show that EZH2 interacts with components of the estrogen and Wnt pathways, integrates estrogen and Wnt signaling, and promotes cell proliferation in breast cancer cells.
MATERIALS AND METHODS
Cell cultures and reagents.
MCF-7 and T47-D cells (American Type Culture Collection, Manassas, VA) were grown in phenol red-free Eagle's minimal essential medium (HyClone, Logan, UT) supplemented with 10% dextran-charcoal-stripped fetal bovine serum (FBS; HyClone). Cells from <50 passages were used for experiments. 17β-Estradiol (E2) was purchased from Sigma (St. Louis, MO). Polyclonal anti-EZH2 antibodies were obtained from Zymed Laboratories Inc. (San Francisco, CA). Polyclonal anti-estrogen receptor α (anti-ERα) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal anti-c-Myc (9E10) antibody and monoclonal anti-cyclin D1 (A-12) antibody were obtained from Santa Cruz Biotechnology, and protein A-Sepharose CL-4B beads were from Amersham Biosciences. Protease inhibitor mixture cocktail was obtained from Roche Applied Science.
Transient transfection and reporter assays.
Cells were plated at a density of 1 × 105 cells per cell in 24-well plates. After 24 h, cells were transfected with c-Myc promoter-driven luciferase constructs (16) (c-Myc-Luc) or cyclin D1 promoter-driven luciferase constructs (50) (cyclin D1-Luc) using Lipofectamine 2000 according to the manufacturer's instruction. Forty-eight hours after transfection, the cells were harvested. Firefly and Renilla luciferase activities were measured using a dual luciferase kit (Promega, Madison, WI). The firefly luciferase data for each sample were normalized based on transfection efficiency as measured by Renilla luciferase activity. Each experiment was performed in triplicate and repeated at least three times.
Real-time quantitative PCR.
The ABI Prism 7700 sequence detector and the TaqMan EZ reverse transcriptase PCR (RT-PCR) kit (Applied Biosystems, Foster City, CA) were used to quantitate gene expression, with glyceraldehyde-3-phosphate dehydrogenase as the internal control. The primers and probes used were as follows: c-Myc forward primer, GCCACGTCTCCACACATCAG; c-Myc reverse primer, TCTTGGCAGCAGGATAGTCCTT; c-Myc probe, 6-carboxyfluorescein-ACGCAGCGCCTCCCTCCACTC-6-carboxytetramethylrhodamine; cyclin D1 forward primer, CACGCGCAGACCTTCGTT; cyclin D1 reverse primer, GCGGATTGGAAATGAACTTCA; cyclin D1 probe, 6-carboxyfluorescein-CCTCTGTGCCACAGAT-6-carboxytetramethylrhodamine.
RNA interference.
A vector-based RNAi method was utilized. For initial experiments, three specific sequences corresponding to different regions of mRNAs were synthesized and tested for efficiency in knockdown of specific protein expression, and the sequence with an optimal efficiency was used in the experiment. Plasmids were constructed by inserting a synthesized 64-mer oligonucleotide containing the specific sequences for open reading frames of the genes into pSUPER or pSILENCER vectors (58). The sequences synthesized were as follows: EZH2 small interfering RNA (siRNA), forward, 5′-GATCCCGCTTCTGTGAGCTCATTGCGTTCAAGAGACGCAATGAGCTCACAGAAGTTTTTGGAAA-3′, and reverse, 5′-AGCTTTTCCAAAAACTTCTGTGAGCTCATTGCGTCTCTTGAACGCAATGAGCTCACAGAAGCGG-3′; β-catenin siRNA, forward, 5′-GATCCCCTGCTTGGTTCACCAGTGGATTCAAGAGATCCACTGGTGAACCAAGCATTTTTGGAAA-3′, and reverse, 5′-AGCTTTTCCAAAAATGCTTGGTTCACCAGTGGATCTCTTGAATCCACTGGTGAACCAAGCACGGG-3′; ERα siRNA, forward, 5′-GATCCCCGCTACTGTTTGCTCCTAACTTCAAGAGAGTTAGGAGCAAACAGTAGCTTTTTGGAAA-3′, and reverse, 5′-AGCTTTTCCAAAAAGCTACTGTTTGCTCCTAACTCTCTTGAAGTTAGGAGCAAACAGTAGCGGG-3′. The oligonucleotides were resuspended in annealing buffer (100 nM potassium acetate, 30 mM HEPES-KOH, pH 7.4, 2 mM acetate) and heated to 95°C for 4 min, 70°C for 10 min, and then cooled to room temperature to generate double-stranded DNA. The double-stranded DNA was then phosphorylated and cloned into the BglII/HindIII site of the pSUPER vector (pSUPER-EZH2) and the BamHI/HindIII site of the pSILENCER vector (pSILENCER-EZH2). The vectors were then transfected into MCF-7 cells with the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA).
GST pull-down experiments.
Glutathione S-transferase (GST) fusion constructs were expressed in BL21 Escherichia coli cells, and crude bacterial lysates were prepared by sonication in 50 mM Tris-HCl, pH 7.4, 1.5 mM EDTA, 1 mM dithiothreitol, 10% (vol/vol) glycerol, 0.4 M NaCl in the presence of the protease inhibitor mixture. The in vitro transcription and translation experiments were done with rabbit reticulocyte lysate (TNT systems; Promega, Madison, WI) and l-[35S]methionine (Amersham Biosciences) according to the manufacturer's recommendation. In GST pull-down assays, about 10 μg of the appropriate GST fusion protein was mixed with 5 to 8 μl of the in vitro-transcribed/translated products and incubated in binding buffer (75 mM NaCl, 50 mM HEPES, pH 7.9) at room temperature for 30 min in the presence of the protease inhibitor mixture. The binding reaction mixture was then added to 30 μl of glutathione-Sepharose beads and mixed at 4°C for 2 h. The beads were washed three times with binding buffer, resuspended in 30 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and resolved on 10% gels. The gels were then fixed in 50% methanol, 10% acetic acid for 30 min and dried. Protein bands were detected by autoradiography at −80°C for 4 to 24 h.
Coimmunoprecipitation assay and Western blotting.
MCF-7 cells were grown in dishes of 100-mm diameter seeded with 1 × 106 cells. For immunoprecipitation experiments, cells were maintained in phenol red-free medium supplemented with 10% dextran-charcoal-stripped FBS for 72 h and then treated with 10 nM E2 or with vehicle (100% ethanol) for 2 h. Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and then incubated in lysis buffer (50 mM Tris, pH 8.0, 120 mM NaCl, 0.5% [vol/vol] Nonidet P-40, and protease inhibitor mixture) for 10 min on ice. Cell extracts were collected using a rubber policeman and incubated with the anti-ERα and anti-EZH2 antibodies or control normal rabbit immunoglobulin G overnight 4°C on a rotator, followed by addition of protein A-Sepharose CL-4B beads for 2 h at 4°C. Beads were washed twice in NETN buffer I (20 mM Tris, pH 8.0, 1 mM EDTA, 900 mM NaCl, 0.5% [vol/vol] Nonidet P-40, and protease inhibitor mixture), and washed twice in NETN buffer II (20 mM Tris, pH 8.0, 1 mM EDTA, 900 mM NaCl, 0.5% [vol/vol] Nonidet P-40, and protease inhibitor mixture). Complexes were released from the protein A-Sepharose CL-4B by boiling for 5 min in 2× SDS-PAGE loading buffer. The immunoprecipitated materials were separated on 10% SDS-PAGE and blotted onto nitrocellulose membrane, and the membrane was probed using appropriate antibodies.
ChIP and re-ChIP assays.
Chromatin immunoprecipitation (ChIP) assays and re-ChIP were performed essentially the same as described previously (43-45, 60). The primers used were the following: for the c-Myc promoter, forward, 5′-CACAGGACAAGGATGCGGTT-3′, and reverse, 5′-CTTCTGCCTCTCGCTGGAAT-3′; for the MYT1 promoter, forward, 5′-GGGTGAATGTGGTGGTTGTG-3′, and reverse, 5′-TGGAGTCCTGTGTCCTGCTCT-3′. Quantitative ChIP was performed using Sybr Green (Molecular Probes, Eugene, OR) as a marker for DNA amplification on the ABI Prism 7700 sequence detector. The amounts of immunoprecipitated DNA were normalized to the input and are reported relative to the amount obtained at about bp −7000, which was set to 1. The bp −7000 site was used to normalize all the ChIP data in this study because it represented a negative control for transcription factor and coactivator binding.
Construction of EZH2 deletion mutants.
EZH2 deletion mutants were constructed via PCR with subsequent ligation of the N-terminal and C-terminal fragments. The PCR primers for creating deletion 1 were the following: N-terminal fragment, 5′ primer (CTCGGATCCATAATCATGGGC) and 3′ primer (TTCTCTATCCAAGTCACTGGTCACCGAACA); C-terminal fragment, 5′ primer (GATAGAGAATGTGGGTTTATAAATGAT) and 3′ primer (CATGCTCGAGGTTCCTGAAGC). The PCR primers for creating deletion 2 were as follows: N-terminal fragment, 5′ primer (CTCGGATCCATAATCATGGGC) and 3′ primer (CTTTGCTCCAGGTGGGCGGCTTTCTTTATC); C-terminal fragment, 5′ primer (GGAGCAAAGGAGTTTGCTGCT) and 3′ primer (CATGCTCGAGGTTCCTGAAGC). The PCR primers for creating deletion 3 were the following: N-terminal fragment, 5′ primer (CTCGGATCCATAATCATGGGC) and 3′ primer (CAATAGATGGTGTGCAGCCCACAACCG); C-terminal fragment, 5′ primer (CATCTATTGCTGGCACCATCTGAC) and 3′ primer (CATGCTCGAGGTTCCTGAAGC). The PCR primers for creating deletion 4 were the following: 5′ primer (CTCGGATCCATAATCATGGGC) and 3′ primer (TGCATGCTCGAGTCAGTCAGATGGTGCCAGCAATAGATG). The PCR primers for creating deletion 5 were 5′ primer (CTCGGATCCATAATCATGGGC) and 3′ primer (TGCATGCTCGAGTCAGTCAGATGGTGCCAGCAATAGATG). The resulting products were inserted into BamHI and XhoI sites of the pcDNA3.1(+) plasmid (Invitrogen, Carlsbad, CA).
Cell cycle analysis.
To prepare cells for fluorescence-activated cell sorter analysis, MCF-7 cells were transfected with empty vector and constructs containing EZH2 and EZH2 siRNA. Forty-eight hours after transfection, the cells were treated with ICI 182 780 for 12 h and were then switched to fresh medium and treated with E2 for different periods of time. The cells were then fixed in 70% ethanol overnight at −20°C. After washing with 1% bovine serum albumin-PBS, cells were incubated with 10 μl (10 mg/ml) of RNase A (Sigma) in PBS-0.25% Triton X-100 for 30 min at room temperature and then stained with 50 mg/ml propidium iodide in 50 mM sodium citrate for 20 min at room temperature in the dark. Cell cycle data were collected with a FACScan (BD Bio-Sciences Immunocytochemistry Systems) and analyzed with CELLQUEST software.
RESULTS
EZH2 enhanced the transactivation of c-Myc and cyclin D1 by estrogen and Wnt signaling pathways.
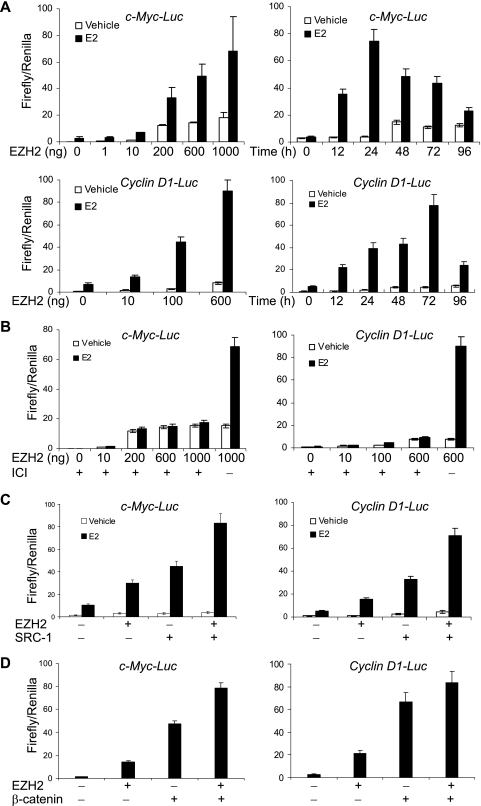
The role of estrogen in breast cancer carcinogenesis is well established (42). As stated above, recent investigations have found that EZH2 is also implicated in promoting the carcinogenesis and aggression of breast cancer (3, 20). In order to investigate whether there is any connection between estrogen signaling and EZH2 activity in breast cancer cells, we first examined what, if any, effects of EZH2 on the transactivation of ER target genes such as c-Myc (10) and cyclin D1 (32), both of which are known to play a crucial role in regulation of cell proliferation. For this purpose, we cotransfected the ER+ breast cancer cell line MCF-7 with a c-Myc promoter-driven luciferase reporter (c-Myc-Luc) or a cyclin D1 promoter-driven luciferase reporter (cyclin D1-Luc) and an EZH2 mammalian expression construct. The cells were then grown in the absence or presence of E2 for different periods of time, and the luciferase activity was measured. As shown in Fig. 1, in contrary to its established role as a transcription repressor, EZH2 drastically enhanced the expression of the reporter genes driven by the c-Myc promoter (Fig. 1A, upper panel) and by the cyclin D1 promoter (Fig. 1A, lower panel), and this effect was time dependent and EZH2 dose dependent. Furthermore, the enhancement activity of EZH2 is dependent on ERs, as the addition of the ER antagonist ICI 182 780 almost abolished the reporter activation (Fig. 1B). Similar results were obtained in another ER+ breast cancer line, T47-D, but not in an ER− breast cancer cell line, MDA-MA 231 (data not shown), further supporting the dependence on ERs of EZH2 in facilitating estrogen downstream gene transactivation.
FIG. 1.
EZH2 enhanced the transactivation of c-Myc and cyclin D1 promoters. (A) EZH2 dose- and time-dependent response of c-Myc (upper panel) and cyclin D1 (lower panel) promoters. MCF-7 cells were grown in the absence of estrogen and were cotransfected with c-Myc-Luc or cyclin D1-Luc reporter, a Renilla construct, along with different amounts of an EZH2 expression construct. Eighteen hours after transfection, cells were treated with 100 nM E2 or ethanol (vehicle) for different times and harvested for a luciferase activity assay. Each bar represents the mean ± standard deviation (SD) for triplicate experiments. (B) EZH2 potentiation of c-Myc and cyclin D1 transactivation was dependent on estrogen receptors. MCF-7 cells were grown in the absence of estrogen and were cotransfected with c-Myc-Luc or cyclin D1-Luc reporter along with different amounts of EZH2 expression construct. Eighteen hours after transfection, cells were treated with 100 nM E2, 1 mM ICI 182 780, or ethanol (vehicle) for different times and harvested for a luciferase activity assay. Each bar represents the mean ± SD for triplicate experiments. (C) EZH2 enhanced the transactivation of c-Myc and cyclin D1 promoters by SRC-1. MCF-7 cells were grown in the absence of estrogen and were cotransfected with c-Myc-Luc or cyclin D1-Luc reporter along with the EZH2 expression construct and a SRC-1 expression construct. Eighteen hours after transfection, cells were treated with 100 nM E2 or ethanol (vehicle) for 24 h and harvested for a luciferase activity assay. Each bar represents the mean ± SD for triplicate experiments. (D) EZH2 enhanced the transactivation of c-Myc and cyclin D1 promoters by β-catenin. MCF-7 cells were grown in the absence of estrogen and were cotransfected with c-Myc-Luc or cyclin D1-Luc reporter along with the EZH2 expression construct and a β-catenin expression construct. Forty-eight hours after transfection, cells were treated with E2 and harvested for a luciferase activity assay. Each bar represents the mean ± SD for triplicate experiments.
ERs regulate gene transcription through recruitment of coactivators that function to remodel the chromatin structure (28, 31, 42). Previously we showed that the p160 family of coactivators are both necessary and sufficient in transducing ER signaling (44). Therefore, it is reasonable to believe that EZH2 and the p160 proteins have an additive or synergistic enhancement effect on ER-regulated gene transcription. Indeed, we observed that cotransfection of EZH2 and SRC-1 induced a higher level of c-Myc-Luc or cyclin D1-Luc reporter activity than transfection of either EZH2 or SRC-1 alone in MCF-7 cells under the treatment of estrogen (Fig. 1C), further supporting a connection between EZH2 and estrogen signaling in gene activation.
Since both c-Myc and cyclin D1 are known targets for Wnt pathways (16, 47, 50), we next investigated the effect of EZH2 on the transactivation of c-Myc and cyclin D1 induced by the Wnt pathway. The EZH2 expression vector and a β-catenin expression vector were either transfected alone or in combination to MCF-7 cells, and the luciferase activation driven by the c-Myc or cyclin D1 promoter was assayed. As shown in Fig. 1D, EZH2 and β-catenin appeared to have an additive effect on the activation of the c-Myc and cyclin D1 promoters, suggesting that there might also be a connection between EZH2 activity and the Wnt pathway in gene activation.
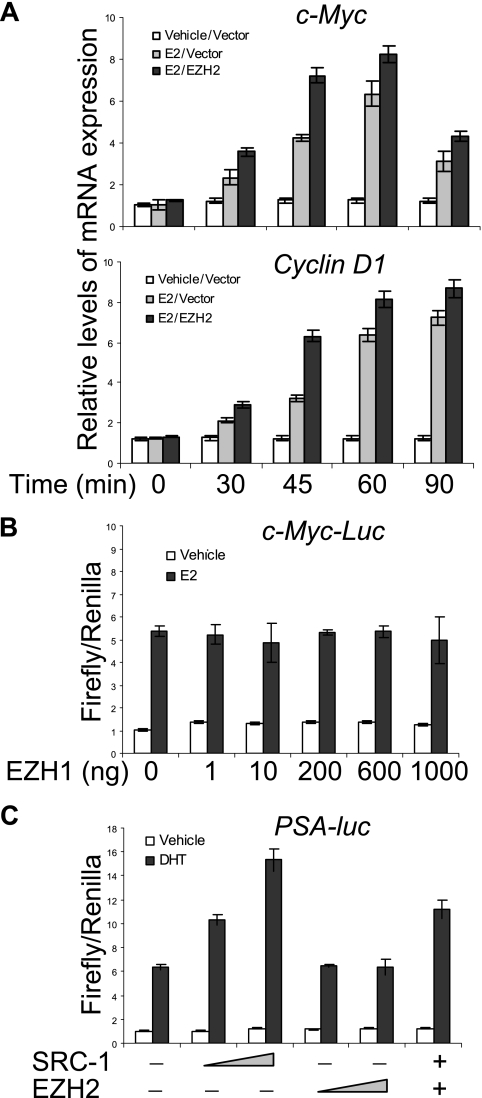
Measurement of the transcription of endogenous c-Myc and cyclin D1 by real-time RT-PCR supported the observation described above (Fig. 2A). In addition, the effect of EZH2 on gene transactivation appeared to be EZH2 specific, as its homolog EZH1 did not have such an effect (Fig. 2B), and the effect of EZH2 also appeared to be specific for ER, as EZH2 had little effect on androgen receptor (AR)-regulated gene transcription in the prostate cancer cell line LnCAP (Fig. 2C).
FIG. 2.
Transcriptional activation of endogenous genes by and functional specificity of EZH2. (A) EZH2 enhanced estrogen-stimulated mRNA expression of c-Myc and cyclin D1. MCF-7 cells were transfected with an empty vector or with an EZH2 expression plasmid. Sixty hours after transfection, the cells were switched to estrogen-deprived medium for 48 h. The cells were then left untreated or treated with 100 nM E2 for different times, as indicated, before the cells were collected for RNA extraction and for gene expression analysis by real-time RT-PCR. (B) EZH1 did not affect the transactivation of c-Myc promoters. MCF-7 cells were grown in the absence of estrogen and were cotransfected with c-Myc-Luc, a Renilla construct, along with different amounts of an EZH1 expression construct. Eighteen hours after transfection, cells were treated with E2 or ethanol (vehicle) for different times and harvested for a luciferase activity assay. Each bar represents the mean ± standard deviation (SD) for triplicate experiments. (C) EZH2 did not affect androgen receptor-regulated gene transcription. LnCAP cells were transfected with a PSA-luc (prostate-specific antigen-luciferase) construct together with 100 ng or 500 ng of SRC-1 or 100 ng or 500 ng of EZH2 expression constructs as indicated. After growing the cells in the absence of steroids for 48 h, cells were treated with 100 nM dihydrotestosterone (DHT) for 12 h, and the reporter activity was measured. Each bar represents the mean ± SD for triplicate experiments.
EZH2 transactivated c-Myc and cyclin D1 promoters.
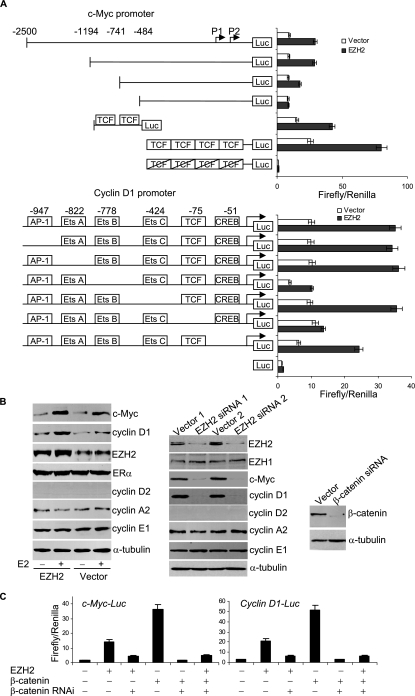
To further support an EZH2 role in the activation of c-Myc and cyclin D1 promoters, we next mapped the cis-elements within the c-Myc and cyclin D1 promoters that are responsible for EZH2 transactivation. The c-Myc promoter and its mutant constructs, a synthetic sequence containing four copies of the TCF-binding site and its mutant (16), and the cyclin D1 promoter and its mutant constructs (50) (Fig. 2A) were transfected into MCF-7 cells with or without coexpression of EZH2. The reporter activity was then measured under these experimental conditions. As shown in Fig. 3A, mutation of TCF-binding sites in either the c-Myc promoter or cyclin D1 promoter abolished EZH2 transactivation activity. In addition, EZH2 also strongly transactivated the reporter gene driven by the synthetic TCF sequence, and mutation of TCF-binding sites abrogated the EZH2 transactivation activity. These experiments clearly demonstrated that the transactivation activity of EZH2 is mediated by the TCF/β-catenin-binding sites in the c-Myc and cyclin D1 promoters, further supporting the observation that EZH2 enhanced β-catenin-regulated transcription of c-Myc and cyclin D1.
FIG. 3.
EZH2 transactivated c-Myc and cyclin D1 expression. (A) Map of cis-acting elements within c-Myc and cyclin D1 promoters which are responsible for EZH2 transactivation. The schematic represents the c-Myc promoter and cyclin D1 promoter, their deletion mutants, synthetic TCF binding sequence (GCTTTGATC), and mutated TCF binding sequence that were fused to a luciferase (Luc) reporter, shown on the left. MCF-7 cells were transfected with the indicated reporter plasmids with or without cotransfection of an EZH2 expression construct in the presence of estrogen. Each bar represents the mean ± standard deviation (SD) for triplicate experiments. (B) Regulation of the expression of endogenous c-Myc and cyclin D1 by EZH2. Left panel: enhancement of c-Myc and cyclin D1 expression by EZH2 overexpression. MCF-7 cells were grown in the absence of estrogen and transfected with the EZH2 expression construct or empty vector. Eighteen hours after transfection, the cells were treated with 100 nM of E2 for another 6 h, and the total proteins were extracted and examined by Western blotting analysis using antibodies against the indicated proteins. Middle panel: abolishment of c-Myc and cyclin D1 expression under EZH2 expression knockdown by RNAi. MCF-7 cells were grown in the presence of estrogen and transfected with the pSUPER-EZH2 siRNA (EZH2 siRNA 1) or pSILENCER-EZH2siRNA (EZH2 siRNA 2) construct. Forty-eight hours after transfection, total proteins were extracted and analyzed for the expression of the indicated proteins by Western blotting. Right panel: confirmation of β-catenin knockdown by RNAi in experiments described below. (C) Knockdown of expression of β-catenin by RNAi affected the transactivation of c-Myc and cyclin D1 promoters by both β-catenin and EZH2. MCF-7 cells were grown in the absence of estrogen and were cotransfected with c-Myc-Luc or cyclin D1-Luc reporter, a Renilla construct, along with the EZH2 expression construct, the β-catenin expression construct, or a β-catenin siRNA construct. Twenty-four hours after transfection, cells were collected for a luciferase activity assay. Each bar represents the mean ± SD for triplicate experiments.
Next, we tested the effect of EZH2 on endogenous c-Myc and cyclin D1 protein expression. As shown in Fig. 3B, overexpression of EZH2 in MCF-7 cells resulted in increased c-Myc and cyclin D1 protein expression in the presence but not in the absence of estrogen treatment (Fig. 3B, left panel). In the meanwhile, RNAi was applied to knock down the expression of EZH2 protein, and the effect of the knockdown on the c-Myc and cyclin D1 protein levels was measured by Western blotting. The results indicated that when the expression of EZH2 was silenced, the protein expression of c-Myc and cyclin D1 was significantly decreased, whereas either overexpression or knockdown of EZH2 had limited effects on the expression of other cell cycle-regulated genes, such as cyclin D2 (which is undetectable in MCF-7 cells due to promoter methylation) (9), cyclin A2, and cyclin E1 (Fig. 3B, middle panel).
To further consolidate the connection of EZH2 with the Wnt pathway in transactivation of c-Myc and cyclin D1 expression, we next investigated the effect of β-catenin knockdown by RNAi (Fig. 3B, right panel) on the transactivation of c-Myc and cyclin D1 by EZH2. As stated before, either EZH2 or β-catenin enhanced the promoter activity of c-Myc and cyclin D1, and EZH2 and β-catenin in combination had an additive effect (Fig. 1D). However, in β-catenin-silenced cells, not only was the β-catenin-enhanced activation of the c-Myc and cyclin D1 promoters diminished and the additive effect of EZH2 and β-catenin no longer observed, but also the enhancement effect of EZH2 was severely affected (Fig. 3C), suggesting that EZH2 might be involved in the Wnt pathway.
Molecular mechanism for transactivation activity of EZH2.
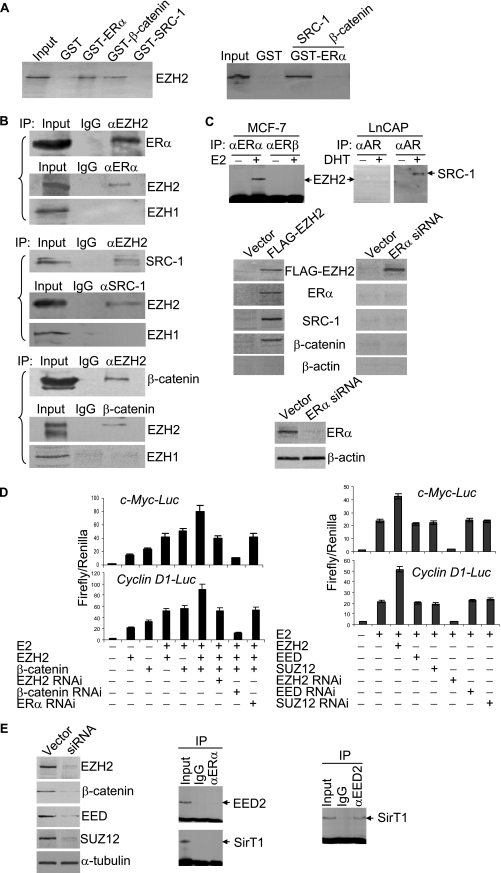
We next investigated the molecular mechanism underlying transcription potentiation of c-Myc and cyclin D1 by EZH2 in response to estrogen and Wnt signaling. Estrogen is believed to initiate its signaling pathway mainly through its binding to ERs (31, 42). Liganded ERs then bind to their cognate gene promoters, where coactivator recruitment occurs by means of protein-protein interaction (28, 31). Since ERα is the isoform that mediates the most current known estrogen biological activities, we wanted to test if there is a physical association between ERα and EZH2. For this purpose, GST pull-down experiments with bacterially expressed GST-ERα and in vitro-transcribed/translated EZH2 and reciprocal coimmunoprecipitation (co-IP) were performed. As shown in Fig. 4, GST pull-down experiments revealed a specific interaction between ERα and EZH2 (Fig. 4A, left panel). Reciprocal co-IP experiments also indicated that ERα and EZH2 interact in vivo (Fig. 4B, upper panels). Notably, co-IP experiments indicated that the interaction between ERα and EZH2 was estrogen dependent and ERα specific, as ERβ and AR did not interact with EZH2 in vivo (Fig. 4C, upper panels). In addition, while reciprocal co-IP experiments with antibodies against EZH2 and SRC-1 revealed that EZH2 and SRC-1 were present in the same protein complex (Fig. 4B, middle panels), no EZH2 and SRC-1 interaction was found in GST pull-down experiments (Fig. 4A, left panel), suggesting that there is not a direct physical interaction between EZH2 and SRC-1. On the other hand, EZH2 was found to interact with β-catenin both in vitro (Fig. 4A, left panel) and in vivo (Fig. 4B, bottom panels). These results strongly suggest that β-catenin-EZH2-ERα-SRC-1 was among the components of the transcription complex formed in activation of the c-Myc and cyclin D1 promoters.
FIG. 4.
EZH2 interacted with ERα and β-catenin in vitro and in vivo. (A) Equal amounts of 35S-labeled EZH2 (left panel) or SRC-1 or β-catenin (right panel) were used in GST pull-down experiments. Inputs represent 10% of fractions. (B) Coimmunoprecipitations for proteins that interacted with EZH2. MCF-7 cells were maintained in DMEM supplemented with 10% normal FBS for 72 h. Total proteins were then extracted, and coimmunoprecipitations (IP) were performed with antibodies against the indicated proteins. Inputs represent 10% of fractions. (C) Coimmunoprecipitations for proteins that interacted with EZH2. Upper panel: protein extracts from MCF-7 cells or LnCAP cells in the presence or absence of receptor ligands were immunoprecipitated with antibodies against ERs or AR. The immunoprecipitates were then immunoblotted with antibodies against EZH2 or SRC-1. Middle panel: FLAG-tagged EZH2 was expressed in MCF-7 cells and immunoprecipitated with anti-FLAG followed by immunoblotting with antibodies against the indicated proteins. Bottom panel: confirmation of ERα knockdown by Western blotting. (D) Effect of knockdown of expression of ERα, β-catenin, and EZH2 (left panel) or EED and SUZ12 (right panel) on the promoter activity of c-Myc and cyclin D1. MCF-7 cells were grown in the absence or presence of estrogen and were cotransfected with c-Myc-Luc or cyclin D1-Luc reporter, along with the indicated gene constructs. Forty-eight four hours after transfection, cells were collected for a luciferase activity assay. Each bar represents the mean ± standard deviation for triplicate experiments. (E) Confirmation of protein knockdown by Western blotting analysis (left panel) and coimmunoprecipitation with anti-ERα (middle panel) or anti-EED2 (right panel) antibody followed by immunoblotting with antibodies against the indicated proteins. Inputs represent 10% of fractions.
In order to further support an in vivo interaction of EZH2 with estrogen-Wnt signaling components, FLAG-tagged EZH2 was expressed in MCF-7 cells. Nuclear extracts were prepared from these cells and were incubated with FLAG antibody-conjugated Sepharaose 4B columns. The elutes were resolved on SDS-PAGE and were immunoblotted with antibodies against ERα, β-catenin, SRC-1, and β-actin. As shown in Fig. 4C (middle left panels), the protein complex that was pulled down with anti-FLAG antibody contained EZH2, ERα, β-catenin, and SRC-1, supporting the argument that these proteins exist in a functional protein complex in vivo. In addition, the formation of this complex was dependent on ERα, as knockdown of the expression of ERα (Fig. 4C, bottom panels) resulted in undetected ERα, β-catenin, and SRC-1 in immunoprecipitates pulled down by anti-FLAG (Fig. 4C, middle right panels).
ERs can bind the genes whose promoters do not contain any estrogen-responsive elements (31). Neither the c-Myc nor cyclin D1 promoter harbors canonical estrogen-responsive elements, but they do contain multiple TCF/β-catenin-binding sites (16, 47, 50). In view of a previous report that ERα coimmunoprecipitated with β-catenin (22), there is a strong possibility that ERα targets c-Myc and cyclin D1 through its interaction with TCF/β-catenin. However, a direct interaction between ERα and β-catenin was not detected by GST pull down in our experiments (Fig. 4A, right panel), and failure to detect a direct ERα-β-catenin interaction was also reported by other groups (57). In light of the observations that EZH2 directly interacts with ERα and β-catenin and EZH2 exerts its transactivation activity via TCF/β-catenin-binding sites (Fig. 3A), these experiments placed EZH2 in the intersection of the estrogen and Wnt pathways, at least in the transactivation of c-Myc and cyclin D1, suggesting EZH2 functions as an integrator of the Wnt pathway and estrogen signaling. This model was supported by our experiments showing that while knockdown of the expression of either EZH2 or ERα decreased the c-Myc and cyclin D1 promoter activity, β-catenin was essential for both estrogen and Wnt signaling in activation of the c-Myc and cyclin D1 promoter (Fig. 4D, left panels).
It is believed that EZH2 exerts its biological activities through formation of PRC protein complexes with other proteins, such as EED and SUZ12 (5). In order to investigate whether the transactivation activity of EZH2 and the association of EZH2 with ERα and β-catenin was a manifestation of EZH2 itself or of a PRC protein complex, we performed reciprocal coimmunoprecipitation with antibodies against EED, SUZ12, ERα, and β-catenin. The results of these experiments indicated that there were no physical associations between ERα and EED/SUZ12 and between β-catenin and EED/SUZ12 (data not shown). In addition, neither overexpression nor knockdown of the expression of EED or SUZ12 affected the transactivation activity of EZH2 (Fig. 4D, right panels). The results of RNAi experiments are shown in Fig. 4E (left panels). Furthermore, it has been reported that overexpression of EZH2 in tissue culture promotes formation of a previously undescribed PRC, PRC4, that contains histone deacetylase SirT1 and isoform 2 of the PRC component Eed (EED2) (24). However, neither EED2 nor SirT1 was detected in coimmunoprecipitation experiments with antibodies against ERα (Fig. 4E, middle panels), whereas SirT1 was detected in coimmunoprecipitations with anti-EED2 (Fig. 4E, right panel). Collectively, these results indicated that the transactivation activity of EZH2 was not from a PRC protein complex.
Recruitment of EZH2 on the promoters of c-Myc and cyclin D1.
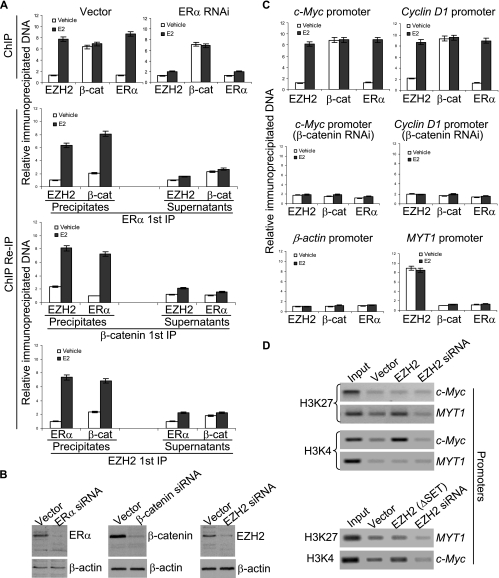
In order to determine whether EZH2 is indeed associated with c-Myc and cyclin D1 promoters and to further investigate the association of β-catenin-EZH2-ERα on c-Myc and cyclin D1 promoters in vivo, we next performed ChIP and re-ChIP according to the procedure described previously (44, 60) by quantitative PCR to examine the recruitment pattern of β-catenin, EZH2, and ERα on c-Myc and cyclin D1 promoters. In these experiments, MCF-7 cells were grown in the absence of estrogen for at least 3 days followed by no treatment or treatment with saturating levels of E2 for 45 min. The presence of β-catenin, EZH2, and ERα on c-Myc and cyclin D1 promoters was first determined using ChIP with antibodies against these proteins. Then, both the precipitates and the supernatants were subjected to re-ChIP. As shown in Fig. 5, the presence of all three, β-catenin, EZH2, and ERα, could be detected on the c-Myc promoter under the experimental conditions and was dependent on ERα (Fig. 5A, upper panels). As we reported earlier for p160 proteins (60), this could be explained by one of the following scenarios: each of β-catenin, EZH2, and ERα could be associated with a different promoter in different cells; pairs of three proteins could be associated with a different promoter in different cells; or the three proteins could exist in the same complex. The re-ChIP experiments demonstrated that, in precipitates, the c-Myc promoter that was immunoprecipitated with antibodies against ERα could be reimmunoprecipitated with antibodies against β-catenin and EZH2; likewise, the c-Myc promoter that was immunoprecipitated with antibodies against β-catenin could be reimmunoprecipitated with antibodies against ERα and EZH2; analogously the c-Myc promoter that was immunoprecipitated with antibodies against EZH2 could be reimmunoprecipitated with antibodies against ERα and β-catenin (Fig. 5A, lower three panels). On the other hand, supernatants from ChIP with antibodies against β-catenin, EZH2, or ERα showed no detection of the other two proteins on the c-Myc promoter (Fig. 5A, lower three panels). Similar results were obtained with the cyclin D1 promoter and in T47-D cells, and there was no detectable EED or SUZ12 on c-Myc and cyclin D1 promoters under the experimental conditions (data not shown). The binding of ERα, β-catenin, and EZH2 on the promoter of cyclin D1 was also confirmed by real-time quantitative PCR in which the associations of EZH2 with the promoter of one of its known target genes, myelin transcription factor 1 (MYT1) (19) (positive control), and with β-actin (negative control) were validated (Fig. 5C). These experiments strongly suggest that β-catenin-EZH2-ERα existed in the same protein complex on c-Myc and cyclin D1 promoters in vivo.
FIG. 5.
(A) Recruitment of ERα, β-catenin, and EZH2 on c-Myc promoters. MCF-7 cells were grown in the absence of estrogen for at least 3 days and left untreated or treated with 100 nM of E2 for 45 min. ChIP assays and ChIP/re-IP experiments were performed using specific antibodies against ERα, β-catenin, and EZH2 by real-time quantitative PCR analysis. The results are means ± standard deviations (SD) from three independent experiments. (B) Confirmation of the silenced expression of the indicated proteins by Western blotting. (C) Recruitment of ERα, β-catenin, and EZH2 on the indicated gene promoters in MCF-7 cells with (middle panel) or without (upper panel) β-catenin knockdown by real-time quantitative PCR analysis. The results are means ± SD from three independent experiments. (C) ChIP analysis of the methylation status of lysine 27 (K27) and lysine 4 (K4) of histone H3 under the indicated experimental conditions.
It is believed that the normal function of EZH2 in transcription repression is associated with the methylation of histone H3 lysine 27 (H3K27) (4, 25). In order to further support the transactivation activity of EZH2, the methylation status of H3K27 around the c-Myc promoter was examined by ChIP assays. The results indicated that neither overexpression nor knockdown of EZH2 affected the methylation status of H3K27 (Fig. 5D). In contrast, the overexpression of EZH2 was associated with the methylation of H3K4, a well-characterized gene activation mark (30), further supporting EZH2's role in gene activation. The knockdown of protein expression was confirmed by Western blotting (Fig. 5B).
The transcription activation function of EZH2 is separated from its SET domain.
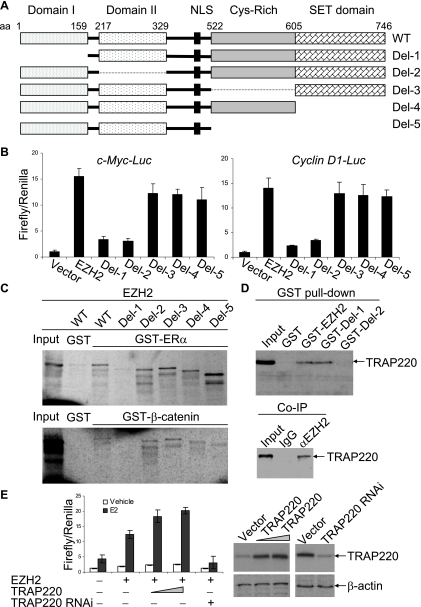
Genetic evolution analysis revealed four conserved domains in human EZH1, human EZH2, mouse EZH1, mouse EZH2, and Drosophila E(Z) (17, 26, 33). The SET domain in the carboxy terminal is the most highly conserved domain, is required for the HMTase activity of several SET-containing proteins, and is believed to mediate the transcription repression activity of EZH2 (5). A cysteine-rich region precedes the SET domain. In the amino-terminal region, EZH2 harbors domain I and domain II with proposed protein-protein interaction functions. In order to further delineate the molecular basis for the EZH2 role in connecting the estrogen and Wnt signaling, we generated EZH2 mutants in which the SET, cysteine-rich region, domain I, or domain II was individually or combinatorially deleted (Fig. 6A). The effects of these mutants on the transactivation of estrogen-induced c-Myc and cyclin D1 promoters were analyzed. As shown in Fig. 6B, while deletion of the SET or the cysteine-rich domain either alone or in combination had minimal effect on the promoter activity of c-Myc and cyclin D1, deletion of either domain I or domain II rendered a diminished effect of EZH2 on c-Myc and cyclin D1 promoter activation, suggesting that both domain I and domain II are essential for the transactivation activity of EZH2.
FIG. 6.
The functional domains of EZH2 that are involved in transactivation of the c-Myc and cyclin D1 promoters. (A) Schematic diagrams of EZH2 deletion mutants. (B) Transactivation activity of EZH2 deletion mutants on the c-Myc or cyclin D1 promoter. MCF-7 cells were grown in DMEM supplemented with 10% normal FBS and were cotransfected with c-Myc-Luc or cyclin D1-Luc reporter, a Renilla construct, along with the EZH2 deletion constructs. Eighteen hours after transfection, cells were collected for a luciferase activity assay. Each bar represents the mean ± standard deviation (SD) for triplicate experiments. (C) GST pull-down experiments for interaction between ERα (upper panel) or β-catenin (lower panel) and the EZH2 deletion mutants. (D) Direct interaction between EZH2 and TRAP220/DRIP205 as detected by GST pull-down experiments (upper panel) and by coimmunoprecipitation with primary antibodies against EZH2 and then blotting with antibodies against TRAP220/DRIP205 (lower panel). Inputs represent 10% of fractions. (E) TRAP220/DRIP205 potentiated EZH2-enhanced transactivation of the c-Myc promoter. MCF-7 cells were grown in the absence of estrogen and were cotransfected with c-Myc-Luc reporter along with EZH2 or a TRAP220/DRIP205 expression construct or with a TRAP220/DRIP205 siRNA construct. Eighteen hours after transfection, cells were treated with 100 nM E2 or ethanol (vehicle) for 16 h and harvested for a luciferase activity assay. Each bar represents the mean ± SD for triplicate experiments. Protein expression was confirmed by Western blotting (right panel).
Because we demonstrated that EZH2 directly interacted with ERα and β-catenin (Fig. 4A and B), we next analyzed the functional domain(s) in EZH2 that may be involved in these interactions by GST pull-down experiments using the above-described EZH2 mutants. As shown in Fig. 6C, while the deletions of the cysteine-rich domain or SET domain still preserved the interaction ability of EZH2 with ERα and β-catenin, deletion of domain I abolished the interaction of EZH2 with both ERα and β-catenin, indicating that both ERα and β-catenin interact directly with EZH2 through domain I of EZH2.
Because domain II of EZH2 was also required for EZH2 transactivation activity (Fig. 6B) and this structure is also proposed for a protein-protein interaction function (17, 26, 33), we next screened for, by GST pull-down assays, a possible transcription cofactor(s) that has been implicated in ER and/or Wnt signaling-mediated transcription and that might directly interact with domain II of EZH2. Since we had already shown that SRC-1 did not interact directly with EZH2 (Fig. 4A), we thus selected histone acetyltransferases CBP, p300, and pCAF, histone methyltransferases CARM1 and PRMT1, human SWI/SNF components Brg1, BAF250, and BAF170, and human Mediator components TRAP220/DRIP205 and TRAP150/DRIP150 to test, individually, whether these transcription regulators could interact with EZH2. Among these proteins, our experiments detected that only TRAP220/DRIP205 directly interacted with EZH2, and we showed that this interaction was dependent on domain II of EZH2 (Fig. 6D, upper panel). In vivo interaction of EZH2 with TRAP220/DRIP205 was also confirmed in MCF-7 cells by coimmunoprecipitation assays (Fig. 6D, lower panel). In addition, we showed that TRAP220/DRIP205 was indeed able to enhance the transactivation activity of EZH2 on the c-Myc promoter in MCF-7 cells (Fig. 6E).
Collectively, these experiments clearly established domain I as the interaction domain for both ERα and β-catenin and suggested that domain II serves as a platform for the Mediator interaction. These data also suggested that the transactivation activity and transrepression activity of EZH2 reside in separated domains and thus provided a mechanism for EZH2's dual function in transcription activation and repression.
Promotion of cell cycle progression by EZH2.
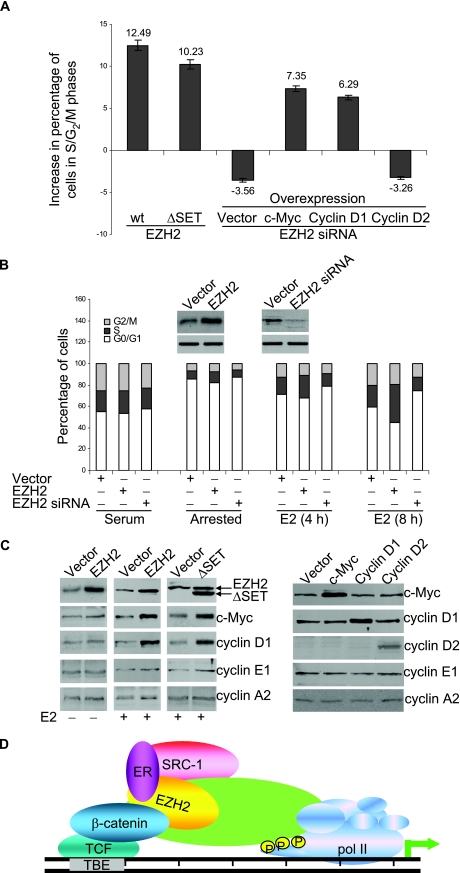
In order to determine whether the integration function of EZH2 for gene activation mediated by the estrogen and Wnt pathways extends to a physiologically relevant response in breast cancer cells, we examined the effect of EZH2 on cell cycle progression. Estrogen is normally required for the G1/S transition of MCF-7 cells, and estrogen deprivation leads to a significant G1 arrest (9). We cotransfected estrogen-deprived MCF-7 cells with EZH2 or an EZH2 siRNA together with a GFP construct. The cell cycle profile of the GFP-expressing population was then analyzed by flow cytometry after estrogen stimulation. As shown in Fig. 7A, overexpression of EZH2 led to a significant increase of cells in the S phase and G2/M phase, whereas knockdown of the expression of EZH2 with siRNA resulted in accumulation of cells in the G0/G1 phases. Promotion of cell cycle progression was also observed in cells transfected with the SET domain-deleted EZH2. Moreover, the inhibitory effect of EZH2 knockdown on the G1/S transition of the cell cycle could be partially rescued by overexpression of c-Myc or cyclin D1 but not cyclin D2.
FIG. 7.
(A) EZH2 promotes cell cycle progression. Serum-starved MCF-7 cells were transfected with an empty vector or the wild-type (wt) or SET domain-deleted (ΔSET) EZH2 expression construct or an EZH2 siRNA construct, plus c-Myc, cyclin D1, or cyclin D2 expression constructs. Twenty-four hours after transfection, cells were treated with estrogen for another 16 h and collected for cell cycle profile analysis by cell flow cytometry. The percentages of cells in G1, S, and G2/M phases are shown. (B) EZH2 promoted the G1/S transition of MCF-7 cells. MCF-7 cells were transfected with an EZH2 expression construct or an EZH2 siRNA construct. Twenty-four hours after transfection, cells were switched to normal medium (serum) or treated with 1 mM ICI 182 780 (arrested) for 24 h before adding E2 for the indicated times. The cells were then collected for cell cycle profile analysis by cell flow cytometry. The percentages of cells in G1, S, and G2/M phases under different experimental conditions are shown. (C) Western blotting analysis of protein expression under the indicated experimental conditions. (D) Model for EZH2 involvement in transactivation. See the text for details.
To further support the role of EZH2 in promoting cell cycle progression, MCF-7 cells were transfected with an EZH2 expression construct or an EZH2 siRNA construct. Twenty-four hours after transfection, cells were switched to normal medium or treated with 1 mM of ICI 182 780 for 24 h before addition of E2 for different times. The cells were then collected for cell cycle profile analysis by cell flow cytometry. The results indicated that EZH2 was able to facilitate the E2-stimulated G1/S transition of MCF-7 cells (Fig. 7B). The expression of EZH2, c-Myc, cyclin D1, and other cell cycle-related genes is shown in Fig. 7C. These experiments indicated that EZH2 promotes the G1/S transition of MCF-7 cells through its transactivation of c-Myc and cyclin D1 in these cells, and the effect of EZH2 on cell cycle progression is independent of its SET domain.
DISCUSSION
There have been intensive studies concerning the oncogenic mechanism underlying EZH2 overexpression. Since EZH2 is associated with transcription repression, silencing the expression of genes that negatively regulate cell proliferation by EZH2 overexpression is a plausible possibility. However, transcriptional profiling in cell culture systems identified numerous solute transporters among the downregulated genes, but otherwise no strong functional themes emerged (52). Alternatively, it has recently been noted that overexpression of EZH2 in tissue culture promotes formation of a previously undescribed PRC, PRC4, that contains the NAD+-dependent histone deacetylase SirT1 and isoform 2 of the PRC component EED (24). EED2 is expressed in cancer and undifferentiated embryonic stem cells but is undetectable in normal and differentiated embryonic stem cells. The distinct PRCs have been shown to exhibit differential histone substrate specificities, and the formation of a transformation-specific PRC may have a major role in resetting patterns of gene expression by regulating chromatin structure (24). However, in our current experiments, we did not detect the association of EZH2 with either SirT1 or EED2. Nevertheless, the observation that overexpression of EZH2 in tissue culture promotes formation of a novel PRC is at least an indication for a dynamic biochemistry of EZH2 in cells in supporting its dynamic and diverse functions. Functional protein complexes are constituted by numerous protein subunits and are formed through evolution by a restricted stoichiometry. A balanced expression and relative abundance of the different subunits must be maintained in order for physiologically functional complexes to be formed. Once this balanced is skewed, either the formation of the normal protein complex is in jeopardy or the increased enrichment of a protein subunit could functionally go awry. It is intriguing to speculate that EZH2 overexpression renders EZH2 participation in biological processes which otherwise do not involve EZH2, and this unsolicited participation, in some cases, could be consequently detrimental to the cell. It is also worth noting that in addition to EZH2, overexpression of other polycomb proteins, such as BMI1, MEL18, CBX, PHC1, RING1, SCML1/2 L3MBTL, EZH1, and SUZ12, have also been implicated in tumorigenesis (37). Although the mechanisms for these proteins' involvement in tumorigenesis may or may not necessarily be similar to the mechanism for EZH2, the maintenance of a balanced expression of polycomb proteins is underscored.
The central involvement of estrogen in the development and carcinogenesis of reproductive organs is well established (1, 8, 9, 42, 48, 55). However, the understanding of the genomic mechanisms of estrogen action continues to evolve (14, 42, 56, 59). Estrogen receptors can function both as transcription factors and transcription cofactors, and the number of proteins capable of coactivating estrogen receptor-mediated gene transcription continues to grow. In addition, recent reports indicated that proteins relevant to cell cycle promotion are posttranslationally stabilized in response to estrogen (39, 40). These mechanisms can be potentially exploited selectively to amplify tissue-specific responses to estrogen. Alternatively, survival pathways in cancer could evolve to alter the entire responsiveness to ER signaling. Over the years, studies of the estrogenic regulation of molecules with known roles in the control of G1/S-phase progression have resulted in significant advances in understanding the links between estrogen action and the cell cycle machinery. Estrogen regulates the expression and function of c-Myc and cyclin D1, and the induction of either c-Myc or cyclin D1 is sufficient to recapitulate the effects of estrogen on cell cycle progression (9, 39, 40). Based on the current understanding of estrogen action and the data of our current experiments, it is reasonable to believe that, when it is overexpressed, the transcription coregulator EZH2 is recruited to the estrogen signaling pathway to function as a coactivator and to amplify estrogen signaling.
Analogously, β-catenin together with TCF/LEF has been recognized to act as a switch to determine cell fate and promote cell survival and proliferation at several stages during mammary gland development (15). Each of the β-catenin-induced phenotypes is accompanied by upregulation of the target genes cyclin D1 and c-Myc (15). Potential cross talk between estrogen and Wnt signaling in vivo has been implicated in physiological studies on tissues as different as brain (6), neurons (36), uterus (18), and transgenic Drosophila (22). While these and other observations suggested the possibility of functional interaction between ER and β-catenin, a molecular basis for such an interaction is lacking. We demonstrated in this report that EZH2 directly interacted with both ERα and β-catenin with its domain I, thus acting as a bridge to link β-catenin and ERα on the c-Myc and cyclin D1 promoters (Fig. 7D). In addition, we demonstrated that EZH2 functions in transactivation of c-Myc and cyclin D1 in TCF/LEF sites. A recent report indicated direct ERα regulation via a novel cell-type-specific enhancer downstream of the cyclin D1-coding region (11). Whether these cis-acting elements act cooperatively or specifically in different cell types or in response to different environmental cues needs to be addressed in future studies.
The question is what, if any, biochemical activity of EZH2 is responsible for its transcription activation activity. The best-characterized domain in EZH2, the SET domain, is believed to be associated with HMTases that are implicated in the transrepression function of EZH2 and is not required for the EZH2 transactivation function. A less-well-characterized domain in EZH2, the cysteine-rich domain, is also dispensable for EZH2 transactivation function. Our experiments demonstrated that EZH2 interacts with the Mediator complex through its domain II. The mammalian Mediator complex is essential for tight regulation of transcription by RNA polymerase II through its interaction with RNA polymerase II (21, 38). Thus, it is reasonable to suggest that EZH2 might enhance transcription through its interaction with the Mediator complex. However, our experiments do not exclude other biochemical activities that are intrinsic to EZH2 and that may be responsible for the transactivation function of EZH2. In addition, what are other genes whose transactivation involves EZH2 and how EZH2 participates in the activation of these genes? Also relevant to our study is the question of what is the genetic/molecular basis for EZH2 overexpression and what are other molecular effectors for EZH2-mediated carcinogenesis? These questions need to be addressed in future studies through molecular and genetic approaches, including a transgenic model.
Acknowledgments
We thank Joanne Balmer Green (Penn State University) for editorial assistance.
This work was supported by grants (30621002, 30393113, and 30470912 to Y.S.) from the National Natural Science Foundation of China and grants (863 Program 2006AA02Z466 and 973 Program 2005CB522404 to Y.S.) from the Ministry of Science and Technology of China.
Footnotes
Published ahead of print on 14 May 2007.
REFERENCES
- 1.Acconcia, F., B. Manavathi, J. Mascarenhas, A. H. Talukder, G. Mills, and R. Kumar. 2006. An inherent role of integrin-linked kinase-estrogen receptor alpha interaction in cell migration. Cancer Res. 66:11030-11038. [DOI] [PubMed] [Google Scholar]
- 2.Attwooll, C., S. Oddi, P. Cartwright, E. Prosperini, K. Agger, P. Steensgaard, C. Wagener, C. Sardet, M. C. Moroni, and K. Helin. 2005. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J. Biol. Chem. 280:1199-1208. [DOI] [PubMed] [Google Scholar]
- 3.Bracken, A. P., D. Pasini, M. Capra, E. Prosperini, E. Colli, and K. Helin. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22:5323-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 5.Cao, R., and Y. Zhang. 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14:155-164. [DOI] [PubMed] [Google Scholar]
- 6.Cardona-Gomez, P., M. Perez, J. Avila, L. M. Garcia-Segura, and F. Wandosell. 2004. Estradiol inhibits GSK3 and regulates interaction of estrogen receptors, GSK3, and beta-catenin in the hippocampus. Mol. Cell. Neurosci. 25:363-373. [DOI] [PubMed] [Google Scholar]
- 7.Dellino, G. I., Y. B. Schwartz, G. Farkas, D. McCabe, S. C. Elgin, and V. Pirrotta. 2004. Polycomb silencing blocks transcription initiation. Mol. Cell 13:887-893. [DOI] [PubMed] [Google Scholar]
- 8.den Hollander, P., and R. Kumar. 2006. Dynein light chain 1 contributes to cell cycle progression by increasing cyclin-dependent kinase 2 activity in estrogen-stimulated cells. Cancer Res. 66:5941-5949. [DOI] [PubMed] [Google Scholar]
- 9.Doisneau-Sixou, S. F., C. M. Sergio, J. S. Carroll, R. Hui, E. A. Musgrove, and R. L. Sutherland. 2003. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr. Relat. Cancer 10:179-186. [DOI] [PubMed] [Google Scholar]
- 10.Dubik, D., and R. Shiu. 1988. Transcriptional regulation of c-myc oncogene expression by estrogen in hormone-responsive human breast cancer cells. J. Biol. Chem. 263:12705-12708. [PubMed] [Google Scholar]
- 11.Eeckhoute, J., J. S. Carroll, T. R. Geistlinger, M. I. Torres-Arzayus, and M. Brown. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 20:2513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, E., C. Marcos-Gutierrez, M. del Mar Lorente, J. C. Moreno, and M. Vidal. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18:3404-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Cuellar, M. P., O. Zilles, S. A. Schreiner, M. Birke, T. H. Winkler, and R. K. Slany. 2001. The ENL moiety of the childhood leukemia-associated MLL-ENL oncoprotein recruits human Polycomb 3. Oncogene 20:411-419. [DOI] [PubMed] [Google Scholar]
- 14.Gururaj, A. E., S. K. Rayala, R. K. Vadlamudi, and R. Kumar. 2006. Novel mechanisms of resistance to endocrine therapy: genomic and nongenomic considerations. Clin. Cancer Res. 12:1001s-1007s. [DOI] [PubMed] [Google Scholar]
- 15.Hatsell, S., T. Rowlands, M. Hiremath, and P. Cowin. 2003. Beta-catenin and Tcfs in mammary development and cancer. J. Mammary Gland Biol. Neoplasia 8:145-158. [DOI] [PubMed] [Google Scholar]
- 16.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 17.Hobert, O., I. Sures, T. Ciossek, M. Fuchs, and A. Ullrich. 1996. Isolation and developmental expression analysis of Enx-1, a novel mouse Polycomb group gene. Mech. Dev. 55:171-184. [DOI] [PubMed] [Google Scholar]
- 18.Hou, X., Y. Tan, M. Li, S. K. Dey, and S. K. Das. 2004. Canonical Wnt signaling is critical to estrogen-mediated uterine growth. Mol. Endocrinol. 18:3035-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18:1592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleer, C. G., Q. Cao, S. Varambally, R. Shen, I. Ota, S. A. Tomlins, D. Ghosh, R. G. Sewalt, A. P. Otte, D. F. Hayes, M. S. Sabel, D. Livant, S. J. Weiss, M. A. Rubin, and A. M. Chinnaiyan. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 100:11606-11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30:235-239. [DOI] [PubMed] [Google Scholar]
- 22.Kouzmenko, A. P., K. Takeyama, S. Ito, T. Furutani, S. Sawatsubashi, A. Maki, E. Suzuki, Y. Kawasaki, T. Akiyama, T. Tabata, and S. Kato. 2004. Wnt/beta-catenin and estrogen signaling converge in vivo. J. Biol. Chem. 279:40255-40258. [DOI] [PubMed] [Google Scholar]
- 23.Kuzmichev, A., T. Jenuwein, P. Tempst, and D. Reinberg. 2004. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14:183-193. [DOI] [PubMed] [Google Scholar]
- 24.Kuzmichev, A., R. Margueron, A. Vaquero, T. S. Preissner, M. Scher, A. Kirmizis, X. Ouyang, N. Brockdorff, C. Abate-Shen, P. Farnham, and D. Reinberg. 2005. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc. Natl. Acad. Sci. USA 102:1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laible, G., A. Wolf, R. Dorn, G. Reuter, C. Nislow, A. Lebersorger, D. Popkin, L. Pillus, and T. Jenuwein. 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaJeunesse, D., and A. Shearn. 1995. Trans-regulation of thoracic homeotic selector genes of the Antennapedia and bithorax complexes by the trithorax group genes: absent, small, and homeotic discs 1 and 2. Mech. Dev. 53:123-139. [DOI] [PubMed] [Google Scholar]
- 28.Lemon, B. D., and L. P. Freedman. 1999. Nuclear receptor cofactors as chromatin remodelers. Curr. Opin. Genet. Dev. 9:499-504. [DOI] [PubMed] [Google Scholar]
- 29.Lund, A. H., and M. van Lohuizen. 2004. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16:239-246. [DOI] [PubMed] [Google Scholar]
- 30.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 31.McDonnell, D. P., and J. D. Norris. 2002. Connections and regulation of the human estrogen receptor. Science 296:1642-1644. [DOI] [PubMed] [Google Scholar]
- 32.Musgrove, E. A., and R. L. Sutherland. 1994. Cell cycle control by steroid hormones. Semin. Cancer Biol. 5:381-389. [PubMed] [Google Scholar]
- 33.O'Connell, S., L. Wang, S. Robert, C. A. Jones, R. Saint, and R. S. Jones. 2001. Polycomb-like PHD fingers mediate conserved interaction with enhancer of zeste protein. J. Biol. Chem. 276:43065-43073. [DOI] [PubMed] [Google Scholar]
- 34.Orlando, V. 2003. Polycomb, epigenomes, and control of cell identity. Cell 112:599-606. [DOI] [PubMed] [Google Scholar]
- 35.Otte, A. P., and T. H. Kwaks. 2003. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr. Opin. Genet. Dev. 13:448-454. [DOI] [PubMed] [Google Scholar]
- 36.Quintanilla, R. A., F. J. Munoz, M. J. Metcalfe, M. Hitschfeld, G. Olivares, J. A. Godoy, and N. C. Inestrosa. 2005. Trolox and 17β-estradiol protect against amyloid β-peptide neurotoxicity by a mechanism that involves modulation of the Wnt signaling pathway. J. Biol. Chem. 280:11615-11625. [DOI] [PubMed] [Google Scholar]
- 37.Raaphorst, F. M. 2005. Deregulated expression of Polycomb-group oncogenes in human malignant lymphomas and epithelial tumors. Hum. Mol. Genet. 14(Spec. 1):R93-R100. [DOI] [PubMed] [Google Scholar]
- 38.Rachez, C., and L. P. Freedman. 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13:274-280. [DOI] [PubMed] [Google Scholar]
- 39.Rodrik, V., E. Gomes, L. Hui, P. Rockwell, and D. A. Foster. 2006. Myc stabilization in response to estrogen and phospholipase D in MCF-7 breast cancer cells. FEBS Lett. 580:5647-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodrik, V., Y. Zheng, F. Harrow, Y. Chen, and D. A. Foster. 2005. Survival signals generated by estrogen and phospholipase D in MCF-7 breast cancer cells are dependent on Myc. Mol. Cell. Biol. 25:7917-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sewalt, R. G., M. J. Gunster, J. van der Vlag, D. P. Satijn, and A. P. Otte. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang, Y. 2006. Molecular mechanisms of oestrogen and SERMs in endometrial carcinogenesis. Nat. Rev. Cancer 6:360-368. [DOI] [PubMed] [Google Scholar]
- 43.Shang, Y., and M. Brown. 2002. Molecular determinants for the tissue specificity of SERMs. Science 295:2465-2468. [DOI] [PubMed] [Google Scholar]
- 44.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 45.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 46.Shearn, A. 1989. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics 121:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh, R. R., and R. Kumar. 2005. Steroid hormone receptor signaling in tumorigenesis. J. Cell Biochem. 96:490-505. [DOI] [PubMed] [Google Scholar]
- 49.Soulez, M., A. J. Saurin, P. S. Freemont, and J. C. Knight. 1999. SSX and the synovial-sarcoma-specific chimaeric protein SYT-SSX co-localize with the human Polycomb group complex. Oncogene 18:2739-2746. [DOI] [PubMed] [Google Scholar]
- 50.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 51.Trimarchi, J. M., B. Fairchild, J. Wen, and J. A. Lees. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98:1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varambally, S., S. M. Dhanasekaran, M. Zhou, T. R. Barrette, C. Kumar-Sinha, M. G. Sanda, D. Ghosh, K. J. Pienta, R. G. Sewalt, A. P. Otte, M. A. Rubin, and A. M. Chinnaiyan. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624-629. [DOI] [PubMed] [Google Scholar]
- 53.Visser, H. P., M. J. Gunster, H. C. Kluin-Nelemans, E. M. Manders, F. M. Raaphorst, C. J. Meijer, R. Willemze, and A. P. Otte. 2001. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br. J. Haematol. 112:950-958. [DOI] [PubMed] [Google Scholar]
- 54.Voncken, J. W., H. Niessen, B. Neufeld, U. Rennefahrt, V. Dahlmans, N. Kubben, B. Holzer, S. Ludwig, and U. R. Rapp. 2005. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J. Biol. Chem. 280:5178-5187. [DOI] [PubMed] [Google Scholar]
- 55.Wu, H., Y. Chen, J. Liang, B. Shi, G. Wu, Y. Zhang, D. Wang, R. Li, X. Yi, H. Zhang, L. Sun, and Y. Shang. 2005. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 438:981-987. [DOI] [PubMed] [Google Scholar]
- 56.Wu, H., L. Sun, Y. Zhang, Y. Chen, B. Shi, R. Li, Y. Wang, J. Liang, D. Fan, G. Wu, D. Wang, S. Li, and Y. Shang. 2006. Coordinated regulation of AIB1 transcriptional activity by sumoylation and phosphorylation. J. Biol. Chem. 281:21848-21856. [DOI] [PubMed] [Google Scholar]
- 57.Yang, F., X. Li, M. Sharma, C. Y. Sasaki, D. L. Longo, B. Lim, and Z. Sun. 2002. Linking beta-catenin to androgen-signaling pathway. J. Biol. Chem. 277:11336-11344. [DOI] [PubMed] [Google Scholar]
- 58.Yin, N., D. Wang, H. Zhang, X. Yi, X. Sun, B. Shi, H. Wu, G. Wu, X. Wang, and Y. Shang. 2004. Molecular mechanisms involved in the growth stimulation of breast cancer cells by leptin. Cancer Res. 64:5870-5875. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, H., L. Sun, J. Liang, W. Yu, Y. Zhang, Y. Wang, Y. Chen, R. Li, X. Sun, and Y. Shang. 2006. The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 25:4223-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, H., X. Yi, X. Sun, N. Yin, B. Shi, H. Wu, D. Wang, G. Wu, and Y. Shang. 2004. Differential gene regulation by the SRC family of coactivators. Genes Dev. 18:1753-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]