Abstract
The Myc oncoprotein family comprises transcription factors that control multiple cellular functions and are widely involved in oncogenesis. Here we report the identification of Myc-nick, a cytoplasmic form of Myc generated by calpain- dependent proteolysis at lysine 298 of full-length Myc. Myc-nick retains conserved Myc Box regions but lacks nuclear localization signals and the bHLHZ domain essential for heterodimerization with Max and DNA binding. Myc-nick induces α-tubulin acetylation and altered cell morphology by recruiting histone acetyltransferase GCN5 to microtubules. During muscle differentiation, while the levels of full-length Myc diminish, Myc-nick and acetylated α-tubulin levels are increased. Ectopic expression of Myc-nick accelerates myoblast fusion, triggers the expression of myogenic markers, and permits Myc deficient fibroblasts to transdifferentiate in response to MyoD. We propose that the cleavage of Myc by calpain abrogates the transcriptional inhibition of differentiation by full-length Myc and generates Myc-nick, a driver of cytoplasmic reorganization and differentiation.
Introduction
The Myc family (c-Myc, N-Myc, and L-Myc) of basic-helix-loop-helix-zipper (bHLHZ) transcription factors controls the expression of a large number of target genes and non-coding RNA loci. These Myc targets mediate the physiological effects of Myc on cell proliferation, metabolism, apoptosis, growth, and differentiation (Eilers and Eisenman, 2008). To promote transcriptional activation at target genes, Myc forms heterodimers with its partner Max and recruits chromatin-modifying complexes to E-box containing promoters. Myc is also involved in transcriptional repression through the inhibition of the transcriptional activator Miz1 (Kleine-Kohlbrecher et al., 2006). Aberrant elevation of Myc levels has been shown to contribute to the genesis of many types of human tumors (Hanahan and Weinberg, 2000).
Myc family proteins contain highly conserved regions termed Myc boxes (MB) that are essential for Myc’s biological activities (see Fig. 1E). A major determinant of Myc transcriptional function is MBII, which is the site of recruitment of co-activator complexes containing histone acetyl transferases (HATs) such as GCN5 (McMahon et al., 2000) and TIP60 (Frank et al., 2003). MBI functions as a phosphorylation-dependent binding site for the ubiquitin ligase Fbw7 (Welcker et al., 2004), while MBII is one of the binding sites for the ligase SKP2 (Kim et al., 2003; von der Lehr et al., 2003). Fbw7 and Skp2 both contribute to the rapid degradation of Myc protein (t1/2 ∼20 minutes). The C-terminus of Myc harbors nuclear localization signals and the bHLHZ motif that mediates dimerization with Max and DNA binding.
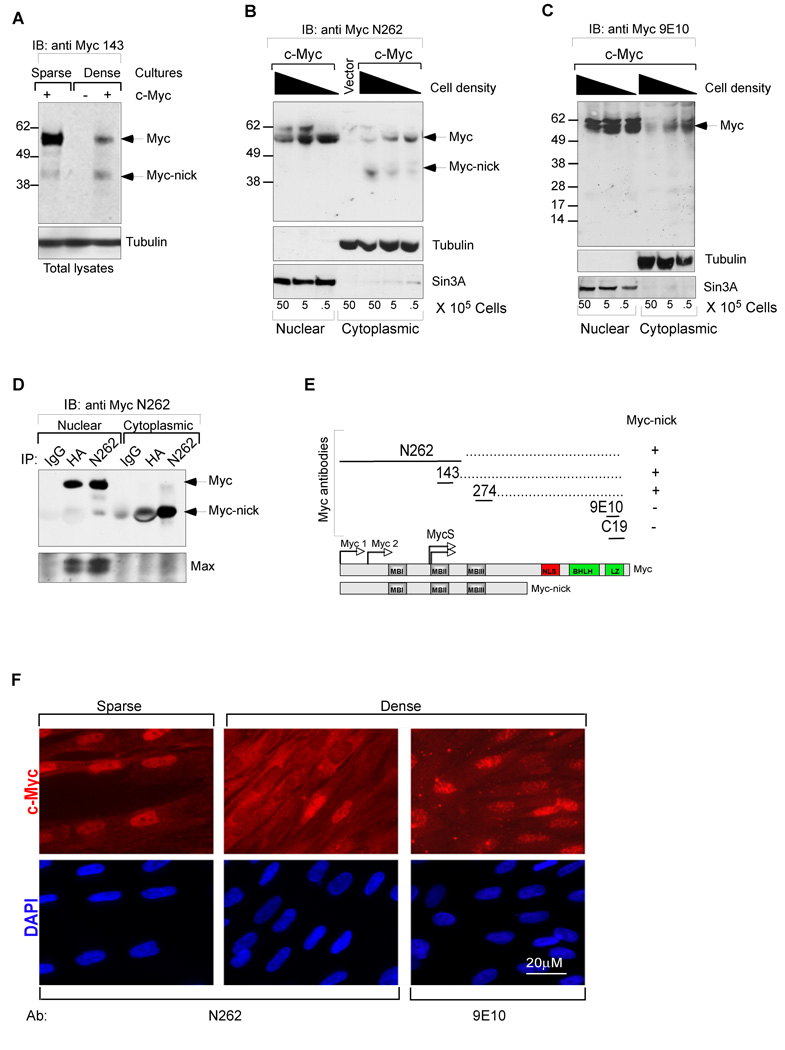
Figure 1. Identification of Myc-nick in the cytoplasm of cells grown at high density.
(A) Total cell lysates of Rat1 myc-null fibroblasts infected with c-Myc, or empty retroviral vectors were prepared for western blot by adding boiling sample buffer. (B–C) Nuclear and cytoplasmic fractions of HFF cells expressing c-Myc were prepared 48h after plating at the indicated increasing densities. (D) Immunoprecipitation of HA-c-Myc (N-terminal tag) with anti-N262, anti-HA, and normal IgG from nuclear and cytoplasmic fractions of HFFs. Note that Max is only co-immunoprecipitated along with nuclear c-Myc. (E) Schematic representation of antibody mapping. (F) HFF cells infected with c-myc expressing retrovirus were cultured for 4 days after reaching confluency (middle and right panels) and compared with a subconfluent culture (left panel) by immunofluorescence using N262 and 9E10 antibodies. Nuclear (N) and cytoplasmic (C) fractions. See also Fig. S1.
Several variant forms of Myc protein have been previously identified. All of them are nuclear localized, low abundance, proteins generated by alternative translation initiation. A weak CUG translational initiation site, upstream and in-frame of the predominant AUG codon, produces an N-terminally extended form of c-Myc called c-Myc1 (Hann et al., 1988). Another Myc protein variant is MycS, generated by internal translational initiations at two AUG codons located ∼100 amino acids from the normal N-terminus (Spotts et al., 1997). MycS lacks MBI but contains MBII and retains much of full-length Myc’s biological activity (Xiao et al., 1998).
As expected, given their broad role as transcriptional regulators, Myc family proteins are predominantly localized to the cell nucleus during proliferation. Surprisingly however, there have been multiple reports of cytoplasmically localized Myc, mostly in differentiated cells. For example, N-Myc localization was shown to change from nuclear to cytoplasmic in differentiating neurons of the neural crest, retinal ganglion cells, neurons of spinal ganglia (Wakamatsu et al., 1997; Wakamatsu et al., 1993) and Purkinje cells (Okano et al., 1999; Wakamatsu et al., 1993). Cytoplasmic Myc was also reported in tumors with diverse origins (Bai et al., 1994; Calcagno et al., 2009; Pietilainen et al., 1995). These studies relied on immunostaining protocols and the form of the Myc protein involved was not characterized.
Interestingly, association of Myc with several cytoplasmic proteins has been reported. The best characterized is the interaction of c-Myc with tubulins (Alexandrova et al., 1995) (Koch et al., 2007; Niklinski et al., 2000). Myc has also been reported to interact with other proteins that are predominantly cytoplasmic such as cdr2 (Okano et al., 1999) and AMY-1 (Taira et al., 1998). However the nature of the cytoplasmic Myc protein and its potential function remains an enigma. Here we report the identification of Myc-nick, a cytoplasmically localized cleavage product of Myc and provide evidence for its role in cytoskeletal organization and cell differentiation.
Results
Myc-nick is a truncated form of Myc localized predominantly in the cytoplasm
While studying regulation of c-Myc degradation we noticed an inverse correlation between the levels of full-length c-Myc and a cytoplasmic 42KDa protein in anti-Myc immunoblots derived from confluent fibroblast cultures (Fig. 1A–B). As described below, this protein, which we have named Myc-nick, is a cytoplasmic cleavage product of full-length c-Myc generated at high cell density (Fig.1B). Myc-nick is recognized by three antibodies against the N-terminal two-thirds of c-Myc (anti-Myc N262, 274, 143; Fig.1A–B and Fig. S1A) but not by anti- C-terminal antibodies (anti-Myc 9E10, C19; Fig. 1C). Furthermore, an anti-HA antibody immunoprecipitates Myc-nick from cytoplasmic extracts of cells expressing N-terminally HA-tagged c-Myc (Fig. 1D). In addition, cytoplasmic Myc bearing N-terminal but not C-terminal epitopes is detected by imunofluorescence in confluent cultures (see below, Fig.1F). Together, these results indicate that Myc-nick is a truncated protein lacking the C-terminal portion of c-Myc while preserving an intact N-terminus comprising Myc boxes I–III (Fig. 1E). This makes Myc-nick distinct from any other previously identified form of Myc (see Introduction).
We have detected endogenously expressed Myc-nick in the cytoplasm of a large number of cell lines including HFF, Wi38, L-cells, HCT116, SW480, Hela (Fig. S1B), C2C12 (Fig. 7D), 293T, A431, Rat1, U2OS, ES cells, mouse neurospheres (not shown), in addition to mouse tissues such as muscle, brain, cerebellum, (Fig. 7A and Fig. S7A). We also observed Myc cytoplasmic localization by immunofluorescence of confluent cultures of HFFs (Fig. 1F, and Fig. S5A) and Rat1 myc-null cells (not shown) expressing c-myc. Therefore a protein with the size and properties of Myc-nick is very widely expressed. In some settings we observe relatively low and variable amounts of Myc-nick in the nucleus (e.g. Fig. 2A).
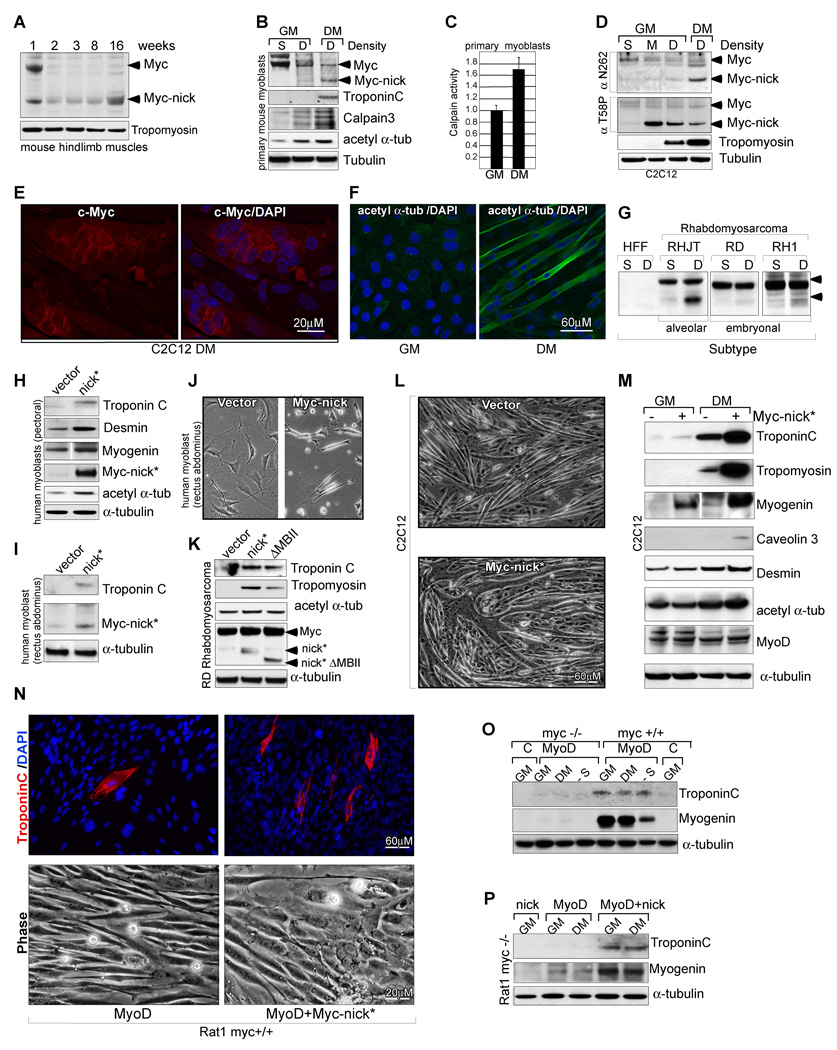
Fig. 7. Myc-nick accelerates muscle cell differentiation.
(A) Hindlimb muscles dissected from 1, 2, 3, 8, and 16 weeks old mice were processed for immunoblotting using N-terminal anti-Myc sera (anti 143+274). (B) Mouse primary myoblasts isolated from hindlimb muscles of 8-week old mice were cultured as sparse cultures for 24h or as dense cultures in the presence of either growth medium (GM) or differentiation medium (DM) for 3 days. Total cell extracts were immunoblotted for c-Myc (anti 143+274) and indicated proteins. (C) Mouse primary myoblasts were cultured in GM or DM for 3 days, lysed in buffer A, and total calpain activity was measured using Suc-LLVY-AMC synthetic substrate. (n=2. Calpain activity in DM was compared with calpain activity in GM (set to 1) +/− SEM). (D) C2C12 mouse myoblasts cultured at sparse (S), medium (M) or high densities (D). Dense cultures were harvested or switched to DM for 7 days. Total cell extracts were immunoblotted for c-Myc using antibodies against total c-Myc (N262) or against phosphorylated T58 Myc, a signal for Myc degradation. (E) C2C12 cells cultured in DM for 7 days and stained for endogenous c-Myc (anti-N262). (F) C2C12 cells grown in the presence of growth medium (GM) or differentiation medium (DM) for 5 days and stained with anti-acetylated α-tubulin. (G) Western blotting for Myc in Rhabdomyosarcoma cell lines grown as dense or sparse cultures for 3 days. (H−I) Human myoblasts expressing vector, or Myc-nick were cultured in DM and processed for immunoblotting after 4 days. (J) Human myoblasts expressing vector, or Myc-nick were cultured in DM and photographed after 2 days. (K) RD Rhabdomyosarcoma cells expressing vector, Myc-nick, or Myc-nick ΔMBII (Δ106–143) were grown in DM for 4 days and processed for immunobloting. (L) C2C12 cells expressing vector, or Myc-nick, were grown as dense cultures and stimulated to differentiate for 4 days and then photographed. (M) C2C12 cells expressing vector, or Myc-nick were grown to confluency and harvested or stimulated to differentiate for 3 days and then harvested. Total cell lysates were immunoblotted with antibodies against the indicated proteins. (N) Rat1 (myc+/+) cells expressing MyoD, or MyoD + Myc-nick were cultured in DM for 3 days and photographed (lower panels), or stained for Troponin C and DAPI (upper panels). (O) Rat1 (myc+/+) or Rat1 myc-null (myc−/−) cells expressing vector (lane C), or MyoD were cultured in GM, DM or serum free medium (lane -S) for 4 days and processed for immunobloting. (P) Rat myc null (myc−/−) cells expressing Myc-nick, MyoD, or MyoD + Myc-nick were grown in GM or DM for 3 days and processed for immunoblotting. See also Fig. S7.
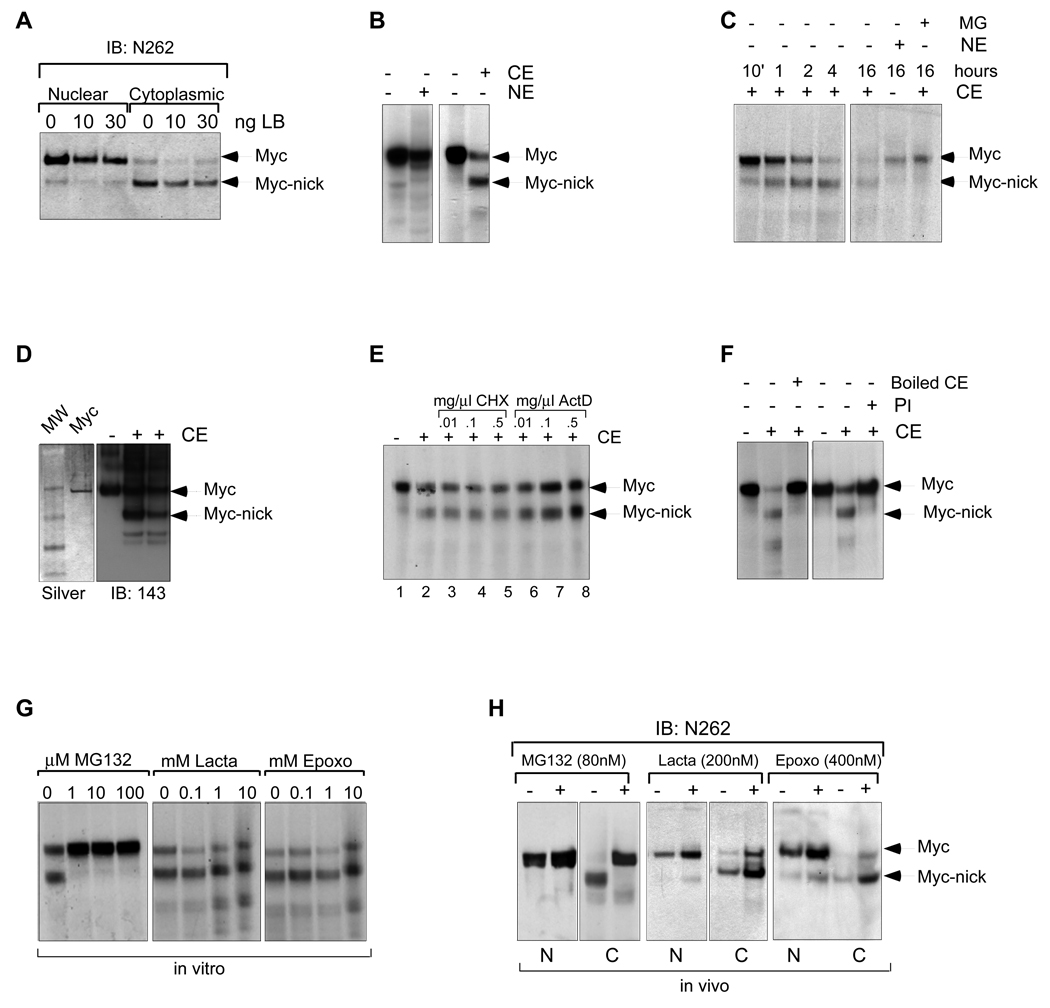
Fig. 2. Myc-nick is product of c-Myc cytoplasmic cleavage independent of the proteasome.
(A) CRM1 dependant nuclear export is not involved in the formation or localization of Myc-nick. Rat1 myc-null cells infected with c-Myc were treated with leptomycin B (LB) for 4h before harvesting. (B) c-Myc gives rise to Myc-nick in vitro when incubated with cytoplasmic extracts. Radiolabeled IVT c-Myc was incubated for 2h with cytoplasmic (CE) or nuclear extracts (NE) from Rat1 c-myc null cells. (C) Time course of in vitro cleavage of c-Myc. (D) Recombinant c-Myc is cleaved in the presence of CE. 1 µg of recombinant c-Myc was incubated with 30µg of CE and processed for Western blot. (E) IVT c-Myc was incubated with CE for 4h in the presence of of Actinomycin D (ActD) or cyclohexamide (CHX). (F) The cleavage of c-Myc is inhibited by protease inhibitors and by heat inactivation. CE was boiled prior to incubation with IVT c-Myc and the Protease Inhibitor (PI) was added to the incubation mixture. (G,H) The cleavage of c-Myc is inhibited by MG132, but not by Lactacystein or Epoxomycin. (G) IVT c-Myc was incubated with CE, in the presence of increasing amounts of MG132, Lactacystein and Epoxomycin for 1h. (H) Rat1 myc-null cells expressing c-Myc were incubated with MG132, Lactacytein or Epoxomycin for 2h prior to harvesting. See also Fig. S2.
Myc-nick is generated by proteolytic cleavage of c-Myc in the cytoplasm
We considered the possibility that Myc-nick is generated in the cytoplasm because the nuclear export inhibitor Leptomycin B had no effect on the production or cytoplasmic localization of Myc-nick (Fig. 2A). To determine whether a cytoplasmic activity could convert full-length c-Myc into Myc-nick we incubated in vitro translated [35S]-methionine labeled c-Myc or purified full-length recombinant c-Myc with nuclear or cytoplasmic extracts from Rat1 myc-null cells. Only cytoplasmic extracts (CE) were capable of producing a protein (Fig. 2B–D) having the same apparent molecular weight as Myc-nick that was recognized by antibodies against the N-terminus, but not C-terminus of c-Myc (Fig. S2A). Increasing the incubation time with CE augmented Myc-nick production and decreased the amounts of full-length c-Myc input (Fig. 2C). In agreement with our observations made in vivo, CE of dense cultures were more efficient in cleaving c-Myc than CE of sparse cultures (Fig. S2B). In experiments designed to characterize the cytoplasmic activity responsible for formation of Myc-nick we found that inhibitors of transcription or translation had no effect on Myc-nick formation (Fig. 2E) while heating or adding protease inhibitors to the CE blocked Myc-nick, consistent with proteolysis (Fig. 2F). While MG132, an inhibitor of proteasome (and other cysteine proteases) blocked Myc nick formation, specific proteasome inhibitors such as Epoxomycin and Lactacystein failed to block the formation of Myc-nick in vitro and in vivo (Fig. 2G,H). In addition, inhibition of either the Fbw7 or Skp2 degradation pathways, known to be responsible for proteasomal Myc turnover, had no effect on Myc-nick formation (data not shown). Moreover, incubation of Myc with 20S and 26S proteasomes failed to produce Myc-nick (not shown). These results indicate that full-length c-Myc is converted into Myc-nick by a cytoplasmic protease that is independent of the proteasome.
Myc is cleaved by a calcium-activated calpain to produce Myc-nick
In a systematic search for the proteases mediating cytoplasmic cleavage of Myc we ruled out both caspases and lysosomal proteases on the basis of inhibitors and cleavage conditions (Fig. S2C). However, we found that all calpain inhibitors tested including calpeptin, calpain inhibitor XII (Fig. 3A) and Calpain inhibitor VI (Fig. 3B) blocked the formation of Myc-nick in vitro and in vivo. Moreover, MG132, while well known as a proteasome inhibitor has also been shown to inhibit calpains (Mailhes et al., 2002). Calpains had been linked to Myc stability earlier, but the antibodies used in those studies would not have detected Myc-nick (Gonen et al., 1997; Small et al., 2002). Calpains comprise a large family of calcium-dependent cytoplasmic cysteine proteases that function at neutral pH and are primarily associated with partial protein cleavage rather than complete protein degradation. The most well studied members of this family are mcalpain and μcalpain. These ubiquitously expressed catalytic subunits form functional heterodimers with a calpain regulatory subunit (calpain r) to bind calcium. To determine whether Myc-cleavage is regulated by calcium-activated calpains, we employed siRNA to knockdown calpain r in cells expressing c-Myc either endogenously or under control of a retroviral vector. In both settings a partial silencing of calpain r correlated with decreased Myc-nick and increased full-length c-Myc (Fig. 3C). Knockdown of m or μcalpains alone did not block the formation of Myc-nick, most likely due to the presence of other redundant calpains (not shown). Because calpain activity is calcium dependent we next examined the effects on Myc-nick of modulating calcium levels. Treatment with the calcium chelators Bapta or EGTA reduced the ability of CEs to cleave c-Myc (Fig. 3D,E). Incubation of either IVT c-Myc (Fig. 3F) or recombinant c-Myc (Fig. 3G) with purified m or μ calpain (together with calpain r) produced Myc-nick. These results demonstrate that Myc-nick is directly generated by calcium dependant calpain cleavage of full-length c-Myc.
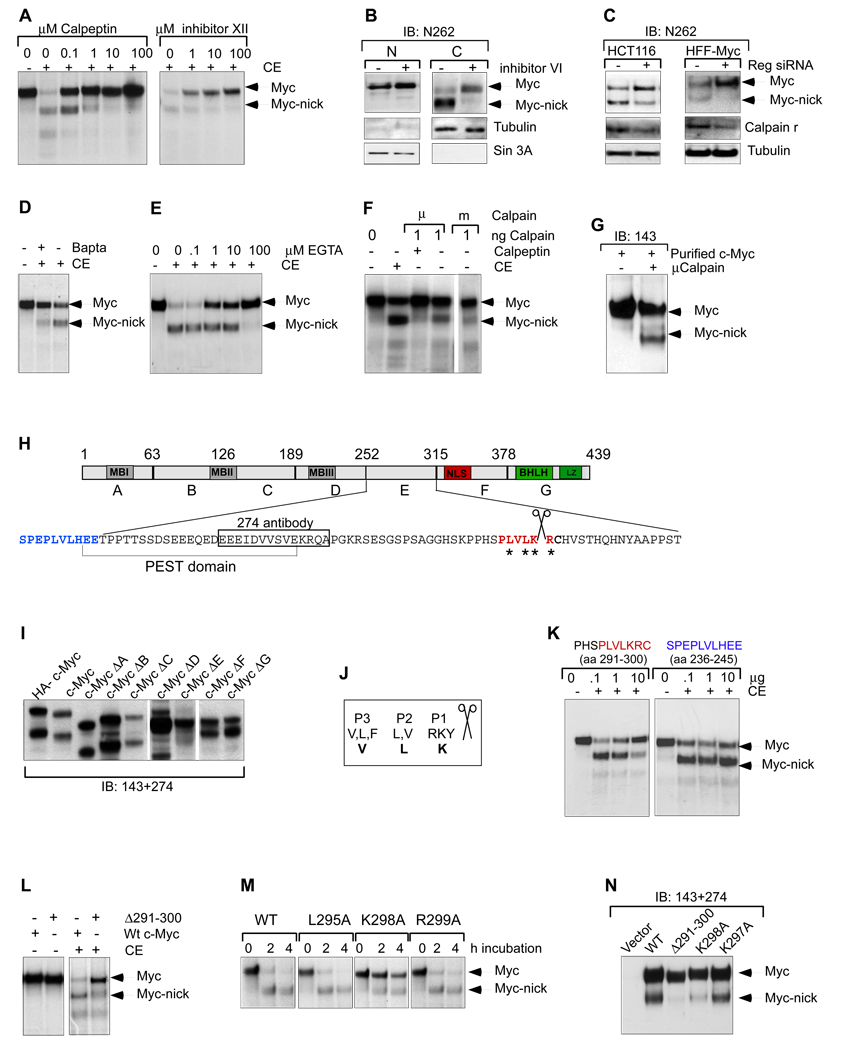
Fig. 3. Myc-nick is generated by calpain cleavage of full-length Myc.
(A) IVT c-Myc was incubated with CE for 1h in the presence of the calpain inhibitors calpeptin and calpain inhibitor XII. (B) Dense cultures of HFF cells infected with a c-Myc retroviral vector were incubated with Calpain inhibitor VI for 2h prior to harvesting. (C) siRNA for calpain regulatory subunit (Reg) reduces the formation of Myc-nick in HCT116 and HFF-Myc cells. (D) Rat1 myc-null cells were incubated for 2h in the presence of the calcium chelant Bapta, and then cytoplasmic extracts were prepared and incubated with IVT c-Myc (as in 2B). (E) IVT c-Myc was incubated with CE for 1h in the presence of increasing amounts of EGTA. (F) IVT c-Myc was incubated with purified recombinant μCalpain or mCalpain and r subunit for 1h on ice. (G) 100ng of purified μcalpain and r subunit was incubated with 2µg of recombinant c-Myc, for 30 min on ice. (H) Schematic representation of c-Myc protein indicating A–G deletion regions (deletion endpoints indicated above), putative calpain cleavage region in red. (I) c-Myc and the deletion mutants ΔA-ΔG were expressed in 293T cells and 48h later the presence of a Myc-nick-like protein in cytoplasmic extracts was determined. (J) Amino acid preference for calpain cleavage region according to Tompa, 2004. P1–P3 indicate the position of preferred residues in relation to the cleavage site, bold letters indicate the c-Myc calpain cleavage site. (K) IVT c-Myc was incubated with CE in the presence of a peptide that contains the potential calpain cleavage site (amino acids 291–300-in red) or a nearby sequence (amino acids 236–245-in blue). (L) WT and Δ291 –300 IVT c-Myc were incubated with CE for 1h (as in 2B). (M) IVT WT, L295A, K298A and K299A c-Myc were incubated with CE for the indicated time points. (N) Cleavage products produced from the indicated c-Myc mutants in 293T cells. See also Fig. S3.
Lysine 298 is the primary calpain cleavage site in c-Myc
To map the calpain cleavage region on c-Myc, we used a series of internal deletion mutants lacking 60 residue segments (c-Myc ΔA-ΔG) (Tworkowski et al., 2002) (Fig. 3H) and determined whether any failed to generate a shorter c-Myc protein. Only the deletion of residues 252–315 (ΔE) resulted in loss of a Myc-nick-like product (Fig. 3I). This region contains a high scoring PEST domain, often present in unstable proteins and frequently associated with calpain cleavage sites. We further narrowed our search to residues 270–315, based on the ability of antibody 274 to detect Myc-nick (Fig. S1A and Fig. 3H). While there is no universal consensus for a calpain cleavage site, a comparison of 106 sites present in 49 known calpain substrates indicated a preference for calpain cleavage after K or R, and to lesser extent Y especially when these amino acids are flanked by P, V, and L (Fig. 3J) (Tompa et al., 2004). We noted a region localized C-terminal to the PEST domain containing the sequence PLVLKRC (Fig. 3H, marked in red). This region is evolutionarily conserved in c-Myc and N-Myc but not L-Myc (Fig. S3A), consistent with the fact that c-Myc and N-Myc, but not L-Myc, are cleaved (Fig. S3B,C). To determine whether this region functions as a calpain cleavage site we synthesized a 10 amino acid peptide corresponding to the putative c-Myc calpain cleavage region (291–300) and to another a nearby region of c-Myc (236–245) (Fig. 3H, in red and blue respectively) and asked whether they could act as competitive inhibitors of Myc-nick formation in vitro. Addition of increasing amounts of the peptide containing the putative calpain cleavage site blocked the formation of Myc-nick in vitro while the control peptide had no effect (Fig. 3K). Next, we generated a deletion mutant of Myc lacking residues 291–300 and found it to be resistant to cleavage by CE (Fig. 3L). When this mutant was ectopically expressed in 293T cells the cleavage was also reduced in comparison to the wild type c-Myc (Fig. 3N), indicating that this is the major calpain cleavage site within c-Myc. Next, we made point mutations in the calpain cleavage region (labeled with asterisks in Fig. 3H), and assessed their cleavage. We found that the K298A mutation reduced the cleavage of c-Myc in vitro (Fig. 3M) and in vivo when ectopically expressed in 293T cells (Fig. 3N). Although K298 appears to be the major calpain cleavage site, a K298A mutant was still cleaved in vivo most likely because when this residue is mutated the cleavage is shifted to R299 (and perhaps L297, V296). We also identified weaker calpain cleavage sites localized in the C-terminus of Myc that become more pronounced in the cleavage deficient mutants.
Expression of Myc-nick alters cell morphology and increases acetylation of α-tubulin
To study Myc-nick function we generated a truncated form of Myc containing amino acids 1–298 (referred to as Myc-nick*). We found that ectopically expressed Myc-nick* is localized predominantly to the cytoplasm and migrates on SDS-PAGE with the same apparent molecular weight as Myc-nick generated by cleavage of full-length c-Myc (Fig. 4A). Myc-nick is degraded at a comparable rate to full-length c-Myc with a half-life of about 30 minutes (data not shown). Because Myc-nick contains the MBI phosphodegron, with its GSK3β phosphorylation and Fbw7 binding sites, we would expect Myc-nick to be degraded through similar proteasomal pathways as full-length Myc. Indeed, blocking proteasome activity by pharmacological inhibitors or by silencing E3 ligases and components of the proteasome induced the stabilization of both full-length Myc and Myc-nick (data not shown). Calpain inhibitors are incapable of inducing accumulation of Myc-nick, indicating that calpains are unlikely to play a major role in Myc-nick turnover in vivo.
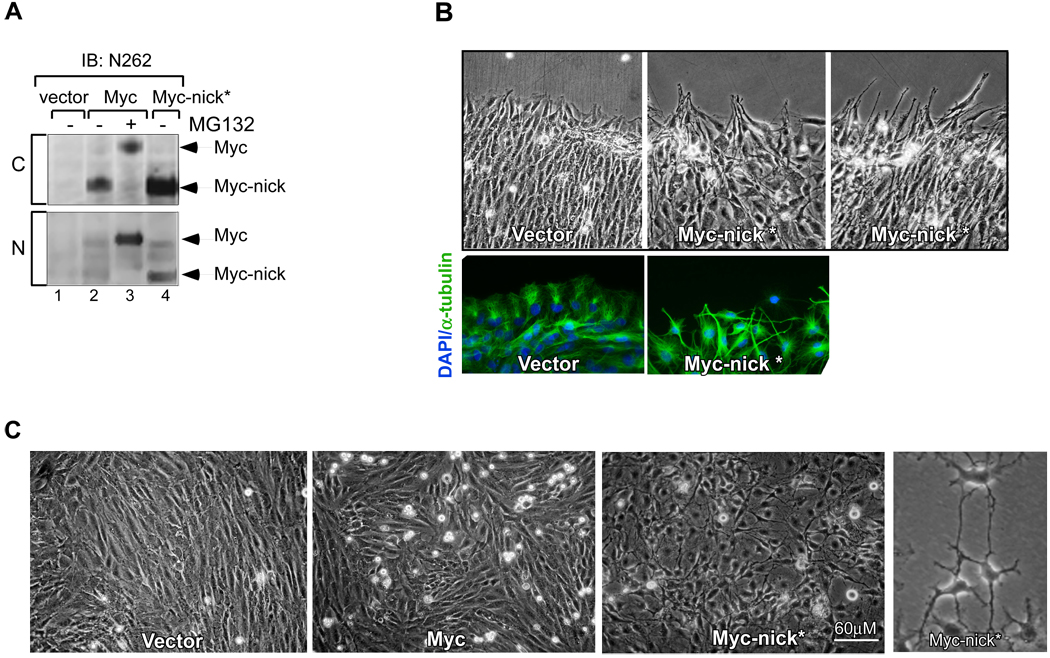
Fig. 4. The expression of Myc-nick* promotes changes in cell morphology.
(A) Myc-nick* (1–298) is cytoplasmic and has the same apparent molecular weight as Myc-nick derived from full-length c-Myc (compare lanes 2 and 4, upper panel). Rat1 myc-null cells expressing empty vector, full-length c-Myc, and Myc-nick were fractionated into nuclear (N) and cytoplasmic (C) fractionations. (B) Myc-nick* expressing cells extend protrusions at the wound edge. A confluent monolayer of Rat1 myc-null cells expressing either vector or Myc-nick was scratched using a 100 µl tip and phase contrast images taken at 12h. (C) Rat1 myc-null cells expressing empty vector, c-Myc and Myc-nick at 14 days after selection. See also Fig. S4.
While overexpression of full-length c-Myc in Rat1a myc-null fibroblasts is associated with increased proliferation and apoptosis, the ectopic expression of Myc-nick* had no detectable effect on either cell doubling time or survival (data not shown). However, Myc-nick* expressing cells displayed dramatic morphological changes - they appeared spindle-like with long protrusions that occasionally formed intercellular contacts (Fig. 4C, Fig. S4, Fig. S5C). This morphology was specific for Myc-nick* expression and could not be produced by expressing the C-terminal 100 residues of c-Myc (not shown). Introducing a scratch wound across a confluent cell monolayer showed that while control cells migrated into the wound by extending lamellipodia, Myc-nick* expressing cells aligned parallel to each other and extended long protrusions into the wound (Fig. 4B).
The morphological changes induced by Myc-nick* are suggestive of altered cell-cell contacts and/or major cytoskeletal reorganization. The elongated cellular protrusions present in Myc-nick expressing cells resemble specialized structures formed by stable microtubules. Because stable microtubules display increased acetylation of α-tubulin on Lysine 40 (Hubbert et al., 2002) we stained Myc-nick* expressing cells with antibodies against α-tubulin and acetylated α-tubulin. Figure 5A shows that while acetylated α-tubulin immunostaining in vector and c-Myc expressing cells is low, Myc-nick* cells display intense staining of elongated structures (Fig. 5A and F). This was confirmed by immunoblotting for acetylated α-tubulin in Rat1 myc-null or 293T cells expressing Myc-nick* (Fig. 5C; Fig. S6B). Importantly neither the levels of total cellular acetylated lysine (Fig. S6A) nor the levels of tyrosinated tubulin (not shown), were affected. To ask whether Myc-nick* expression increased microtubule stability, we treated cells expressing Myc-nick*, c-Myc or vector with nocodozole for 15’. This treatment disrupts unstable microtubules with a half-life of about 10 minutes, but not stable microtubules with a half-life >2 hours. Only Myc-nick* expressing cells possessed microtubules that survived nocodozole treatment and these stained strongly with anti-acetylated α-tubulin (Fig. 5B).
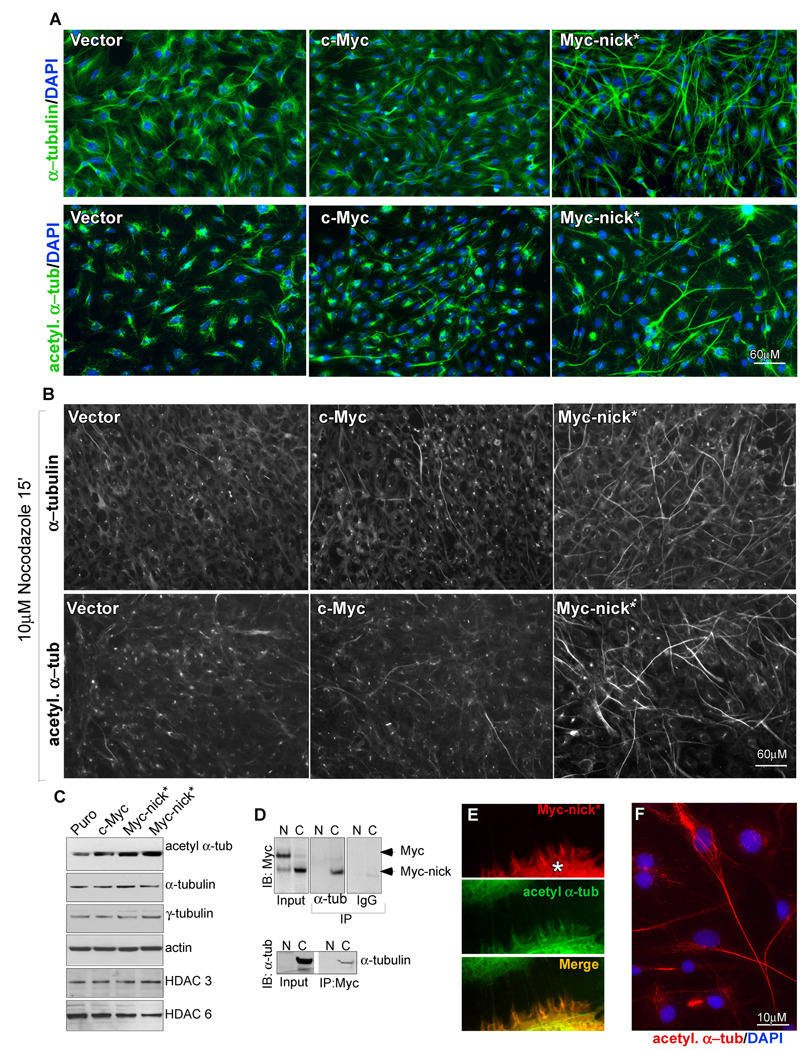
Fig. 5. Myc-nick* cells display increased levels of acetylated (-tubulin and microtubule stabilization.
(A) Immunofluorescence for α-tubulin (upper panels) and acetylated α-tubulin (lower panels) of Rat1 myc-null cells infected with empty vector, c-Myc or Myc-nick.* (B) As for (A) but incubated in the presence of nocodazole for 15 minutes prior to fixation. (C) Immunoblotting of Rat1 cell extracts using antibodies against the indicated proteins. (D) NE or CE of Rat1 myc-null cells expressing c-Myc were immunoprecipitated with anti α-tubulin or normal mouse IgG and immunoblotted for Myc (top panel), or immunoprecipitated with anti c-Myc N262 antibody, and immunoblotted with anti α-tubulin. (E) Immunofluorescence for Myc and acetylated α-tubulin in A431 lung epithelial cells cell transfected with Myc-nick (*). (F) Detail of Myc-nick expressing cell stained for acetylated α-tubulin. See also Fig. S5.
Myc-nick interacts with α- and β- tubulins
We observed partial cytoplasmic co-localization between Myc and α-tubulin in several cell types expressing c-Myc or Myc-nick (Fig. 5E; Fig. S5A–C). Using 293T cells expressing GFP-tubulin and Myc-nick we performed immunoprecipitation with anti-GFP and immunobloted with anti-Myc. Myc-nick was co-immunoprecipitated with GFP-tubulin but not GFP-EB1, a microtubule binding protein (Fig. S5D). In addition, immunoprecipitation of either c-Myc or endogenous α-tubulin from cytoplasmic extracts indicated that Myc-nick interacts with α-tubulin in vivo (Fig. 5D). To examine in vitro interactions we incubated 35[S]-methionine labeled IVT c-Myc with purified brain tubulins for 1h, then immunoprecipitated α, β, γ, pIII and acetylated α-tubulin and exposed the gel to detect radioactive Myc. Full-length c-Myc was co-immunoprecipitated mostly with β-tubulin (Fig. S5E middle panel). We also incubated IVT c-Myc with CE for 1h to produce Myc-nick and then performed immunoprecipitation for tubulins as above. The results showed that Myc-nick interacts with α- and β-tubulin (Fig. S5E upper panel). The binding of Myc-nick to tubulin is consistent with the previous report of an N-terminal region of Myc capable of associating with tubulin (Alexandrova et al., 1995).
Myc-nick directly regulates α-tubulin acetylation through GCN5
Because Myc-nick lacks the Myc C-terminal dimerization and DNA binding domains, we surmised that the increase in acetylated α-tubulin induced by Myc-nick* expression is independent of Myc transcriptional activity. One possibility is that Myc-nick recruits an acetyltransferase microtubules. We observed marked acetylation of α-tubulin in cytoplasmic extracts incubated in the presence of either recombinant c-Myc (Fig. 6A) or of IVT c-Myc (Fig. 6B and Fig. S6C). Importantly, Myc proteins are known to associate with the acetyltransferases GCN5, and TIP60 (Frank et al., 2003; Sterner and Berger, 2000) via TRRAP that interacts with MBII (residues 106–143) (McMahon et al., 1998). We therefore tested a Myc-nick deletion mutant lacking Myc box II (ΔMBII) and found that its ability to promote α-tubulin acetylation was reduced (Fig. 6 C, D). In addition, the ΔMBII Myc-nick* mutant failed to induce the cell morphological changes that we had detected with WT Myc-nick* while a comparably sized deletion in a region adjacent to MBII was similar to wild type (Fig. 6E).
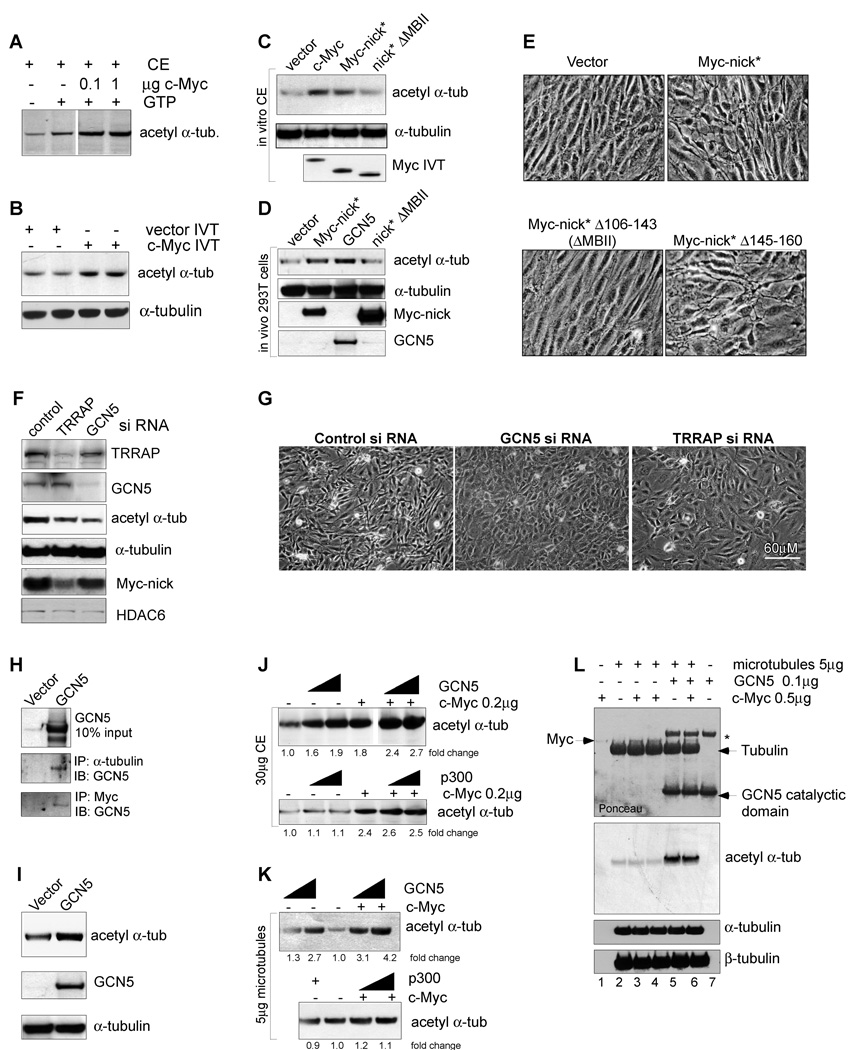
Fig. 6. c-Myc and the HAT GCN5 promote α-tubulin acetylation.
(A) CE was incubated with recombinant c-Myc and c-Myc dilution buffer; or (B) with c-Myc IVT or vector IVT for 1h at 37C. The samples were immunoblotted as indicated. (C) CE was incubated with IVT vector, c-Myc, Myc-nick and Myc-nick ΔMBII (Δ106–143) for 30’ at 37°C, then processed for for immunoblotting. (D) 293T cells were transfected with empty vector, GCN5, Myc-nick, Myc-nick ΔMBII and processed for immunoblotting after 48h. (E) Rat1 myc-null cells were infected as indicated and photographed 10 days after selection. (F–G) Rat1 myc-null cell expressing Myc-nick were transfected with 100nM of control, TRRAP or GCN5 siRNA and 76h later processed for immunoblotting (F) or photographed (G). (H) CE of 293T cells transfected with empty or GCN5 vectors were immunoprecipitated with anti-α-tubulin or anti-Myc (143+274) and immunoblotted for GCN5. (I) 293T cells were transfected with control or GCN5 expressing vectors and 48h later processed for immunoblotting. (J–L) GCN5 induces acetylation of α-tubulin. (J) CE were incubated with 200ng of Myc, 100ng or 300ng of GCN5 (upper panel) or 100ng or 500ng of p300 (lower panel). (K) Assembled microtubules were incubated with 100ng or 500ng of recombinant full-length GCN5 (upper panel) or p300 (lower panel) in the presence or absence of 200ng of purified c-Myc. (L) Purified assembled microtubules were incubated with recombinant c-Myc and GCN5-catalytic domain. The asterisk indicates a nonspecific bacterial band copurified with GCN5-catalytic domain. See also Fig. S6.
The dependence on MBII for tubulin acetylation and altered cell morphology suggests involvement of the acetyltranferases known to bind this region. We detected substantial amounts of GCN5 and TRRAP in the cytoplasm (Fig. S6D), while Tip60 was predominantly nuclear (not shown). To test GCN5’s involvement in α-tubulin acetylation we performed siRNA-mediated knockdown of TRRAP and GCN5 in Myc-nick* expressing cells and found that decreasing either one of these proteins reduced the levels of acetylated α-tubulin (Fig. 6F) and reverted the changes in cell morphology induced by Myc-nick to that of vector infected cells (Fig. 6G). Moreover, GCN5 co-immunoprecipated with α-tubulin and Myc in cytoplasmic extracts of 293T cells transfected with GCN5 (Figure 6H).
Both ectopic expression of GCN5 in 293T cells (Fig. 6D,I), and addition of full-length recombinant GCN5 to cytoplasmic extracts (Fig. 6J upper panel), resulted in increased levels of acetylated α-tubulin. The addition of Myc further increased the levels of α- tubulin acetylation induced by GCN5 in cytoplasmic extracts (Fig. 6J). Full-length recombinant GCN5 also induced the acetylation of α-tubulin present in assembled microtubules (Fig. 6K) and synergized with c-Myc to promote further acetylation (Fig. 6K, compare lanes 1 to 4 and 2 to 5). The GCN5 catalytic domain alone was sufficient to induce α- tubulin acetylation, but no synergy with Myc was detected, most likely because association between these two proteins occurs outside of GCN5’s active site (Fig. 6L). As a further control we tested p300, a HAT that binds the C-terminus of Myc (Faiola et al., 2005). P300 neither induced tubulin acetylation nor did it synergize with Myc (Fig. 6 J, K bottom panels). These data indicate that GCN5 can specifically acetylate tubulin and that Myc augments tubulin acetylation by binding to tubulin and recruiting GCN5.
Myc-nick is produced in differentiating cells and tissues
When examining the expression of Myc-nick in adult mouse tissues, we found that brain, cerebellum and skeletal muscle express significantly higher levels of Myc-nick than any other tissue (Fig. 7A, Fig. S7A). Interestingly, both neuronal and muscle differentiation require major cytoskeletal rearrangements that have been associated with increased microtubule stability and elevated levels of acetylated α-tubulin. For example it has been demonstrated that the acetylation of α-tubulin by the Elp3 acetyltransferase is required for proper cortical neuronal migration and differentiation (Creppe et al., 2009). Increased levels of acetylated α-tubulin are found during myogenic differentiation (Gundersen et al., 1989). Importantly, inhibiting the activity of the deacetylases HDAC6 and Sirt2 (known to deacetylate α-tubulin) generates augmented levels of acetylated α-tubulin, promotes differentiation of myoblasts (Iezi et al., 2004). Furthermore, the presence of the primary cilium, a microtubule based antenna-like structure composed of acetylated α-tubulin, is essential for cardiomiocyte differentiation (Clement et al., 2009).
The extensive cytoskeletal changes that occur during muscle differentiation are regulated by calcium and calpains (Dedieu et al., 2004). The inhibition of calpain activity either by pharmacological inhibitors or by overexpression of Calpastatin (an endogenous inhibitor of calpains) blocks muscle cell differentiation (Dedieu et al., 2004). In addition, both types of ubiquitous calpains (m and μ) were shown to regulate muscle cell differentiation in vitro (Moyen et al., 2004). Moreover, mutations in the Calpain 3 gene (the muscle specific Calpain) cause limb girdle muscle dystrophy 2A (LGMD2A) (Richard et al., 1995). Finally, knocking out Calpain r (the regulatory subunit shared by all calcium dependent calpains) is lethal due to impaired cardiovascular development (Arthur et al., 2000).
To study the cleavage of Myc to Myc-nick during the process of muscle differentiation, we employed human primary myoblasts purified from pectoral girdle, mouse myoblasts purified from hindlimb muscles, and C2C12 cultured mouse myoblasts. When stimulated to differentiate, these cells displayed low levels of full-length c-Myc and high levels of Myc-nick when compared to undifferentiated cycling cells (Fig. 7B, D; Fig. S7B). Interestingly, Myc-nick levels were higher in a Rhabdomyosarcoma cell line from the alveolar subtype (Fig. 7G), a very aggressive tumor derived from partially differentiated muscle cells.
The increased levels of Myc-nick in differentiated primary mouse myoblasts correlates with an increase in Calpain 3 levels (Fig. 7B; Fig. S7B) and in total calpain activity (Fig. 7C). Similarly, when C2C12 cells were stimulated to differentiate they also displayed an increase in total calpain activity (Fig. S7C) and in the ability to cleave Myc in vitro (Fig. S7D). Importantly, the levels of acetylated α-tubulin were also elevated during the process of differentiation (Fig. 7B, D; Fig. S7B, F). Acetylation of α-tubulin is accompanied by myoblast fusion to form multinucleated myotubes (Fig. 7F; Fig. S7F). When myoblasts are stimulated to differentiate they fuse into multinucleated myotubes. In addition to displaying increased levels of acetylated α-tubulin, these myotubes show decreased immunostaining for nuclear Myc and increased cytoplasmic Myc staining (Fig. 7E). Conversely, undifferentiated satellite cells in the same culture display nuclear staining for Myc (Fig. S7E). This is consistent with our results showing conversion of full-length Myc into predominantly cytoplasmic Myc-nick.
In C2C12 cells, concomitant with increased Myc-nick abundance we observed a decrease in Myc-nick phosphorylation at threonine 58 (T58) after the switch to differentiation conditions (Fig. 7D). Phospho-T58 mediates Fbw7-dependent degradation of Myc by the proteasome. Decreased phosphorylation at this site is consistent with the notion that stabilization of Myc-nick contributes to its elevated levels. Interestingly we found that Myc-nick phosphorylation at T58 is also reduced in adult muscle, brain and cerebellum (Fig. S7A and data not shown).
In summary, we found that during muscle differentiation there is an increase in Myc-nick, concomitant with an elevation in calpain activity and tubulin acetylation (Fig. 7B–D; Fig. 7F; Fig. S7B–D; Fig. S7F).
Myc-nick accelerates muscle differentiation
We examined the effects of ectopic expression of Myc-nick* in human primary myoblasts isolated from the pectoral girdle (Fig. 7H) and rectus abdominus (Fig, 7I–J), the human rhabdomyosarcoma cell line RD (Fig. 7K), and in C2C12 cells (Fig. 7L–M). In all cells we found that Myc-nick* expression accelerated muscle cell differentiation, augmented α-tubulin acetylation, and elevated expression of muscle specific proteins (Fig. 7H–M). The expression of Myc-nick* in C2C12 myoblasts promoted an increase in cell fusion not only in confluent cultures (Fig. 7L; Fig. S7H), but also in sparse cultures (Fig. S7G). The role of Myc-nick in promoting muscle cell differentiation is partially dependent on MBII, since the differentiation of RD and C2C12 cells by Myc-nick can be delayed, but not inhibited by the deletion of MBII (Fig. 7K; Fig. S7I).
To examine the requirement for Myc-nick production during differentiation further, we first tested calpain inhibitor XII and found that it reduces differentiation and tubulin acetylation (Fig. S7J). Next we employed c-Myc mutant (Δ291–300) which is deficient in cleavage to Myc-nick. Similar to full-length wt c-Myc, this mutant can induce proliferation, apoptosis, fibrillarin expression, and phosphorylated H2AX in Rat1 myc-null cells (Fig. S7L–M). However the Δ291–300 c-Myc mutant significantly reduces C2C12 cell differentiation when compared to c-Myc, even though the proteins are expressed at equal levels. While full-length c-Myc only blocks the fusion of C2C12 myoblasts, the c-Myc mutant (Δ291–300) with reduced ability to generate Myc-nick, dramatically reduces the expression of muscle markers in addition to blocking fusion (Fig. S7N–O).
Myc-nick renders Rat1 myc-null cells competent to differentiate
MyoD has been long known to induce the transdifferentiation in diverse cell types. We found that MyoD induces expression of muscle-specific markers in Rat1 fibroblasts but not in Rat1 myc-null fibroblasts (Fig. 7O), indicating that myc is necessary for the transdifferentiation process. Expressing Myc-nick together with MyoD in myc+/+ Rat1 fibroblasts further elevated the levels of muscle specific markers and promoted cell fusion compared to MyoD alone (Fig. 7N). Importantly, we find that in Rat1 myc-null cells, the co-expression of Myc-nick (Fig. S7K) permitted transdifferentiation to muscle in response to MyoD (Fig. 7P). This experiment indicates that Myc-nick is sufficient for Myo-D induced transdifferentiation supporting the idea that conversion of full-length Myc to Myc-nick is important in MyoD induced differentiation.
Discussion
Recent studies have demonstrated that Myc directly regulates transcriptional activation through the three RNA polymerases, as well as transcriptional repression, and DNA replication (Eilers and Eisenman, 2008). Here we identified and characterized, Myc-nick, a novel form of Myc performing transcription-independent functions in the cytoplasm. One of these functions is to regulate α-tubulin acetylation in cooperation with the HAT GCN5. Moreover, Myc-nick levels are elevated in differentiated muscle tissues and Myc-nick overexpression accelerates muscle cell differentiation.
Despite its widespread expression Myc-nick has not been previously characterized. There are several likely reasons for this. First, the most commonly used antibodies against Myc (such as 9E10) recognize its C-terminus and would not detect Myc-nick. Second, the proportion of Myc-nick to Myc increases when cells are grown as confluent cultures, conditions that are not commonly employed when studying Myc proteins. Third, most studies use total protein or nuclear extracts and have not analyzed the cytoplasmic pool of Myc. Fourth, Myc-nick may have been mistaken for the similarly sized MycS protein (Hann et al., 1988), a nuclear localized product of internal translation initiation.
Calpain cleavage as a post-translational functional switch
We found that Myc-nick is generated in the cytoplasm by calpain mediated cleavage of full-length Myc. Cytoplasmic cleavage by calpains has been reported for over 100 proteins including many cytoskeletal proteins, membrane receptors, and transcription factors (Tompa et al., 2004). This number is probably an underestimate because the lack of a clearly defined consensus cleavage site for calpains makes the identification of novel calpain substrates difficult. For most substrates the role of calpain cleavage is not known. However, there are several noteworthy examples of calpain cleavage operating as a functional switch (Abe and Takeichi, 2007);(Yousefi et al., 2006). Here we have shown that the cleavage of Myc by calpain converts this predominantly nuclear transcription factor into Myc-nick, a cytosolic factor that regulates α-tubulin acetylation. Based on our findings and the examples in the literature, we surmise that the partial proteolytic cleavage of proteins by calpains functions as an irreversible post-translational modification.
A role for Myc-nick in α-tubulin acetylation
We have shown that Myc-nick mediates the acetylation of α-tubulin by forming a complex with microtubules and the histone acetyl transferase (HAT) GCN5. GCN5 has been long known to associate with nuclear Myc through the highly conserved Myc Box II, a region retained in Myc-nick. Although tubulin acetylation was first described 20 years ago, little is known about the enzymes that catalyze this reaction. Recently Elp3, a histone acetyl transferase was demonstrated to be critical in acetylation of α-tubulin in cortical projection neurons, an event linked to neural differentiation and migration (Creppe et al., 2009). We surmise that GCN5 represents another acetyltransferase targeting tubulin. Deacetylation of α-tubulin is mediated by both HDAC6 and Sirt2 (Hubbert et al., 2002; Matsuyama et al., 2002; North et al., 2003). However the increase in acetylated α-tubulin by Myc-nick does not occur through modulation of HDAC activity (Fig. S 6E, F).
A connection between α-tubulin acetylation and calpain activation was previously suggested by two independent studies. First, calcium depletion, which inactivates calpains, was shown to decrease the levels of acetylated α-tubulin in epithelial cells (Ivanov et al., 2006). Second, the ectopic expression of calpain 6 causes microtubule stabilization, elevates the levels of acetylated α-tubulin and impairs cytokinesis in HeLa cells (Tonami et al., 2007). Our findings suggest that the induction of α-tubulin acetylation by calpains could be mediated at least in part by Myc-nick and GCN5.
The role of Myc-nick in terminal differentiation
The role of Myc family members in differentiation is complex. Endogenous Myc is strongly downregulated during terminal differentiation of many cell types. Moreover, ectopic expression of Myc has been shown to block terminal differentiation. The ability of Myc to negatively regulate differentiation is consistent with its role in maintaining pluripotency in ES cells and in the generation of iPS cells (Cartwright et al., 2005; Takahashi et al., 2007). However, Myc has also been implicated in promoting both proliferation and differentiation in specific cellular contexts such as in progenitor cells of the skin (Gandarillas and Watt, 1997) (Gebhardt et al., 2006), B lymphocytes (Habib et al., 2007) and hematopoietic stem cells (Wilson et al., 2004). Our data suggest that while full-length Myc blocks differentiation at the transcriptional level, Myc-nick may be involved in promoting differentiation through transcription independent mechanisms. In agreement with this hypothesis there have been numerous reports of Myc antigenicity localized predominantly in the cytoplasm of differentiated cells (see introduction). In addition, we show that ectopic expression of Myc-nick accelerates muscle cell differentiation and renders Rat1 myc-null cells competent to differentiate into muscle following introduction of MyoD.
A number of studies have shown a strong correlation between muscle cell differentiation, calcium influx and calpain activation, (Dedieu et al., 2004; Kumar et al., 1992). In addition, limb-girdle muscular dystrophy 2A (LGMD2A) is caused by mutations in calpain 3 that affect its activity (Richard et al., 1995). During the process of normal differentiation, myoblasts elongate and fuse into syncytial myotubes. An early event during this process is the remodeling of the microtubule cytoskeleton, involving disassembly of the centrosome and the alignment of stable microtubules into a parallel array along the long axis of the cell. We observed an increase in the levels of acetylated α-tubulin (an indicator of microtubule stabilization and cytoskeletal reorganization) and an elevation in Myc-nick abundance during muscle cell differentiation. Induction of α-tubulin acetylation and microtubule stabilization are likely to be important events in terminal differentiation especially when cells must establish and maintain a new shape. Acetylation of α-tubulin was also demonstrated to regulate cortical neuron differentiation and migration (Creppe et al., 2009).
We propose a model for the regulation of Myc by calpains and for the function of Myc-nick. During proliferation, full-length Myc is rapidly synthesized and transported to the nucleus where it transcriptionally activates growth and proliferation related genes and represses genes involved in differentiation. External cues that stimulate differentiation, such as cell-cell contact, and calcium influx can lead to the activation of calpains. Activated calpains interact with and may retain Myc in the cytoplasm where it is cleaved to produce Myc-nick. Since the cleavage removes the NLS and the DNA and Max binding domains, Myc-nick is predominantly cytoplasmic and transcriptionally inactive. Indeed we have shown that Myc-nick, when expressed in cells devoid of full-length Myc, does not stimulate proliferation, growth or apoptosis – the transcription dependent functions of full-length Myc. In such Myc-nick expressing cells we instead observe morphological changes and increased tubulin acetylation which are unique to Myc-nick. During muscle differentiation Myc-nick appears to be stabilized by dephosphorylation of threonine 58 within the Myc Box I phosphodegron. We propose that one of the functions carried out by Myc-nick in the cytoplasm involves binding tubulin and recruiting GCN5 to mediate α-tubulin acetylation. Myc-nick is also likely to have additional functions and partners in the cytoplasm since the deletion of MBII in Myc-nick, which should block the interaction with GCN5, only partially reduced the ability of Myc-nick to promote muscle differentiation. We predict that the functions carried out by Myc-nick in the cytoplasm cooperate to promote cytoskeletal changes that can further drive terminal differentiation. In our model, the cleavage of Myc by calpains has two roles. First, it helps diminish nuclear Myc abundance preventing newly synthesized Myc from entering the nucleus, therefore eliminating the transcriptional blockade to differentiation caused by Myc. Second, the production of Myc-nick influences cytoskeletal organization and facilitates terminal differentiation. We predict that Myc-nick will play a general role linking myc to both nuclear functions and cytoplasmic organization during differentiation of a wide variety of cell types.
Experimental Procedures
Retroviral infection and transfection
For retroviral production, 293T cells were cotransfected with pBabe-puro vector expressing Myc clones and amphotropic helper. Infected cells were selected with 2µg/ml puromycin for 4 days. Rat1 myc-null cells were harvested 10–14 days after infection and C2C12 cells were stimulated to differentiate after selection. For overexpression experiments 293T (transfected with Fugene-Roche) cells were harvested 3 days after transfection with a change in culture medium 24 h before harvesting. Sparse cultures (S) are about 20% confluent, medium cultures (M) are about 40–60% confluent and dense cultures (D) are grown as confluent cultures for 2–4 days.
Total cell lysates and nuclear and cytoplasmic fractionation
For total extracts cells were lysed in either boiling sample buffer or in RIPA buffer. For cellular fractionation cells were lysed in buffer A on ice for 20 minutes, centrifuged for 3 minutes and the supernatant employed as the cytoplasmic fraction. The pellet was resuspended in buffer B, rotated for 20 minutes, sonicated, and centrifuged for 10 minutes. The supernatant employed as the nuclear fraction. 10–20 µg of total extracts or 10µg of nuclear and 30 µg of cytoplasmic extracts were probed overnight with indicated antibodies. See Supplementary Experimental Procedures for composition of buffers.
Immunofluorescence
Cells were grown on glass coverslips and fixed with 4% paraformaldehyde for 20 minutes, permeabilized with 0.5% triton X100 for 5 minutes and blocked with Image IT FX signal enhancer (Invitrogen) for 30 minutes. Primary and secondary antibodies were diluted in PBS at 1:200 and incubated for 1h.
In vitro cleavage of Myc
All cDNAs were cloned into pCS2+ and were transcribed from the SP6 promoter using the Promega wheat germ system, in the presence of cold (Promega) or 35[S]- labeled methyonine (Perkin Elmer) according to the manufacturer’s instructions. For the in vitro cleavage experiments Myc (1 µl IVT Myc or 1 µg recombinant Myc) was incubated with 30µg of nuclear or cytoplasmic extracts in 20µl of buffer G at 37°C and the samples were processed for autoradiography or Western blot as specified. Nuclear and cytoplasmic fractions were dissolved in the same buffer and adjusted to identical salt concentration for these experiments. For Myc cleavage using recombinant calpains, 1 µl IVT c-Myc or 0.25µg of purified c-Myc were incubated with the indicated amounts of calpains in the presence of 20µl buffer G. Total calpain activity was measured using a Calbiochem kit.
In vitro tubulin acetylation Assays
CE (30µg) or purified assembled microtubules (1µg) were incubated with 1 µl of IVT c-Myc or IVT vector, or with recombinant Myc for 1h in the presence of acetylation buffer. Assembled microtubules were incubated at 30°C while CE at 37°C.
See Supplementary Experimental Procedures for buffer composition, constructs, siRNA sequence, pharmacological inhibitors, antibodies, cell lines and culture media.
Acknowledgements
We are grateful to Nao Ikegaki, William Tansey, John Sedivy and Peter Hurlin for reagents essential for this work. We also thank Stephen Tapscott, Maura Parker, Hector Rincon, Bill Carter, Linda Wordeman, and members of the Eisenman lab for advice and discussion. M. Conacci-Sorrell was a recipient of an EMBO Long-Term Fellowship. This work was supported by NIH/NCI grant CA20525 (R.N.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe K, Takeichi M. NMDA-receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron. 2007;53:387–397. doi: 10.1016/j.neuron.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Alexandrova N, Niklinski J, Bliskovsky V, Otterson GA, Blake M, Kaye FJ, Zajac-Kaye M. The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol Cell Biol. 1995;15:5188–5195. doi: 10.1128/mcb.15.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JS, Elce JS, Hegadorn C, Williams K, Greer PA. Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol Cell Biol. 2000;20:4474–4481. doi: 10.1128/mcb.20.12.4474-4481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MK, Costopoulos JS, Christoforidou BP, Papadimitriou CS. Immunohistochemical detection of the c-myc oncogene product in normal, hyperplastic and carcinomatous endometrium. Oncology. 1994;51:314–319. doi: 10.1159/000227356. [DOI] [PubMed] [Google Scholar]
- Calcagno DQ, Guimaraes AC, Leal MF, Seabra AD, Khayat AS, Pontes TB, Assumpcao PP, De Arruda Cardoso Smith M, Burbano RR. MYC insertions in diffuse-type gastric adenocarcinoma. Anticancer Res. 2009;29:2479–2483. [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Clement CA, Kristensen SG, Mollgard K, Pazour GJ, Yoder BK, Larsen LA, Christensen ST. The primary cilium coordinates early cardiogenesis and hedgehog signaling in cardiomyocyte differentiation. J Cell Sci. 2009;122:3070–3082. doi: 10.1242/jcs.049676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- Dedieu S, Poussard S, Mazeres G, Grise F, Dargelos E, Cottin P, Brustis JJ. Myoblast migration is regulated by calpain through its involvement in cell attachment and cytoskeletal organization. Exp Cell Res. 2004;292:187–200. doi: 10.1016/j.yexcr.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, Farina A, Martinez E. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25:10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas A, Watt FM. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 1997;11:2869–2882. doi: 10.1101/gad.11.21.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Frye M, Herold S, Benitah SA, Braun K, Samans B, Watt FM, Elsasser HP, Eilers M. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol. 2006;172:139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen H, Shkedy D, Barnoy S, Kosower NS, Ciechanover A. On the involvement of calpains in the degradation of the tumor suppressor protein p53. FEBS Lett. 1997;406:17–22. doi: 10.1016/s0014-5793(97)00225-1. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Khawaja S, Bulinski JC. Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J Cell Biol. 1989;109:2275–2288. doi: 10.1083/jcb.109.5.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib T, Park H, Tsang M, de Alboran IM, Nicks A, Wilson L, Knoepfler PS, Andrews S, Rawlings DJ, Eisenman RN, et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol. 2007;179:717–731. doi: 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Iezzi S, Di Padova M, Serra C, Caretti G, Simone C, Maklan E, Minetti G, Zhao P, Hoffman EP, Puri PL, et al. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev Cell. 2004;6:673–684. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- Ivanov AI, McCall IC, Babbin B, Samarin SN, Nusrat A, Parkos CA. Microtubules regulate disassembly of epithelial apical junctions. BMC Cell Biol. 2006;7:12. doi: 10.1186/1471-2121-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR, 3rd, Menssen A, Hermeking H. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle. 2007;6:205–217. doi: 10.4161/cc.6.2.3742. [DOI] [PubMed] [Google Scholar]
- Kumar A, Shafiq S, Wadgaonkar R, Stracher A. The effect of protease inhibitors, leupeptin and E64d, on differentiation of C2C12 myoblasts in tissue culture. Cell Mol Biol. 1992;38:477–483. [PubMed] [Google Scholar]
- Mailhes JB, Hilliard C, Lowery M, London SN. MG-132, an inhibitor of proteasomes and calpains, induced inhibition of oocyte maturation and aneuploidy in mouse oocytes. Cell Chromosome. 2002;1:2. doi: 10.1186/1475-9268-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyen C, Goudenege S, Poussard S, Sassi AH, Brustis JJ, Cottin P. Involvement of micro-calpain (CAPN 1) in muscle cell differentiation. Int J Biochem Cell Biol. 2004;36:728–743. doi: 10.1016/S1357-2725(03)00265-6. [DOI] [PubMed] [Google Scholar]
- Niklinski J, Claassen G, Meyers C, Gregory MA, Allegra CJ, Kaye FJ, Hann SR, Zajac-Kaye M. Disruption of Myc-tubulin interaction by hyperphosphorylation of c-Myc during mitosis or by constitutive hyperphosphorylation of mutant c-Myc in Burkitt's lymphoma. Mol Cell Biol. 2000;20:5276–5284. doi: 10.1128/mcb.20.14.5276-5284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Okano HJ, Park WY, Corradi JP, Darnell RB. The cytoplasmic Purkinje onconeural antigen cdr2 down-regulates c-Myc function: implications for neuronal and tumor cell survival. Genes Dev. 1999;13:2087–2097. doi: 10.1101/gad.13.16.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilainen T, Lipponen P, Aaltomaa S, Eskelinen M, Kosma VM, Syrjanen K. Expression of c-myc proteins in breast cancer as related to established prognostic factors and survival. Anticancer Res. 1995;15:959–964. [PubMed] [Google Scholar]
- Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- Small GW, Chou TY, Dang CV, Orlowski RZ. Evidence for involvement of calpain in c-Myc proteolysis in vivo. Arch Biochem Biophys. 2002;400:151–161. doi: 10.1016/S0003-9861(02)00005-X. [DOI] [PubMed] [Google Scholar]
- Spotts GD, Patel SV, Xiao Q, Hann SR. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira T, Maeda J, Onishi T, Kitaura H, Yoshida S, Kato H, Ikeda M, Tamai K, Iguchi-Ariga SM, Ariga H. AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC. Genes Cells. 1998;3:549–565. doi: 10.1046/j.1365-2443.1998.00206.x. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilagyi A, Banoczi Z, Hudecz F, Friedrich P. On the sequential determinants of calpain cleavage. J Biol Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- Tonami K, Kurihara Y, Aburatani H, Uchijima Y, Asano T, Kurihara H. Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol Cell Biol. 2007;27:2548–2561. doi: 10.1128/MCB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworkowski KA, Salghetti SE, Tansey WP. Stable and unstable pools of Myc protein exist in human cells. Oncogene. 2002;21:8515–8520. doi: 10.1038/sj.onc.1205976. [DOI] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Watanabe Y, Nakamura H, Kondoh H. Regulation of the neural crest cell fate by N-myc: promotion of ventral migration and neuronal differentiation. Development. 1997;124:1953–1962. doi: 10.1242/dev.124.10.1953. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Watanabe Y, Shimono A, Kondoh H. Transition of localization of the N-Myc protein from nucleus to cytoplasm in differentiating neurons. Neuron. 1993;10:1–9. doi: 10.1016/0896-6273(93)90236-k. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann SR. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]