Abstract
In a follow-up to our previously reported genome-wide association study of cutaneous basal cell carcinoma (BCC)1, we describe here several new susceptibility variants. SNP rs11170164, encoding a G138E substitution in the keratin 5 (KRT5) gene, affects risk of BCC (OR = 1.35, P = 2.1 × 10−9). A variant at 9p21 near CDKN2A and CDKN2B also confers susceptibility to BCC (rs2151280[C]; OR = 1.19, P = 6.9 × 10−9), as does rs157935[T] at 7q32 near the imprinted gene KLF14 (OR = 1.23, P = 5.7 × 10−10). The effect of rs157935[T] is dependent on the parental origin of the risk allele. None of these variants were found to be associated with melanoma or fair-pigmentation traits. A melanoma- and pigmentation-associated variant in the SLC45A2 gene, L374F, is associated with risk of both BCC and squamous cell carcinoma. Finally, we report conclusive evidence that rs401681[C] in the TERT-CLPTM1L locus confers susceptibility to BCC but protects against melanoma.
Cutaneous BCC is the most common cancer among people of European ancestry. The primary environmental risk factor for BCC is sun exposure, but genetics also has a substantial role. Some of the sequence variants that confer susceptibility seem to operate through their association with fair-pigmentation traits, common among Europeans, resulting in reduced protection from the damaging effects of UV radiation. Other sequence variants have no obvious role in pigmentation or UV susceptibility but instead seem to operate in the contexts of growth and differentiation of the basal layers of the skin1–4.
We previously conducted a genome-wide SNP association scan (GWAS) for common BCC risk variants. We discovered susceptibility variants at 1p36, 1q42 and 5p15 (TERT-CLPTM1L)1,4. Here we investigated 30 more high-ranking SNPs from the GWAS by genotyping them in an additional 903 Icelandic BCC cases and a case-control sample from Eastern Europe. SNPs at three loci were then selected for further investigation: rs11170164 in the keratin 5 (KRT5) gene, rs2151280 near the CDKN2A and CDKN2B genes and rs157935 on 7q32. These SNPs were further genotyped in case-control samples from Spain and the United States and proved to be significantly (P < 6 × 10−7) associated with BCC risk. An overview of the samples used in the study is presented in Supplementary Table 1; data for the 27 SNPs that were not studied further at this stage are listed in Supplementary Table 2. We also examined whether the three new BCC variants are concomitantly associated with squamous cell carcinoma (SCC), cutaneous melanoma (CM) and fair-pigmentation traits. We then used the same approach to investigate recently described variants in SLC45A2 and TERT-CLPTM1L4–6.
After genotyping the Icelandic and non-Icelandic replication samples, a combined OR of 1.35 (P = 2.1 × 10−9) was detected for rs11170164[A], specifying a G138E substitution in KRT5 (Table 1). Because this P value is below the Bonferroni threshold for genome-wide significance (P = 1.6 × 10−7) and the association replicated consistently in different populations (OR = 1.49, P = 4.6 ×10−6 in the non-Icelandic samples), we conclude that the G138E substitution confers susceptibility to BCC. The KRT5 gene product K5 and its heterodimeric partner K14 are the major keratins of basal epithelial cells, forming the intermediate filament cytoskeletal network. This network is crucial for the structural integrity of the basal cell layer7.
Table 1.
Association of SNPs in KRT5 and the 9p21 and 7q32 loci with BCC, SCC and CM
| SNP | Allele | Locus | Sample group | Number |
Frequency |
OR | 95% CI | P | Pheta | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||||||
| rs11170164 | A | KRT5 (G138E) | Iceland BCC | 1,833 | 34,845 | 0.109 | 0.086 | 1.29 | (1.14, 1.46) | 4.4 × 105 | |
| Eastern Europe BCC | 525 | 532 | 0.122 | 0.073 | 1.75 | (1.31, 2.35) | 1.6 × 10−4 | ||||
| US BCC | 930 | 849 | 0.097 | 0.071 | 1.40 | (1.10, 1.77) | 6.1 × 10−3 | ||||
| Spain BCC | 185 | 1,689 | 0.065 | 0.052 | 1.28 | (0.81, 2.01) | 0.29 | ||||
| All non-Icelandic BCC | 1,640 | 3,070 | NA | NA | 1.49 | (1.26, 1.77) | 4.6 × 10−6 | 0.38 | |||
| All BCC combined | 3,473 | 37,915 | NA | NA | 1.35 | (1.23, 1.50) | 2.1 × 10−9 | 0.29 | |||
| Iceland SCCb | 434 | 34,845 | 0.111 | 0.086 | 1.31 | (1.05, 1.64) | 1.8 × 10−2 | ||||
| US SCC | 710 | 849 | 0.082 | 0.071 | 1.16 | (0.89, 1.51) | 0.27 | ||||
| All SCC combined | 1,144 | 35,694 | NA | NA | 1.25 | (1.05, 1.48) | 1.2 × 10−2 | 0.49 | |||
| All CMc combined | 3,932 | 39,643 | NA | NA | 1.03 | (0.92, 1.15) | 0.58 | 0.11 | |||
| rs2151280 | C | 9p21 CDKN2A/B | Iceland BCC | 1,831 | 34,998 | 0.581 | 0.531 | 1.22 | (1.13, 1.31) | 7.6 × 10−8 | |
| Eastern Europe BCC | 525 | 524 | 0.548 | 0.517 | 1.13 | (0.95, 1.34) | 0.16 | ||||
| US BCC | 871 | 826 | 0.537 | 0.505 | 1.13 | (0.99, 1.30) | 6.8 × 10−2 | ||||
| Spain BCC | 185 | 1,687 | 0.514 | 0.486 | 1.12 | (0.90, 1.38) | 0.32 | ||||
| All non-Icelandic BCC | 1,581 | 3,037 | NA | NA | 1.13 | (1.03, 1.24) | 1.2 × 10−2 | 0.99 | |||
| All BCC combined | 3,412 | 38,035 | NA | NA | 1.19 | (1.12, 1.26) | 6.9 × 10−9 | 0.65 | |||
| Iceland SCCb | 437 | 34,998 | 0.534 | 0.531 | 1.01 | (0.90, 1.13) | 0.87 | ||||
| US SCC | 642 | 826 | 0.532 | 0.505 | 1.11 | (0.96, 1.29) | 0.15 | ||||
| All SCC combined | 1,079 | 35,824 | NA | NA | 1.05 | (0.96, 1.15) | 0.31 | 0.31 | |||
| All CMc combined | 3,897 | 39,779 | NA | NA | 1.01 | (0.95, 1.07) | 0.74 | 0.88 | |||
| rs157935 | T | 7q32 | Iceland BCC | 1,832 | 34,963 | 0.722 | 0.675 | 1.25 | (1.15, 1.35) | 3.6 × 10−8 | |
| Eastern Europe BCC | 526 | 530 | 0.723 | 0.693 | 1.16 | (0.96, 1.40) | 0.13 | ||||
| US BCCd | 723 | 611 | 0.740 | 0.691 | 1.27 | (1.07, 1.50) | 5.6 × 10−3 | ||||
| Spain BCC | 183 | 1,703 | 0.730 | 0.718 | 1.06 | (0.83, 1.35) | 0.63 | ||||
| All non-Icelandic BCC | 1,432 | 2,844 | NA | NA | 1.18 | (1.06, 1.32) | 3.2 × 10−3 | 0.47 | |||
| All BCC Combined | 3,264 | 37,807 | NA | NA | 1.23 | (1.15, 1.31) | 5.7 × 10−10 | 0.55 | |||
| Iceland SCCb | 433 | 34,963 | 0.666 | 0.675 | 0.96 | (0.79, 1.17) | 0.66 | ||||
| US SCCd | 513 | 611 | 0.712 | 0.691 | 1.11 | (0.92, 1.34) | 0.28 | ||||
| All SCC combined | 946 | 35,574 | NA | NA | 1.03 | (0.90, 1.18) | 0.63 | 0.29 | |||
| All CMc combined | 3,921 | 39,770 | NA | NA | 1.00 | (0.94, 1.06) | 0.94 | 0.35 | |||
| BCC paternal allelee | 1,118f | 34,869g | 0.745 | 0.676 | 1.40 | (1.22, 1.60) | 1.5 × 10−6 | ||||
| BCC maternal allelee | 1,118f | 34,869g | 0.694 | 0.675 | 1.09 | (0.96, 1.25) | 0.18 | ||||
BCC, cutaneous basal cell carcinoma. SCC, cutaneous squamous cell carcinoma. CM, cutaneous melanoma (malignant or in situ).
I value for heterogeneity.
Cases diagnosed with SCC without BCC.
Contributing CM sample groups (and approximate numbers) were from Iceland (589 cases, 34,998 controls), Holland (749 cases, 1,831 controls), Sweden (1,065 cases, 540 controls), Austria (152 cases, 376 controls), Spain (816 cases, 1,703 controls) and Italy (564 cases, 368 controls).
SNP rs157935 could not be typed in the US BCC and SCC samples so a surrogate SNP, rs125124, was used (r2 = 1.0 in HapMap CEU). Frequency and OR are given for the rs125124[C] allele.
Analysis based on Icelandic cases and controls who were typed by an Illumina chip and for whom the Icelandic genealogy and long-range phasing haplotypes were used to determine parental origins of alleles.
Number of paternal/maternal alleles examined in cases.
Number of paternal/maternal alleles examined in controls.
Some sequence variants, particularly those controlling pigmentation trait variation, simultaneously affect risks of BCC, SCC and CM1,3. Combining the Icelandic and US data for SCC revealed an OR of 1.25 (P = 0.012) for rs11170164[A] (Table 1), providing suggestive evidence of a concomitant risk of SCC. We also searched for an association with CM in 3,932 cases and 39,276 controls (Supplementary Table 1). Despite the large sample size, there was no evidence to indicate that G138E affects risk of CM (Table 1). We looked for evidence of association with eye color, hair color, freckles and sun sensitivity using self-reported data from approximately 6,200 Icelanders8,9. There was no evidence of an association with any pigmentation trait (Supplementary Table 3) nor was there any difference in frequency of rs11170164[A] between individuals with BCC lesions at typically sun-exposed (head and arms) versus those with lesions at unexposed (trunk and legs) body sites (P = 0.09). Together, these data suggest that the G138E variant affects BCC susceptibility through mechanisms other than those related to obvious pigmentation characteristics.
We expanded our investigation of KRT5 to include six more exonic polymorphisms that are common in Europeans, including three additional nonsynonymous SNPs (nsSNPs). Two of the nsSNPs showed nominally significant signals: rs641615 (D197E) and rs11549949 (G543S) (Supplementary Table 4). For both, BCC risk was associated with the reference allele. The signal from G543S lost significance when multiple testing was taken into consideration. D197E gave a combined OR of 1.21 (P = 2.2 × 10−5) (Table 2). SNP rs641615 is in the same linkage disequilibrium (LD) block as rs11170164 (G138E) although the frequencies differ markedly (D′ = 1.00, r2 = 0.03). In a multivariate analysis, the signal from D197E remained significant when adjusted for the effect of G138E (Table 2), indicating that the two substitutions affect risk independently. However, it remains possible that another unobserved variant that is in LD with both substitutions is responsible for the BCC susceptibility. The signal from G543S also retained its nominal significance when adjusted for the joint effects of G138E and D197E (Supplementary Table 4), raising the possibility, as yet unproven, of a third independent risk variant in KRT5.
Table 2.
KRT5 rs641615 D197E is associated with BCC independently of rs11170164 G138E
| Sample group | Number |
Frequency |
rs641615[C]a unadjusted |
rs641615[C] adjusted for rs11170164 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | OR | 95% CI | P | OR | Residual P | |
| Iceland BCC | 1,674 | 2,462 | 0.79 | 0.76 | 1.17 | (1.05, 1.30) | 3.8 × 10−3 | 1.13 | 3.5 × 10−2 |
| Eastern Europe BCC | 528 | 532 | 0.77 | 0.72 | 1.31 | (1.08, 1.60) | 6.9 × 10−3 | 1.24 | 3.7 × 10−2 |
| Spain BCC | 182 | 1,687 | 0.76 | 0.71 | 1.26 | (0.99, 1.62) | 6.2 × 10−2 | 1.24 | 8.7 × 10−3 |
| All BCC combinedb | 2,384 | 4,681 | NA | NA | 1.21 | (1.11, 1.32) | 2.2 × 10−5 | 1.17 | 1.0 × 10−3 |
Note that increased BCC risk is associated with the reference (Asp) allele of the D197E substitution encoded by rs641615.
Phet = 0.55 for unadjusted ORs.
Gly138 is highly conserved in vertebrates, and the Gly-to-Glu change is physicochemically nonconservative. We used a battery of predictive tests to evaluate whether G138E has an impact on K5 structure and function (Supplementary Table 5). Not all tests agreed, but the consensus was that G138E is probably damaging. Panther, for example, returned a probability of 96.6% that the substitution is deleterious. Evidence that D197E affects protein structure was less clear cut, whereas evidence for an effect of G543S was weak (Supplementary Table 5).
Some rare mutations affecting the basal cell keratins K5 and K14 cause epidermolysis bullosa (EB), a blistering condition in which the basal cell layer ruptures and breaks away from the underlying dermis10. Various forms of EB exist, ranging from mild blistering conditions to lethal failures of dermatogenesis. As with all keratins, K5 consists of a central α-helical rod flanked by nonhelical head and tail domains. EB mutations tend to cluster around the helix-initiating and helix-terminating regions at either end of the rod10. The G138E and D197E variants both occur in the helix-initiating region (Supplementary Fig. 1). The two variants were originally discovered during linkage searches for EB mutations but were discounted as neutral polymorphisms11. However, subsequent reports indicate that when G138E and D197E occur as compound heterozygotes with high-penetrance EB mutations, they may be associated with a more extreme EB phenotype12,13.
A second BCC predisposition locus was identified in the LD block on 9p21 containing the cyclin-dependent kinase inhibitor genes CDKN2A and CDKN2B, the ARF tumor suppressor encoded by CDKN2A and the noncoding RNA ANRIL14,15 (Fig. 1 and Table 1). CDKN2A is well known for its involvement in familial melanoma16. Curiously, no risk of CM was associated with rs2151280, and we saw no evidence of an association with risk of SCC (Table 1). There was no association between rs2151280 and any of the pigmentation traits tested (Supplementary Table 3).
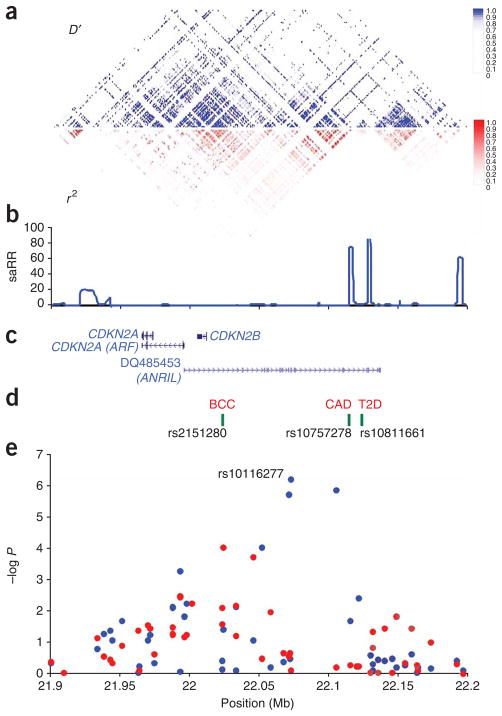
Figure 1.
Schematic view of the LD structure of the 9p21 CDKN2A/B region, locations of relevant genes and genome-wide association data for BCC and coronary artery disease (CAD). (a) Pairwise correlation structure of a 300-kb interval from 21.9 Mb to 22.2 Mb (NCBI Build 36) on 9p21. The upper plot (blue) shows the pairwise D′ values for SNPs with minor allele frequencies >5% from the HapMap v22 CEU dataset. The lower plot shows the corresponding r2 values. (b) Estimated recombination rates (saRR) in cM/Mb from the HapMap Phase II data. (c) Location of RefSeq genes and the ANRIL transcript. (d) Locations of the SNPs giving the most significant signals for BCC, CAD and T2D. (e) Illumina chipderived genome-wide association data for BCC (red dots) and CAD (blue dots). Note that the overall most significant CAD SNP rs10757278 is not represented on the Illumina chip but is highly correlated with rs10116277 (D′ = 0.96, r2 = 0.90 in HapMap CEU).
Previous reports indicated that rs10757278 in the CDKN2A/B LD block predisposes to coronary artery disease (CAD) (Fig. 1)17. Another SNP nearby (rs10811661) is associated with type 2 diabetes (T2D)18–20. The LD relationships between these SNPs and the BCC SNP rs2151280 are shown in Supplementary Table 6. We genotyped rs10757278 and rs10811661 in the Icelandic BCC case-control set. Similarly, we investigated our CAD and T2D GWAS data17,21 for associations with rs2151280. The CAD SNP rs10757278 and the T2D SNP rs10811661 showed no association with BCC risk, and the BCC SNP rs2151280 was not associated with risk of T2D (Supplementary Table 7). The rs2151280[C] allele showed a nominal association with reduced risk of CAD (OR = 0.91, P = 1.3 × 10−3) but this signal disappeared after adjustment for rs10757278 (residual P = 0.42; Supplementary Table 7). The simplest interpretation of these findings is that three distinct signals exist conferring separate risks of BCC, CAD and T2D. This implies that, in addition to the high-penetrance melanoma susceptibility variants in CDKN2A, at least three independent causative variants are present at 9p21, each having a quite distinct phenotypic effect.
A third new BCC susceptibility locus was tagged by rs157935 on 7q32 within fragile site FRA7H22. On replication, the T allele gave an OR of 1.23 (P = 5.7 × 10−10) (Table 1). The SNP showed no association with CM, SCC or pigmentation traits (Table 1 and Supplementary Table 3). The closest RefSeq gene is KLF14, encoding a Kruppel-like transcription factor that shows monoallelic maternal expression23. KLF14 is 167 kb proximal to rs157935 and is separated from it by a region of high recombination. To investigate a possible effect of imprinting on the pattern of risk, we used the Icelandic genealogy and long-range phasing24 to determine the parent-of-origin of the rs157935 alleles inherited by the Icelandic subjects who had been typed on the Illumina platform (see Online Methods). The relative risk of the T allele was estimated as 1.40 (P = 1.5 × 10−6) when inherited paternally but only 1.09 (and not significant) when inherited maternally (Table 1). As a direct test of whether the effect of the Tallele depends significantly on parent-of-origin, we observed that 237 heterozygous cases had inherited the T allele paternally whereas only 182 had inherited the T allele maternally (P = 9.6 × 10−3; see Online Methods). These results suggest that the increased BCC risk occurs mainly or exclusively through paternal transmission of the risk allele and is probably related to the imprinting at this locus. The rs157935 LD block contains microRNAs miR-29a and miR-29b-1, which include DNA methyltransferases among their targets25. Further investigation of this locus is warranted, and any mechanistic model should consider that, although KLF14 is expressed from the maternal allele, the increased BCC risk seems to associate with the paternal allele.
A nsSNP in SLC45A2 (previously MATP) has recently been shown to confer susceptibility to melanoma in Spanish and French populations5,6. The risk variant is the common reference allele of L374F (rs16891982) and is highly associated with fair pigmentation in Europeans26. Here we confirmed the association with fair pigmentation using samples from Iceland, Eastern Europe, Spain and the United States, observing strong associations with all traits except red hair and freckles (Supplementary Table 3). We also confirmed the association with CM risk, noting that the OR was lower in Iceland than abroad (Phet for Icelandic versus non-Icelandic samples = 0.059, I2 = 72%; Table 3). We have previously remarked that other pigmentation-associated variants tend to confer less relative risk of CM in Iceland than in other countries3. This may be a result of little sun exposure among Icelanders. In line with the notion that variants associated with both CM and pigmentation traits are expected to confer cross-risk of nonmelanoma skin cancers, we found that L374F also confers significant risk of BCC and SCC (Table 3). Again, we noted that the effect of the variant on BCC risk in Iceland was less than abroad (Phet for Icelandic versus non-Icelandic samples = 5.0 × 10−3, I2 = 87%; Table 3).
Table 3.
Association of SLC45A2 L374F rs16891982[G] with BCC, SCC and CM
| Sample group | Number |
Frequency |
OR | 95% CI | P | Phet | ||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||||
| Iceland BCC | 1,690 | 2,456 | 0.987 | 0.982 | 1.38 | (0.97, 1.96) | 7.3 × 10−2 | |
| US BCC | 927 | 843 | 0.984 | 0.953 | 2.99 | (2.00, 4.47) | 8.9 × 10−8 | |
| Eastern Europe BCC | 523 | 522 | 0.967 | 0.945 | 1.67 | (1.09, 2.55) | 1.8 × 10−2 | |
| Spain BCC | 186 | 1,672 | 0.917 | 0.829 | 2.27 | (1.61, 3.20) | 3.0 × 10−6 | |
| All BCC combined | 3,326 | 5,493 | NA | NA | 1.97 | (1.63, 2.38) | 1.6 × 10−12 | 0.026 |
| Iceland SCCb | 420 | 2,456 | 0.992 | 0.982 | 2.22 | (1.11, 4.42) | 2.3 × 10−2 | |
| US SCC | 699 | 843 | 0.984 | 0.953 | 2.94 | (1.90, 4.54) | 1.2 × 10−6 | |
| All SCC combined | 1,119 | 3,299 | NA | NA | 2.71 | (1.88, 3.92) | 1.0 × 10−7 | 0.5 |
| Iceland CM | 555 | 2,456 | 0.988 | 0.982 | 1.58 | (0.90, 2.76) | 0.11 | |
| Holland CM | 747 | 1,777 | 0.990 | 0.975 | 2.56 | (1.56, 4.20) | 2.0 × 10−4 | |
| Sweden CM | 1,061 | 541 | 0.988 | 0.957 | 3.58 | (2.23, 5.75) | 1.4 × 10−7 | |
| Austria CM | 152 | 376 | 0.984 | 0.964 | 2.23 | (0.92, 5.42) | 7.7 × 10−2 | |
| Italy CM | 560 | 367 | 0.949 | 0.900 | 2.06 | (1.44, 2.95) | 7.7 × 10−5 | |
| Spain CM | 805 | 1,672 | 0.935 | 0.829 | (2.42, 3.60) | 2.95 | 8.5 × 10−27 | |
| All CM combined | 3,880 | 7,189 | NA | NA | 2.95 | (2.42, 3.60) | 8.3 × 10−39 | 0.15 |
Note that increased risk is associated with the reference (Leu) allele of the L374F variant encoded by rs16891982.
Cases diagnosed with SCC without BCC.
We previously reported that rs401681 in the TERT-CLPTM1L locus affects risk of BCC and other cancers4. We also discovered evidence that the BCC risk allele rs401681[C] confers protection against CM. Here we sought to confirm this observation by typing additional CM case-control samples from Holland, Austria and Italy and larger control sample numbers from Iceland, Sweden and Spain (Table 4). Combined data from only the three new case-control sample sets revealed an OR for CM of 0.83 (P = 9.4 × 10−5) for the rs401681[C] allele, providing an independent confirmation of the initial finding (Table 4). Combining all the samples tested, the OR for CM was estimated as 0.86 (P = 5.0 × 10−8). Typing additional BCC samples from Spain and the United States also provided independent confirmation of the association with BCC, albeit with a considerably lower OR than observed in Iceland (Table 4). There was no evidence from the combined Icelandic and US sample data to support an association of rs401681 with SCC.
Table 4.
Association of TERT-CLPTM1L rs401681 with BCC, SCC and CM
| Sample group | Risk allele | Number |
Frequency |
OR | 95% CI | P | Phet | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||||
| Iceland BCCa | C | 1,850 | 34,998b | 0.60 | 0.55 | 1.23 | (1.14, 1.33) | 6.0 × 10−8 | |
| Eastern Europe BCCa | C | 525 | 525 | 0.62 | 0.57 | 1.19 | (1.00, 1.42) | 4.5 × 10−2 | |
| US BCC | C | 908 | 826 | 0.59 | 0.56 | 1.12 | (0.98, 1.28) | 0.10 | |
| Spain BCC | C | 185 | 1,758 | 0.55 | 0.54 | 1.08 | (0.86, 1.35) | 0.50 | |
| All non-Icelandic BCC | C | 1,618 | 3,109 | NA | NA | 1.13 | (1.03, 1.25) | 1.1 × 10−2 | 0.74 |
| All BCC combined | C | 3,468 | 38,107 | NA | NA | 1.20 | (1.13, 1.27) | 4.8 × 10−9 | 0.53 |
| Iceland SCCa | C | 438 | 34,998b | 0.57 | 0.55 | 1.08 | (0.94, 1.24) | 0.28 | |
| US SCC | C | 665 | 826 | 0.57 | 0.56 | 1.01 | (0.87, 1.17) | 0.92 | |
| All SCC combined | C | 1,103 | 35,824 | NA | NA | 1.04 | (0.94, 1.16) | 0.39 | 0.51 |
| Iceland CMa | C | 591 | 34,998b | 0.52 | 0.55 | 0.90 | (0.80, 1.01) | 7.9 × 10−2 | |
| Sweden CMa | C | 1,056 | 2,631 | 0.49 | 0.54 | 0.85 | (0.77, 0.94) | 1.2 × 10−3 | |
| Spain CMa | C | 748 | 1,758 | 0.51 | 0.54 | 0.90 | (0.80, 1.02) | 9.4 × 10−2 | |
| Holland CM | C | 736 | 1,832 | 0.53 | 0.57 | 0.83 | (0.73, 0.94) | 3.9 × 10−3 | |
| Austria CM | C | 152 | 376 | 0.53 | 0.53 | 0.98 | (0.75, 1.27) | 0.88 | |
| Italy CM | C | 560 | 368 | 0.49 | 0.56 | 0.74 | (0.62, 0.89) | 1.2 × 10−3 | |
| All new CMc | C | 1,448 | 2,576 | NA | NA | 0.83 | (0.76, 0.91) | 9.4 × 10−5 | 0.21 |
| All CM combined | C | 3,843 | 41,963 | NA | NA | 0.86 | (0.81, 0.91) | 5.0 × 10−8 | 0.41 |
Updated data from Rafnar et al.4
Skin cancer-free controls.
Combined CM data from Holland, Austria and Italy.
The rs401681[C] allele may be associated with an acceleration of the gradual shortening of telomeres with age4. Shorter telomeres in peripheral blood cells have been associated with an increased risk of BCC whereas longer telomeres are a risk factor for CM and nevi27,28. This is probably related to the different roles of replicative senescence in basal keratinocytes and melanocytes27. The inverse relationship between rs401681[C] and risks of BCC and CM is perhaps reflective of this difference.
To evaluate the impact of known common variants on the risk of BCC, we determined the joint population attributable risk (PAR; see Online Methods). Even without including the high-frequency SLC45A2 variant, we estimate a joint PAR of 74% for the BCC susceptibility loci we report here together with the previously known BCC susceptibility variants in ASIP, TYR, MC1R, 1p36, 1q42 and TERT-CLPTM1L1,3,4 (Supplementary Table 8). Thus, these sequence variants have some role in most cases of BCC.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
Acknowledgments
We thank G.H. Olafsdottir and L. Tryggvadottir of the Icelandic Cancer Registry for assistance with the ascertainment of affected individuals, S. Sveinsdottir for assistance with subject recruitment, and H. Sigurdsson for assistance with the figures. We are grateful to I. Saaf for permission to reproduce a section of the Human Intermediate Filament Database in Supplementary Figure 1. This study was supported in part by the Jubilaumsfonds of the Austrian National Bank (project numbers 11946 and 12161), the Swedish Cancer Society, the Radiumhemmet Research Funds and the Swedish Research Council, the US National Institute of Environmental Health Sciences (T32E007155), the US National Institutes of Health (R01CA082354 and R01CA57494), and a Research Investment Grant of the Radboud University Nijmegen Medical Centre.
Footnotes
Accession codes. GenBank: KRT5, NP_000415; KRT5 mRNA, NM_000424; SLC45A2, NP_001012527; ANRIL, DQ485453.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
The study was designed and results were interpreted by S.N.S., P.S., J.R.G., T.R., A.K., J.H.O., U.T. and K.S. Patient ascertainment and recruitment was organized and carried out by S.N.S., B. Sigurgeirsson, K.R.B., K.T., R.R., D.S., K.H., P.R., E.G., K. Koppova, R.B.-E., V. Soriano, P.J., B. Saez, Y.G., V.F., C.C., M.G., V.H., A.L., J.J.B., M.M.v.R., K.K.H.A., E.d.V., M.S., M.G.D.M., A.M., J.W., P.H., H.P., J.G., S.G., H.H., V. Steinthorsdottir, K. Kristjansson, G.B., I.O., L.R., M.R., L.A.K., J.H., E.N., J.I.M., R.K., M.R.K., H.H.N., J.H.O. and U.T. Biological material collection and handling was supervised by S.N.S., D.S., P.R., E.G., K. Koppova, B. Saez, V.H., A.L., K.K.H.A., J.S., H.B., I.O., M.R., M.R.K., H.H.N. and U.T. Genotyping was supervised by S.N.S., M.J., A.S., J.G., D.N.M., S.G., H.H., V. Steinthorsdottir, T.B., T.R. and U.T. Statistical analysis was carried out by P.S., G.T., D.F.G., M.L.F. and A.K. Bioinformatic analysis was carried out by S.N.S., S.A.G. and G.M. Authors S.N.S., P.S., A.K. and K.S. drafted the manuscript. All authors contributed to the final version of the paper. Principal collaborators for the case-control population samples were J.H.O. (Iceland), H.H.N. and M.R.K. (USA), R.K. (Eastern Europe), J.I.M. and E.N. (Spain), J.H. (Sweden), L.A.K. (The Netherlands), M.R. (Italy) and I.O. (Austria).
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/.
References
- 1.Stacey SN, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40:1313–1318. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 2.Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer. 2008;8:743–754. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudbjartsson DF, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 4.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guedj M, et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum Mutat. 2008;29:1154–1160. doi: 10.1002/humu.20823. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez LP, et al. SLC45A2: a novel malignant melanoma-associated gene. Hum Mutat. 2008;29:1161–1167. doi: 10.1002/humu.20804. [DOI] [PubMed] [Google Scholar]
- 7.Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulem P, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 9.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 10.Uitto J, Richard G, McGrath JA. Diseases of epidermal keratins and their linker proteins. Exp Cell Res. 2007;313:1995–2009. doi: 10.1016/j.yexcr.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan YM, Yu QC, Fine JD, Fuchs E. The genetic basis of Weber-Cockayne epidermolysis bullosa simplex. Proc Natl Acad Sci USA. 1993;90:7414–7418. doi: 10.1073/pnas.90.15.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalewski C, et al. A novel autosomal partially dominant mutation designated G476D in the keratin 5 gene causing epidermolysis bullosa simplex Weber-Cockayne type: a family study with a genetic twist. Int J Mol Med. 2007;20:75–78. [PubMed] [Google Scholar]
- 13.Shurman D, et al. Epidermolysis Bullosa Simplex with mottled pigmentation: mutation analysis in the first reported Hispanic pedigree with the largest single generation of affected individuals to date. Eur J Dermatol. 2006;16:132–135. [PubMed] [Google Scholar]
- 14.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Pasmant E, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Hocker TL, Singh M, Tsao H. Genetics of melanoma predisposition. Br J Dermatol. 2008;159:286–291. doi: 10.1111/j.1365-2133.2008.08682.x. [DOI] [PubMed] [Google Scholar]
- 17.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 18.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 19.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 22.Mishmar D, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc Natl Acad Sci USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker-Katiraee L, et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3:e65. doi: 10.1371/journal.pgen.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 2005;25:278–284. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- 27.Han J, et al. A prospective study of telomere length and the risk of skin cancer. J Invest Dermatol. 2008;129:415–421. doi: 10.1038/jid.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bataille V, et al. Nevus size and number are associated with telomere length and represent potential markers of a decreased senescence in vivo. Cancer Epidemiol Biomarkers Prev. 2007;16:1499–1502. doi: 10.1158/1055-9965.EPI-07-0152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.