Abstract
The Polycomb group (PcG) proteins are epigenetic suppressors of gene expression that function through modification of histones to change chromatin structure and modulate gene expression and cell behavior. Recent studies show that PcG proteins are expressed in epidermis, that their levels change during differentiation and in disease states, and that PcG expression is regulated by agents that influence cell proliferation and survival. The results indicate that PcG proteins regulate keratinocyte cell-cycle progression, apoptosis, senescence, and differentiation. These proteins are expressed in progenitor cells, in the basal layer, and in suprabasal keratinocytes, and the level, timing, and distribution of expression suggest that the PcG proteins have a central role in maintaining the balance between cell survival and death in multiple epidermal compartments. Additional studies indicate an important role in skin cancer progression.
INTRODUCTION
Polycomb group genes
The role of epigenetic regulation in modulating cell function is an area of intense interest. The Polycomb group (PcG) genes encode a family of evolutionarily conserved epigenetic regulators that were discovered in Drosophila as repressors of the genes that control body segmentation. In mammalian systems PcG proteins regulate genes involved in development, differentiation, and survival through epigenetic (for example, chromatin modification) mechanisms (Orlando, 2003; Valk-Lingbeek et al., 2004). Gene silencing by PcG proteins involves the sequential action of two polycomb repressor complexes—PRC2 and PRC1. The PRC2 protein complex includes four core proteins—Ezh2, Suz12, eed, and RBAP48 (Figure 1). The catalytic subunit of this complex is Ezh2, a methyltransferase that methylates H3-K27 through its SET domain-encoded catalytic site (Simon and Lange, 2008). The complex contains three noncatalytic subunits, including Suz12, eed (embryonic ectoderm development), and RBAP48 (retinoblastoma-binding protein p48). Suz12 and eed are required for optimal Ezh2 histone methyltransferase activity. The complex is not invariant and alternative subunits can be substituted including Ezh1 for Ezh2, RBAP46 for RBAP48, and there are several eed variants derived from a common gene (Simon and Kingston, 2009). Interaction of Suz12 and eed with Ezh2 results in a 1,000-fold increase in Ezh2 catalytic activity, showing that the full complex is required for optimal trimethylated histone H3 lysine K27 (H3K27me3) formation (Cao and Zhang, 2004; Pasini et al., 2004).
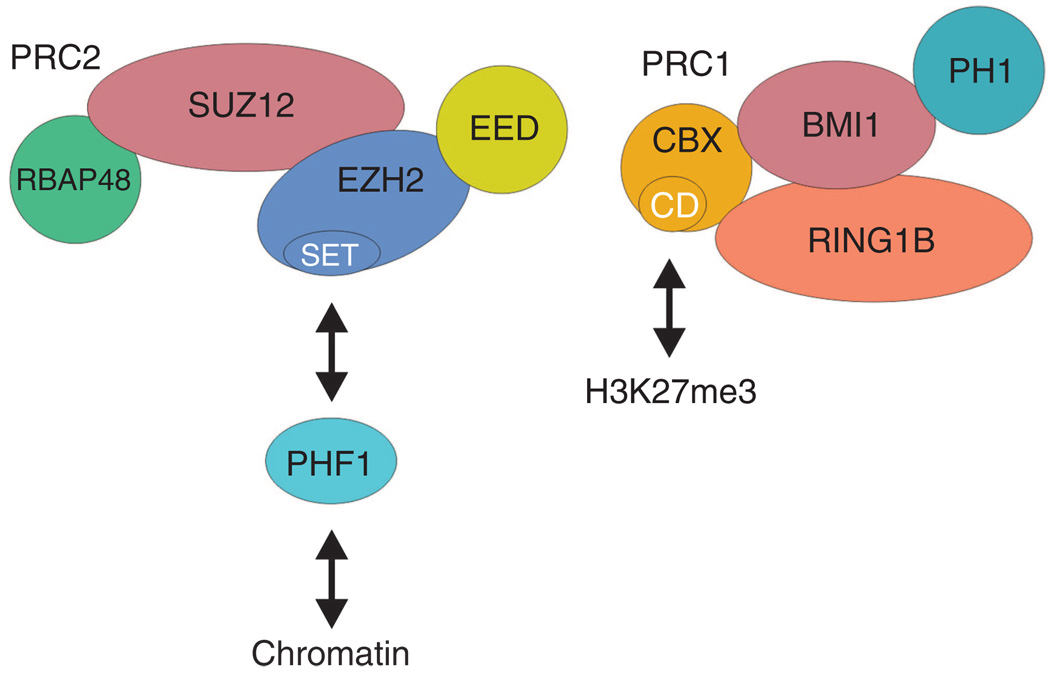
Figure 1. Polycomb complexes.
The PRC2 complex consists of four core proteins including RBAP48, SUZ12, EZH2, and EED. EZH2 is the methyltransferase responsible for methylation of lysine 27 of histone H3, and this methylation activity resides in the SET domain. Interaction with other proteins in the complex is required for EZH2 activity. PHF1 is a polycomb protein that is not part of the core complex, but functions to increase EZH2 activity and is also involved in PRC2 complex interaction with chromatin. The PRC1 complex comprises four core proteins including RING1B, CBX, Bmi-1, and PH1. RING1B is an E3 ubiquitin ligase that specifically ubiquitinylates lysine 119 of histone H2A. Bmi-1 interacts directly with RING1B to enhance Ring1B ubiquitin ligase activity. CBX encodes the domain (chromodomain, CD) responsible for anchoring the PRC1 complex to methylated lysine 27 of histone H3.
The PRC1 protein complex includes a core of four proteins including Ring1B, Bmi-1, PH1, and CBX (Simon and Kingston, 2009). The catalytic subunit of this complex, Ring1B, ubiquitinylates H2A-K119 and is optimally active in association with Bmi-1 (Li et al., 2006). An important role of the CBX protein is interaction with H3K27me3 to anchor the PRC1 complex to chromatin (Hatano et al., 2010). The PRC1 complex is also not invariant and proteins can be substituted including Ring1 for Ring1B, MEL18, or NSPC1 for Bmi-1, CBX2, 6, 7, or 8 for CBX, and PH2 for PH1 (Orlando, 2003; Levine et al., 2004; Sparmann and van Lohuizen, 2006; Simon and Kingston, 2009). PRC1 has been proposed to ubiquitinylate H2A-K119 as part of the process leading to a closed chromatin state. However, recent findings suggest that a second complex, BCOR, which consists of Ring1B, Ring1, NSPC1, FBXL10, and BCOR, may also have an important role in H2A-K119 ubiquitinylation (Simon and Lange, 2008; Simon and Kingston, 2009).
Modification of chromatin by PRC2 and PCR1 is a coordinated sequential process. It is initiated by interaction of PRC2 with chromatin through a mechanism that is not well understood. Polycomb response elements have been identified in Drosophila chromatin. Polycomb response elements serve as DNA binding sites for “recruiter proteins”, including PHO and PHOL, to recruit the PRC2 complex to chromatin (Brown et al., 1998, 2003; Mihaly et al., 1998). Polycomb response elements have not been well described in mammalian cells, but YY1 and Oct4 have been proposed to serve as recruiter proteins in specific contexts (Caretti et al., 2004; Endoh et al., 2008), and noncoding RNA may also function as part of the recruiting complex (Rinn et al., 2007; Pandey et al., 2008).
The first step in polycomb complex-mediated chromatin modification is performed by PRC2. The Ezh2 protein of the PRC2 complex catalyzes trimethylation of Lys27 of histone H3 (H3K27me3). In the next step, the CBX protein of the PRC1 complex binds to H3K27me3 to anchor the PRC1 complex at this site and the PRC1 Ring1B protein catalyzes ubiquitinylation of histone H2A at lysine K119 (Figure 2; Fischle et al., 2003; Cao et al., 2005). These events lead to chromatin compaction-mediated suppression of transcription. The mechanism of transcription repression is not well defined, but the chromatin compaction may block transcription factor–DNA interaction or inhibit transcript elongation. Recent findings suggest that inhibition of transcript elongation may be a key mechanism (Stock et al., 2007; Zhou et al., 2008). The combination of PRC2/PRC1 action results in the stable suppression of gene expression and the resulting gene silencing is associated with increased cell proliferation/survival and decreased senescence and differentiation (Jacobs and van Lohuizen, 2002; Orlando, 2003).
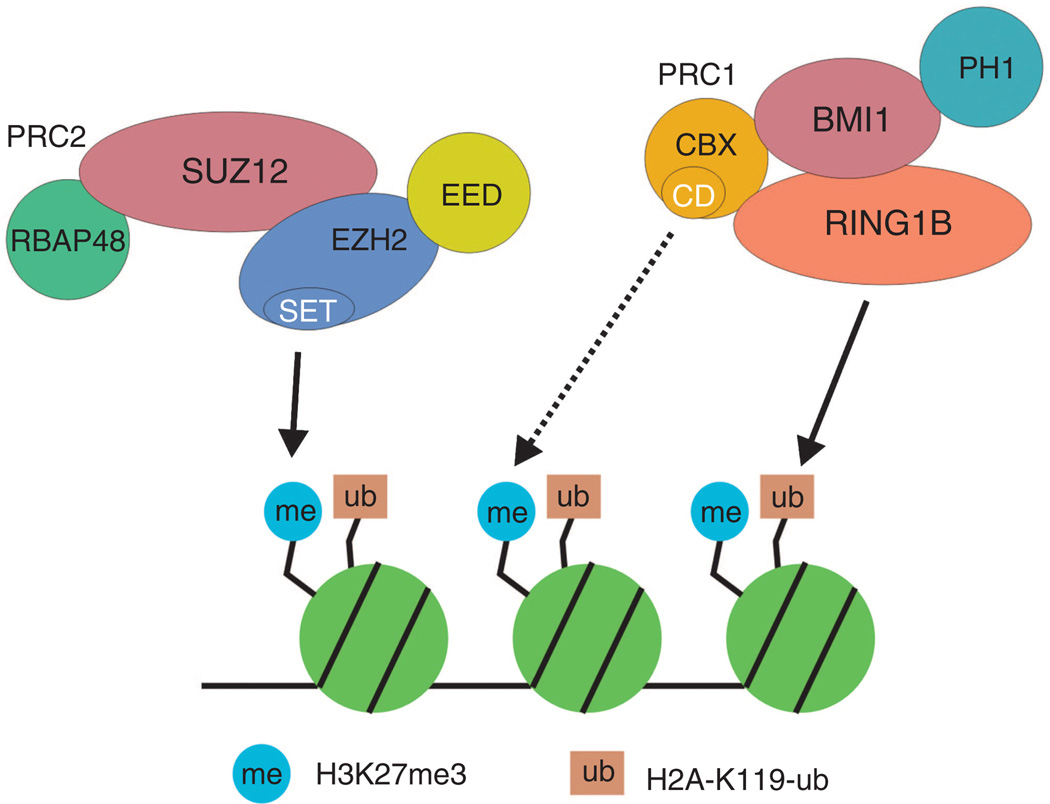
Figure 2. Simplified description of PcG regulation of transcription.
The PRC2 complex is recruited to chromatin, and the Ezh2 protein of this complex, a histone methyltransferase, catalyzes formation of H3K27me3 through its SET catalytic site. H3K27me3 then functions as a binding site for the CBX protein of the PRC1 complex. CBX encodes a chromodomain site that has a high affinity for H3K27me3. The interaction of CBX with H3K27me3 anchors the PRC1 complex to chromatin, and the RING1B subunit of the complex, an E3 ubiquitin ligase, catalyzes formation of H2A-K199-ub. Ultimately these events lead to chromatin folding and compaction and cessation of transcription. The dashed line indicates binding interaction and the solid lines indicate covalent modifications.
PcG proteins are important in maintaining stem cell survival during development (Lessard and Sauvageau, 2003; Molofsky et al., 2003; Iwama et al., 2004), but they also function in adult organisms. For example, knockout mice lacking the PRC1 complex Bmi-1 protein lose stem cells after birth (Lessard and Sauvageau, 2003; Molofsky et al., 2003; Leung et al., 2004), Bmi-1-deficient fibroblasts show slowed proliferation and enhanced senescence (Jacobs et al., 1999b) and cells with elevated Bmi-1 expression show increased cell proliferation (Lessard and Sauvageau, 2003; Yu et al., 2007). Bmi-1 is also important in disease, as it is overexpressed in many tumor types (Vonlanthen et al., 2001; Breuer et al., 2004; Kim et al., 2004a, b) and collaborates with various oncogenes to enhance cell proliferation and survival (Haupt et al., 1993; Jacobs et al., 1999a, b; Dimri et al., 2002; Datta et al., 2007; Haga et al., 2007; Brunner et al., 2008).
The prototypic mode of Bmi-1 action is suppression of gene expression. The Ink4a/Arf locus was one of the first targets identified (Bracken et al., 2007). This locus encodes two tumor suppressor genes called p16Ink4a and p14Arf (p19Arf in mouse) that are transcribed in independent reading frames (Jacobs et al., 1999b). This locus has a key role in control of cell-cycle progression. p16ink4a inhibits the kinase activity of the cyclin D-dependent kinases, Cdk4 and Cdk6, and prevents their association with cyclins. This ultimately leads to reduced pRB phosphorylation and reduced cell-cycle progression through G1 (Serrano et al., 1993). p16ink4a levels are normally reduced under conditions of cell-cycle progression. Consistent with this role, cells in p16Ink4a knockout mice are more susceptible to malignant progression (Krimpenfort et al., 2001; Sharpless et al., 2001). p14Arf promotes cell-cycle arrest by interacting with MDM2 and reducing its ability to target p53 for degradation. This also ultimately leads to cell-cycle arrest (Weber et al., 1999; Lowe and Sherr, 2003). Thus, in the presence of Bmi-1, p16Ink4a and p14Arf levels are reduced thereby permitting the cells to proliferate. As noted below, the Ink4a/Arf locus is an important target gene in keratinocytes, but other target genes have also been identified.
PcG genes in epidermis
Recent studies have begun to focus on the role of the PcG gene products in epidermis, with a particular emphasis on Bmi-1. The epidermis is a multilayered tissue that includes three major functional compartments—undifferentiated cells that retain proliferative potential, differentiating cells that are viable but not able to proliferate, and terminally differentiated dead cells. Maintaining the relative size of these compartments requires that the cell proliferation rate in the basal layer, the tendency of cells to survive versus differentiate in the suprabasal layers, and terminal cell death in the cornified layer be coordinately controlled and balanced (Eckert et al., 1997). Recent studies suggest that the PcG gene products regulate these processes. As noted above, Bmi-1 is a component of the PRC1 PcG complex that functions in the nucleus to modify chromatin and suppress gene expression (Cohen et al., 1996; Aoto et al., 2008). Indeed, Bmi-1 is localized in the nucleus of proliferating keratinocytes in culture (Silva et al., 2006; Lee et al., 2008) and has been detected in the cytoplasm of quiescent keratinocytes (Silva et al., 2006). These findings are consistent with the fact that Bmi-1 has a nuclear localization signal and that inactive Bmi-1 can shuttle to the cytoplasm (Cohen et al., 1996). Bmi-1 was originally characterized as a stem cell maintenance protein that is required for efficient renewal of hematopoietic stem cells (Jacobs et al., 1999a; Molofsky et al., 2003; Park et al., 2003), and leukemic and neuronal stem cells (Lessard and Sauvageau, 2003; Molofsky et al., 2003). However, Bmi-1 does not appear to function exclusively as a stem cell regulator in epidermis. If this were the case, one would expect expression to be restricted to epidermal stem cells compartments. However, the available evidence suggests that Bmi-1 is expressed in multiple epidermal layers, both basal and suprabasal, and is not strictly confined to stem cells (Ressler et al., 2006; Reinisch et al., 2007; Balasubramanian et al., 2008; Lee et al., 2008; Shaw and Martin, 2009). These findings suggest that Bmi-1 is likely to function in multiple epidermal cell layers.
Bmi-1: role in cell survival, proliferation, and senescence
Recent studies have investigated the role of Bmi-1 in regulating keratinocyte senescence. Senescence is a process whereby aging cells gradually lose proliferative potential (Barrandon and Green, 1987). Senescence has the physiological role of limiting the long-term survival of self-renewing cells during aging. A hallmark of senescence is increased expression of products of the Ink4a/Arf locus, p16Ink4a and p14Arf (p19Arf in mouse cells) (Krishnamurthy et al., 2004). Dellambra and co-workers, using the holoclone/meraclone/paraclone clonogenic assay system (Barrandon and Green, 1987), have measured the expression of Bmi-1 as keratinocytes undergo progressive restriction in replicative potential. These studies reveal that Bmi-1 level is high in holoclones (cells with high proliferative potential) and low in paraclones (cells with limited potential) and that there is an inverse relationship between p16Ink4a level and Bmi-1 content. Thus, cells that have high proliferative potential have high Bmi-1 and low p16Ink4a (Silva et al., 2006; Cordisco et al., 2010). This relationship is also observed in aging epidermis, as Bmi-1 levels are high in keratinocytes isolated from young individuals and lower in cells isolated from older individuals (Ressler et al., 2006; Cordisco et al., 2010). Bmi-1 levels are also reduced in keratinocytes from young xeroderma pigmentosum group c, trichothiodystrophy, and progeria patients as compared to keratinocytes isolated from healthy young individuals (Cordisco et al., 2010). In addition, keratinocytes derived from photo-aged epidermis have lower Bmi-1 level than keratinocytes collected from nonexposed epidermis of the same patient (Cordisco et al., 2010). Treatment with trichostatin A results in replicative senescence and this is associated with reduced Bmi-1 level (Cordisco et al., 2010). These studies imply that Bmi-1 has a role in controlling keratinocyte senescence via regulation of the Ink4a/Arf locus.
Keratinocyte survival and proliferation are also influenced by Bmi-1. Treatment with chemopreventive agent or keratinocyte differentiating/apoptosis-inducing agent reduces Bmi-1 expression and the expression of other PcG proteins, and this is associated with reduced survival of normal and transformed keratinocytes (Lee et al., 2008; Balasubramanian et al., 2010). Bmi-1 overexpression in keratinocytes enhances cell number which is associated with increased expression of cdk2 (Lee et al., 2008), Bmi-1 expression extends the replicative life span of keratinocyte-like cells derived from human embryonic stem cells (Dabelsteen et al., 2009) and Bmi-1 siRNA reduces proliferation of the SCC-13 skin cancer cells (Balasubramanian et al., 2010). In this context, Bmi-1 may serve as a prosurvival protein (Lee et al., 2008; Balasubramanian et al., 2010).
Agents that regulate Bmi-1 level in keratinocytes
Given that Bmi-1 regulates keratinocyte survival, it is perhaps not surprising that various extrinsic and intrinsic processes such as cell aging (Silva et al., 2006) and treatment with growth regulatory agents may influence Bmi-1 level (Balasubramanian et al., 2010; Cordisco et al., 2010). Treating with agents that alter keratinocyte survival modulates Bmi-1 level. Hydrogen peroxide treatment reduces Bmi-1 level (Cordisco et al., 2010) as does expression of Ha-Ras(G12V), a constitutively active form of Ha-ras (Cordisco et al., 2010). Potential therapeutic agents also influence Bmi-1 expression. Treatment of keratinocytes with (−)-epigallocatechin-3-gallate (EGCG), which is the major bioactive chemopreventive agent in green tea (Ahmad and Mukhtar, 1999), reduces Bmi-1 level in keratinocytes (Lee et al., 2008; Balasubramanian et al., 2010). This reduction in Bmi-1 level is associated with reduced cell survival and proliferation, and reduced levels of cdk1, cdk2, cdk4, cdk6, cyclin D1, and cyclin E. In addition to reducing Bmi-1 level, EGCG treatment also reduces expression of the PRC2 complex proteins, Ezh2 and Suz12, and reduces histone H3 lysine K27 trimethylation (H3K27me3; Balasubramanian et al., 2010). Ezh2, a component of the PRC2 complex, is the methyltransferase that specifically methylates H3-K27 to form H3K27me3 (Kirmizis et al., 2004; Wang et al., 2004; Kuzmichev et al., 2005). These findings indicate that treatment with EGCG reduces function of both the PRC2 and PRC1 complexes. An interesting finding is that overexpression of Bmi-1, a PRC1 complex protein, is associated with increased expression of Ezh2, a PRC2 complex protein, suggesting that Bmi-1 somehow functions to maintain expression of the enzyme that prepares its chromatin (H3K27me3) binding site (Balasubramanian et al., 2010).
Impact of Bmi-1 on apoptosis
It has been appreciated for several years that Bmi-1 functions to enhance cell survival by regulating cell-cycle-related events. However, less has been studied regarding the impact of Bmi-1 on apoptosis. Recent studies show that challenging keratinocytes with okadaic acid or EGCG results in activation of apoptosis as measured by accumulation of sub-G1 cells and accumulation of cleaved caspases 3 and 9 and PARP (Lee et al., 2008; Balasubramanian et al., 2010). Forced expression of Bmi-1 reverses the okadaic acid/EGCG-associated activation of apoptosis (Lee et al., 2008; Balasubramanian et al., 2010). This finding indicates that Bmi-1 influences a broad range of responses in keratinocytes and the effects cannot be solely explained by Bmi-1-dependent enhancement of cell proliferation or inhibition of senescence. The mechanism of regulation of apoptosis by Bmi-1 is likely to be indirect but has not been investigated.
Other PcG proteins in epidermis
Other PcG proteins have also been studied in epidermis including the PRC2 complex components, Ezh2 and Suz12. Ezh2 expression is inversely correlated with the differentiation status of keratinocytes (Ezhkova et al., 2009). Ezhkova et al. observed high-level Ezh2 expression in keratinocytes maintained in low calcium medium and reduced levels in cells differentiated by treatment with high calcium. Moreover, strong Ezh2 expression is observed in epidermal progenitor cells and the level gradually declines as the cells differentiate. It is interesting that Ezh2 levels, although reduced, are still detected in suprabasal epidermal layers (Ezhkova et al., 2009). These findings suggest that Ezh2 may participate in regulating the survival/differentiation balance in multiple epidermal compartments. Chromatin immuno-precipitation analysis reveals extensive H3K27me3 modification of chromatin in differentiation marker genes in basal progenitor cells, which is associated with gene silencing. Thus, Ezh2 appears to have a role in suppressing differentiation-associated gene expression in basal epidermis (Ezhkova et al., 2009). In addition, loss of Ezh2 from the promoter region of differentiation-associated genes is inversely associated with accumulation of AP1 transcription factors at these sites and gene activation (Ezhkova et al., 2009). Additional studies show increased Ezh2, Suz12 level, and H3K27me3 formation in immortalized/transformed skin cancer cell lines including A431, SCC-13, and HaCaT cells (Balasubramanian et al., 2010), and that Ezh2 knockdown reduces H3K27me3 formation and this is associated with reduced SCC-13 cell proliferation and survival. Ezh2 is an example of a PRC2 protein that is overexpressed in tumors (Varambally et al., 2002; Raaphorst et al., 2003; Raman et al., 2005; Sudo et al., 2005; Saramaki et al., 2006).
In addition to PcG proteins, which are involved in H3K27me3 formation, a class of demethylases have recently been described that specifically erase this mark (Lan et al., 2008; Nottke et al., 2009; Mosammaparast and Shi, 2010). Thus, the PcG protein Ezh2 promotes H3K27me3 formation, whereas the Jumanji C domain-containing H3K27me3-specific demethylases, UTX and JMJD3, remove this mark. A recent study by Khavari and colleagues show that H3K27me3 marks the promoter of many genes that are expressed during differentiation and that loss of this mark is observed when these genes are expressed during calcium-dependent differentiation (Sen et al., 2008). The authors suggest that epigenetic derepression by JMJD3 enhances keratinocyte differentiation. These findings again emphasize the importance of epigenetic modification in regulating the balance between keratinocyte survival and differentiation.
PcG proteins in skin cancer
Several studies have examined the role of PcG proteins in the pathogenesis of skin cancer. Bmi-1 overexpression in HaCaT cells causes malignant transformation as measured by colony formation in soft agar and tumor formation in severe combined immunodeficient mice (Wang et al., 2009). Moreover, Bmi-1 expression is observed at elevated levels in squamous cell carcinoma (SCC) tumors (Reinisch et al., 2007; Balasubramanian et al., 2010). Cultured skin cancer cells express elevated Bmi-1, Suz12, and Ezh2 levels as compared to normal cells and this is associated with increased H3K27me3 (Balasubramanian et al., 2010). Chemopreventive agents suppress proliferation of tumor cells and this is associated with reduced expression of cdk1, cdk2, cdk4, and cyclins D1, E, and A. The level of the cyclindependent kinase (cdk) inhibitors p21 and p27 is increased in these cells (Balasubramanian et al., 2010). As noted above, these chemopreventive agent-associated changes are reversed by forced overexpression of Bmi-1. Chemopreventive agent treatment also activates apoptosis in skin cancer cells as evidenced by increased caspase and PARP cleavage and appearance of sub-G1 cells. It is interesting that these effects are also reversed by forced Bmi-1 expression (Balasubramanian et al., 2010). A potentially meaningful observation is that forced Bmi-1 expression restores Ezh2 levels in EGCG-treated SCC-13 cells, suggesting that feedback regulation functions to assure an appropriate balance of PRC1 and PRC2 complex proteins in cells (Balasubramanian et al., 2010). This may be important as Bmi-1 is not likely to be functional in the absence of Ezh2 and Bmi-1 chromatin binding site is synthesized by Ezh2. Bmi-1 also provides protection when normal keratinocytes are challenged with the differentiation/apoptosis promoting agent, okadaic acid, by inhibiting okadaic acid-dependent apoptosis (Lee et al., 2008). It is also important to note that PcG protein function is also altered in other skin cancer types. For example, PcG protein levels are elevated in basal cell carcinoma (Reinisch et al., 2007) and melanoma (Bachmann et al., 2006; Mihic-Probst et al., 2007).
Processes in wound healing are related to processes that control development and abnormal regulation observed in cancer cells. It is interesting that three major components of the PRC2 complex, eed, Ezh2, and Suz12, are reduced in epidermis during wound healing (Shaw and Martin, 2009). Reexpression of these proteins is observed approximately 10 cells back from the wound edge, and it is proposed that PcG proteins may be involved in silencing the repair genes after completion of wound healing. The loss of these PcG proteins, as expected, is associated with reduced levels of H3K27me3 in the healing epidermis (Shaw and Martin, 2009). Epidermal growth factor receptor and the c-myc genes are markers of the wound healing process. Using a cell culture model, Shaw and Martin (2009) showed that forced expression of eed resulted in reduced epidermal growth factor receptor and c-myc gene expression. These results appear to conflict with the idea that PcG protein expression correlates with enhanced cell survival and increased expression of survival genes. These findings, however, highlight the potential complexity of this regulation, and suggest that the role of the PcG proteins may be context dependent.
CONCLUSION
The findings described above support several conclusions regarding the function of PcG proteins in epidermis. First, expression of Bmi-1 and other PcG proteins is not strictly confined to stem cells in normal epidermis or in cancer cells. A gradient appears to exist such that basal cells express more Bmi-1 (for example) than suprabasal cells; however, expression is observed in a wide range of cells in the progenitor, basal, and suprabasal layers. This suggests that although Bmi-1 expression, and the expression of other PcG proteins, correlates inversely with survival potential, they are not strictly stem-cell-specific proteins (Lee et al., 2008; Ezhkova et al., 2009; Cordisco et al., 2010). Second, Bmi-1 and other PcG proteins enhance expression of pro-proliferation cell-cycle regulatory proteins and suppress differentiation and apoptosis (Silva et al., 2006; Lee et al., 2008; Ezhkova et al., 2009). Moreover, in general, loss of the H3K27me3 mark is associated with expression of differentiation marker genes and reduced cell proliferation/survival. Third, Bmi-1 and other PcG proteins are expressed at higher levels in tumors, both SCC (Reinisch et al., 2007; Balasubramanian et al., 2010), basal cell carcinoma (Reinisch et al., 2007) and melanoma (Bachmann et al., 2006; Mihic-Probst et al., 2007), although the Bmi-1 finding is somewhat controversial for melanoma, as one report describes reduced expression with clinical progression in malignant melanoma (Bachmann et al., 2008). Moreover, forced Bmi-1 expression in HaCaT cells causes transformation (Wang et al., 2009). This suggests that overexpression of these proteins may be one mechanism whereby tumor cells escape death through a mechanism that involves modulation of cell cycle and apoptotic processes (Lee et al., 2008; Balasubramanian et al., 2010). Thus, although it is possible that Bmi-1 could be restricted to maintaining survival of tumor stem cells, the ubiquitous distribution of Bmi-1 in skin tumor cells suggests a broader role in skin cancer (Balasubramanian et al., 2008). Fourth, PcG protein expression declines in aging epidermis, showing that loss of PcG protein expression is associated with keratinocyte senescence both in vivo and in cell culture models (Ressler et al., 2006; Cordisco et al., 2010). Fifth, Bmi-1 and other PcG proteins are likely important targets for cancer prevention, as the level of these proteins is reduced in cells treated with chemopreventive agent and the action of chemopreventive agent is antagonized by overexpression of these proteins (Balasubramanian et al., 2008, 2010).
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 AR053851 and R01 CA131074 to R Eckert.
Abbreviations
- cdk
cyclin-dependent kinase
- H2A-K119-Ub
ubiquitinylated histone H2A lysine 119
- H3K27me3
tri-methylated histone H3 lysine K27
- PcG
Polycomb group genes
- SCC
squamous cell carcinoma
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- Ahmad N, Mukhtar H. Green tea polyphenols and cancer: biologic mechanisms and practical implications. Nutr Rev. 1999;57:78–83. doi: 10.1111/j.1753-4887.1999.tb06927.x. [DOI] [PubMed] [Google Scholar]
- Aoto T, Saitoh N, Sakamoto Y, et al. Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression. J Biol Chem. 2008;283:18905–18915. doi: 10.1074/jbc.M709322200. [DOI] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Bachmann IM, Puntervoll HE, Otte AP, et al. Loss of BMI-1 expression is associated with clinical progress of malignant melanoma. Mod Pathol. 2008;21:583–590. doi: 10.1038/modpathol.2008.17. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Adhikary G, Eckert RL. The Bmi-1 polycomb protein antagonizes the (−)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis. 2010;31:496–503. doi: 10.1093/carcin/bgp314. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Balasubramanian S, Lee K, Adhikary G, et al. The Bmi-1 polycomb group gene in skin cancer: regulation of function by (−)-epigallocate-chin-3-gallate. Nutr Rev. 2008;66 Suppl 1:S65–S68. doi: 10.1111/j.1753-4887.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer RH, Snijders PJ, Smit EF, et al. Increased expression of the EZH2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia. 2004;6:736–743. doi: 10.1593/neo.04160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Fritsch C, Mueller J, et al. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, et al. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Brunner M, Thurnher D, Pammer J, et al. Expression of VEGF-A/C, VEGF-R2, PDGF-alpha/beta, c-kit, EGFR, Her-2/Neu, Mcl-1 and Bmi-1 in Merkel cell carcinoma. Mod Pathol. 2008;21:876–884. doi: 10.1038/modpathol.2008.63. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di PM, Micales B, et al. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen KJ, Hanna JS, Prescott JE, et al. Transformation by the Bmi-1 oncoprotein correlates with its subnuclear localization but not its transcriptional suppression activity. Mol Cell Biol. 1996;16:5527–5535. doi: 10.1128/mcb.16.10.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordisco S, Maurelli R, Bondanza S, et al. Bmi-1 reduction plays a key role in physiological and premature aging of primary human keratinocytes. J Invest Dermatol. 2010;130:1048–1062. doi: 10.1038/jid.2009.355. [DOI] [PubMed] [Google Scholar]
- Dabelsteen S, Hercule P, Barron P, et al. Epithelial cells derived from human embryonic stem cells display p16INK4A senescence, hypermotility, and differentiation properties shared by many P63+ somatic cell types. Stem Cells. 2009;27:1388–1399. doi: 10.1002/stem.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hoenerhoff MJ, Bommi P, et al. Bmi-1 cooperates with H-Ras to transform human mammary epithelial cells via dysregulation of multiple growth-regulatory pathways. Cancer Res. 2007;67:10286–10295. doi: 10.1158/0008-5472.CAN-07-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Martinez JL, Jacobs JJ, et al. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 2002;62:4736–4745. [PubMed] [Google Scholar]
- Eckert RL, Crish JF, Robinson NA. The epidermal keratinocyte as a model for the study of gene regulation and cell differentiation. Physiol Rev. 1997;77:397–424. doi: 10.1152/physrev.1997.77.2.397. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Ohno S, Yugawa T, et al. Efficient immortalization of primary human cells by p16INK4a-specific short hairpin RNA or Bmi-1, combined with introduction of hTERT. Cancer Sci. 2007;98:147–154. doi: 10.1111/j.1349-7006.2006.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano A, Matsumoto M, Higashinakagawa T, et al. Phosphorylation of the chromodomain changes the binding specificity of Cbx2 for methylated histone H3. Biochem Biophys Res Commun. 2010;397:93–99. doi: 10.1016/j.bbrc.2010.05.074. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Bath ML, Harris AW, et al. bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene. 1993;8:3161–3164. [PubMed] [Google Scholar]
- Iwama A, Oguro H, Negishi M, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999a;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, et al. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999b;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- Kim JH, Yoon SY, Jeong SH, et al. Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast. 2004a;13:383–388. doi: 10.1016/j.breast.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Kim JH, Yoon SY, Kim CN, et al. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004b;203:217–224. doi: 10.1016/j.canlet.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, et al. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Adhikary G, Balasubramanian S, et al. Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol. 2008;128:9–17. doi: 10.1038/sj.jid.5700949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Leung C, Lingbeek M, Shakhova O, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- Levine SS, King IF, Kingston RE. Division of labor in polycomb group repression. Trends Biochem Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Li Z, Cao R, Wang M, et al. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Mihaly J, Mishra RK, Karch F. A conserved sequence motif in Polycomb-response elements. Mol Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- Mihic-Probst D, Kuster A, Kilgus S, et al. Consistent expression of the stem cell renewal factor BMI-1 in primary and metastatic melanoma. Int J Cancer. 2007;121:1764–1770. doi: 10.1002/ijc.22891. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, et al. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, Meijer CJ, Fieret E, et al. Poorly differentiated breast carcinoma is associated with increased expression of the human polycomb group EZH2 gene. Neoplasia. 2003;5:481–488. doi: 10.1016/s1476-5586(03)80032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman JD, Mongan NP, Tickoo SK, et al. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin Cancer Res. 2005;11:8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- Reinisch CM, Uthman A, Erovic BM, et al. Expression of BMI-1 in normal skin and inflammatory and neoplastic skin lesions. J Cutan Pathol. 2007;34:174–180. doi: 10.1111/j.1600-0560.2006.00587.x. [DOI] [PubMed] [Google Scholar]
- Ressler S, Bartkova J, Niederegger H, et al. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5:379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramaki OR, Tammela TL, Martikainen PM, et al. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, et al. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Garcia JM, Pena C, et al. Implication of polycomb members Bmi-1, Mel-18, and Hpc-2 in the regulation of p16INK4a, p14ARF, h-TERT, and c-Myc expression in primary breast carcinomas. Clin Cancer Res. 2006;12:6929–6936. doi: 10.1158/1078-0432.CCR-06-0788. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Sudo T, Utsunomiya T, Mimori K, et al. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer. 2005;92:1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van LM. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vonlanthen S, Heighway J, Altermatt HJ, et al. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84:1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li WL, You P, et al. Oncoprotein BMI-1 induces the malignant transformation of HaCaT cells. J Cell Biochem. 2009;106:16–24. doi: 10.1002/jcb.21969. [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, et al. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Yu Q, Su B, Liu D, et al. Antisense RNA-mediated suppression of Bmi-1 gene expression inhibits the proliferation of lung cancer cell line A549. Oligonucleotides. 2007;17:327–335. doi: 10.1089/oli.2007.0070. [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]